Abstract
Clonal evolution occurs during the course of chronic lymphocytic leukemia (CLL) and activation-induced deaminase (AID) could influence this process. However, this possibility has been questioned in CLL because the number of circulating AID mRNA+ cells is exceedingly low; synthesis of AID protein by blood CLL cells has not been demonstrated; the full range of AID functions is lacking in unmutated CLL (U-CLL), and no prospective analysis linking AID expression and disease severity has been reported. The results of the present study show that circulating CLL cells and those within secondary lymphoid tissues can make AID mRNA and protein. This production is related to cell division because more AID mRNA was detected in recently divided cells and AID protein was limited to the dividing fraction and was up-regulated on induction of cell division. AID protein was functional because AID+ dividing cells exhibited more double-stranded DNA breaks, IGH class switching, and new IGHV-D-J mutations. Each of these actions was documented in U-CLL and mutated CLL (M-CLL). Furthermore, AID protein was associated with worse patient outcome and adverse cytogenetics. We conclude that the production of fully functional AID protein by U-CLL and M-CLL cells could be involved in clonal evolution of the disease.
Introduction
Chronic lymphocytic leukemia (CLL) follows either an indolent or an aggressive course1 and clinical decompensation is often accompanied by the appearance of new or increasing numbers of genetic aberrations associated with shorter survival, “clonal evolution.”2 The mechanism(s) responsible for the generation of these genetic abnormalities are not defined in CLL, which is not the case in certain other human cancers, especially lymphoid malignancies of germinal center (GC) origin, in which activation-induced deaminase (AID) appears to be pathogenic.3-5
AID is required for the beneficial generation of Ab diversity in normal B lymphocytes by inducing IGV somatic hypermutation (SHM) and helps in the development of protective effector mechanisms by mediating IGH class-switch recombination (CSR).6,7 These beneficial on-target AID activities occur primarily during a GC reaction and involve conversion of cytidine to uridine on single-stranded DNA at the IG locus. Such on-target actions in CLL B cells have been a matter of interest for several years, primarily because the presence or absence of IGHV mutations (which require AID) in CLL cells is closely linked to clinical outcome. Patients with leukemic clones with minimal (< 2% difference from germline) or no mutation in the IGHV (unmutated CLL [U-CLL]) have a far worse prognosis than patients with IGHV-mutated CLL (M-CLL).8,9 Despite this SHM-based subcategorization of CLL cases, some clones exhibit ongoing IGHV diversification in vivo and in vitro,10-12 with an antigen-driven pattern present in some cases,13 and up to 50% of patients exhibit molecular evidence for intraclonal isotype CSR.14-18
AID activity focused elsewhere (“aberrant” or “off-target” SHM3,19 ) can lead to mutations, deletions, or translocations outside of the IG locus, as in GC-derived lymphomas.3-5 However, such a role for AID in CLL has been questioned for several reasons: (1) although circulating CLL cells can express AID mRNA,20-22 the number of such cells is exceedingly low (0.01%-0.2%)22 ; (2) AID protein synthesis by these same cells has not been demonstrated18,20-23 ; (3) demonstration of the full range of AID functions is lacking in CLL, for example, by failure of cells to demonstrate SHM, especially for U-CLL clones, even on stimulation and induction of AID mRNA,21 thereby creating the apparent paradox that U-CLL patients express more AID mRNA than M-CLL patients yet exhibit no or minimal SHM; and (4) despite association with several prognostic markers,20,21,24-26 no prospective analysis linking AID expression and disease severity has been performed.
In the present study, we aimed to address these issues as a means of determining whether AID could be involved in the evolution of CLL to a more aggressive disease. We report that CLL cells are able to produce AID protein, but synthesis is restricted to that minor subset of the clone that is dividing and/or has recently divided. We also demonstrate that AID from both U-CLL and M-CLL patients can be fully functional in CLL cells by associating protein expression with double-stranded DNA (dsDNA) breaks, Ig class switching, and de novo SHM. Finally, we relate AID expression to genomic aberrations and patient outcome in 2 large patient cohorts, one prospectively.
Methods
CLL patient samples and characterization
The institutional review board of the North Shore-Long Island Jewish Health System sanctioned these studies. After obtaining informed consent in accordance with the Declaration of Helsinki, we collected blood from CLL patients for whom clinical information, laboratory data, and IGH variable region gene DNA sequences were available (supplemental Tables 1-3, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).27 PBMCs were isolated by density gradient centrifugation (Ficoll-Hypaque; Pharmacia). Lymph nodes (LNs), removed for diagnostic purposes from 10 CLL patients, were formalin fixed, paraffin embedded, and sectioned. One additional LN sample was available as dispersed cells. Patients were treated uniformly by the members of our CLL research and treatment program using standardized protocols. Patient AID status was not known by the care givers.
Abs, flow cytometry, and microscopy
Abs used for flow cytometry, immunofluorescent microscopy, and immunohistochemistry are listed in supplemental Table 4. All flow cytometry data were acquired with an LSR II flow cytometer (BD Biosciences) and analyzed with FlowJo Version 7.2.4 software (TreeStar). Light microscopy was performed using an Olympus BX40 microscope fitted with UplanFl 10×/0.30 and 60×/1.25 objectives. Immunofluorescent microscopy was performed using an Olympus IX70 microscope fitted with a UPlanAPO 60×/1.40 objective, one 30-mW argon laser exciting at 488 nm, one 1-mW helium-neon laser exciting at 543 nm, and one 5-mW helium-neon laser exciting at 633 nm. Microscopy was performed as described previously.28,29 Confocal microscopy data were collected using proprietary image acquisition software and images edited for optimal color contrast using Adobe Creative Suite 2 premium (Adobe Systems).
For immunofluorescence of cultured cells, cells were settled onto poly-L-lysine–coated slides (Fisher Scientific) and fixed with 4% formaldehyde (Ted Pella) before staining.
Cell sorting
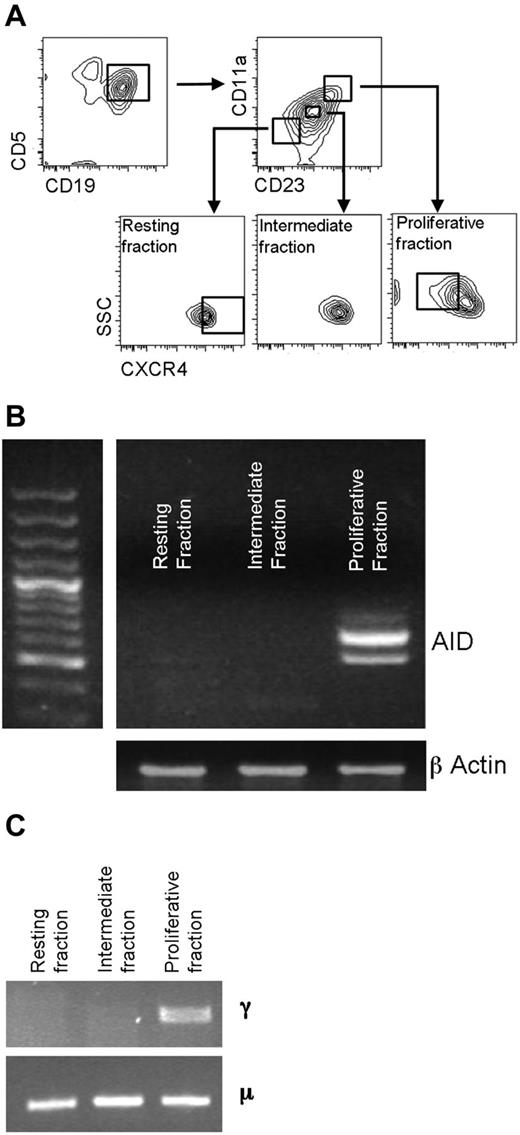
Cell sorting was performed using a FACSAria (BD Biosciences). For recently divided and resting CLL fractions, previously cryopreserved CLL PBMCs were thawed with gated CD19+CD5+ cells sorted into recently divided (CD23BrCD11a+CXCRDim), intermediate (CD23ModCD11a+/−CXCRMod), and resting reentry (CD23DimCD11a−CXCR4Br) fractions (Figure 1A) and sorted into bulk fractions. For both 20-cell and single-cell IGHV-D-J sequencing, sorting was performed after 7 or 14 days of culture. CFSE-labeled unstimulated and stimulated CD5+CD19+ cells were sorted as 20-cell and single-cell aliquots yielding pure populations of either undivided cells or cells that had undergone 5-6 divisions, and cells were sorted into 96-well PCR plates.
AID mRNA expression relates to recent cell division. (A) CD5+CD19+ cells were sorted into recently divided (CD5+CD19+CD23BrCD11a+CXCRDim), intermediate (CD5+CD19+CD23ModCD11a+/−CXCR4Mod), and resting (CD5+CD19+ CD23DimCD11a−CXCR4Br) fractions before analysis of mRNA transcripts for AID. (B) Representative agarose gel of mRNA for AID and β-actin in sorted fractions. Both full-length and a spliced AID transcripts can be seen. (C) In this example, CLL patient-specific μ transcripts were found in all 3 cell fractions by RT-PCR, but switched γ transcripts were only detected in the proliferative fraction. DNA sequencing confirmed that the γ and μ transcripts were of patient origin.
AID mRNA expression relates to recent cell division. (A) CD5+CD19+ cells were sorted into recently divided (CD5+CD19+CD23BrCD11a+CXCRDim), intermediate (CD5+CD19+CD23ModCD11a+/−CXCR4Mod), and resting (CD5+CD19+ CD23DimCD11a−CXCR4Br) fractions before analysis of mRNA transcripts for AID. (B) Representative agarose gel of mRNA for AID and β-actin in sorted fractions. Both full-length and a spliced AID transcripts can be seen. (C) In this example, CLL patient-specific μ transcripts were found in all 3 cell fractions by RT-PCR, but switched γ transcripts were only detected in the proliferative fraction. DNA sequencing confirmed that the γ and μ transcripts were of patient origin.
AID and IGHV-D-J mRNA measurements
For nested PCR measurement of AID expression, the complete coding region (GenBank number AB040431) was amplified from cDNA by PCR, as described previously,22 with β-actin amplified simultaneously. A sample was considered AID mRNA+ if a PCR product of the correct size for full-length AID or its splice variants was observed at least once. μ-IGHV and γ-IGHV mRNA were amplified with IGHC- and IGHV-family specific framework 1 primers, as described previously.27 AID quantitative PCR methodology is described in supplemental Methods.
Flow cytometry for intracellular AID protein
Intracellular AID protein was detected by surface labeling with CD5 and CD19, followed by permeabilizing and fixing with Cytofix/Cytoperm reagent (BD Biosciences). Before AID labeling, blocking was performed with PBS-HEPES containing 30% human AB serum with 0.1% saponin. Anti-AID Abs, secondary reagents, and the appropriate controls used are listed in supplemental Table 4. The fold change in AID mean fluorescence intensity staining was calculated as a ratio of AID mean fluorescence intensity compared with either isotype controls or staining with a specific blocking peptide.
In vitro culture
For short-term culture (up to 3 days), CLL PBMCs were suspended at 1 × 106/mL in RPMI-1640 (Invitrogen) supplemented with 10% heat-inactivated FCS and antibiotics. Alternatively, for long-term culture, PBMCs were first labeled with CFSE (Invitrogen) and then cultured in an enriched medium containing RPMI 1640, 15% FCS, 5 × 10−5M 2-mercaptoethanol, 50 μg/mL of gentamicin, 40 μg/mL of apotransferrin, 1mM sodium pyruvate, 1× nonessential amino acids, and 20mM HEPES. For all cultures, a total of 3 × 106 PBMCs were incubated with irradiated CD32-transfected murine L cells (ATCC) with stimulated cultures receiving anti-CD40 mAb (200 ng/mL, clone MAB89; Beckman Coulter) and IL-4 (10 ng/mL; Sigma-Aldrich) every 3 days (referred to as the “CD40 + IL-4 system”). This stimulation system was chosen because it is most representative of the CLL tissue microenvironment, although we have found that phorbol 12-myristate 13-acetate and ionomycin and CpG 2006 + IL-15 also up-regulate AID expression in CLL cells (not shown). Unstimulated cultures received no anti-CD40 mAb or IL-4. Cells were harvested at 3, 7, and 14 days after initiation; for some experiments, cells were assessed before culture (ie, before stimulation).
20-cell and single-cell IGHV-D-J sequencing
The methodology used for 20-cell and single-cell IGHV-D-J sequencing is described in supplemental Methods.
IGHV-D-J NGS
The methodology used for IGHV-D-J next-generation sequencing (NGS) is described in supplemental Methods.
Statistical analysis
Statistics were calculated using Prism Version 5.0a (GraphPad) or SAS Version 9.2 software. The tests used are indicated in the text.
Results
AID mRNA expression is most marked in recently divided circulating CLL cells. In the present study, we sorted CD5+CD19+ CLL cells into recently divided, intermediate, and resting fractions to determine a relationship between cell division and AID expression (Figure 1A). The markers we used to define these fractions have been shown to identify such fractions by analysis of the in vivo uptake of 2H into replicating DNA of dividing CLL cells.30 AID mRNA transcripts were detected in the recently divided fraction, but not in the resting fraction in 4 of 4 cases (Figure 1B). Fractions were tested for IGHC-switched (γ) transcripts by RT-PCR; these γ transcripts exhibited IGHV-D-J rearrangements identical to the μ transcripts derived from the total unseparated CLL population. No γ IGHV-D-J transcripts were detected in the resting or intermediate fractions (Figure 1C). Therefore, AID mRNA expression is preferentially found in circulating CLL cells that have recently divided.
Infiltrated lymphoid tissues contain proliferating CLL cells that express AID protein and have the same phenotype as AID mRNA+ cells in the blood. Despite the presence of AID mRNA in the proliferative fraction of CLL clones, flow cytometry did not reproducibly detect AID protein in these same fractions (data not shown), a finding consistent with the very low percentage of AID mRNA+ cells in the blood.22 Therefore, we next assessed CLL-infiltrated LN specimens for AID, a site of known CLL proliferation. Fifty percent of specimens (5 of 10) had clearly identifiable AID+ cells, many with the morphology of paraimmunoblasts (Figure 2A). Confocal microscopy demonstrated cytoplasmic AID protein in cells with a CLL phenotype (CD23+) that frequently contained Ki-67, indicating cell-cycle entry (Figure 2B). Intriguingly, only a subset of the Ki-67+ cells in the tissues produced AID protein. In contrast to circulating CLL cells, in the one specimen for which dispersed cells were available for flow cytometric analysis, AID protein was apparent in the minor subset of CD5+CD19+ cells (Figure 2C). These cells had the phenotype of recently divided cells found in blood, with higher expression of CD23, CD11a, and CD5 and lower expression of CXCR4 compared with the entire CD5+CD19+ cell population30 (Figure 2D).
CLL cells in infiltrated LNs express AID protein. (A) Representative low-power (100× original magnification) and high-power (600× original magnification, inset) views of AID+ cells in an infiltrated CLL LN. Scattered AID+ cells (black arrows), which are larger than the majority of cells, are present and have the morphology of paraimmunoblasts (inset). (B) Representative high-power confocal photomicrograph of CLL LN (600× original magnification) showing cells expressing the CLL marker CD23 (red). Several cells are Ki-67+ (blue). An AID+Ki-67+ cell (green) is indicated by the white arrow. (C) Flow cytometry of dispersed LN cells. The percentage of AID+ cells was determined by comparison of samples with and without the use of a peptide that specifically blocks binding of the AID Ab. At least 1% CD5+CD19+ were AID+. (D) Graph showing the mean fluorescence intensity (MFI) of CXCR4, CD5, CD23, and CD11a in AID+ cells compared with the total CD5+CD19+ sorted population using the same gating as in panel C.
CLL cells in infiltrated LNs express AID protein. (A) Representative low-power (100× original magnification) and high-power (600× original magnification, inset) views of AID+ cells in an infiltrated CLL LN. Scattered AID+ cells (black arrows), which are larger than the majority of cells, are present and have the morphology of paraimmunoblasts (inset). (B) Representative high-power confocal photomicrograph of CLL LN (600× original magnification) showing cells expressing the CLL marker CD23 (red). Several cells are Ki-67+ (blue). An AID+Ki-67+ cell (green) is indicated by the white arrow. (C) Flow cytometry of dispersed LN cells. The percentage of AID+ cells was determined by comparison of samples with and without the use of a peptide that specifically blocks binding of the AID Ab. At least 1% CD5+CD19+ were AID+. (D) Graph showing the mean fluorescence intensity (MFI) of CXCR4, CD5, CD23, and CD11a in AID+ cells compared with the total CD5+CD19+ sorted population using the same gating as in panel C.
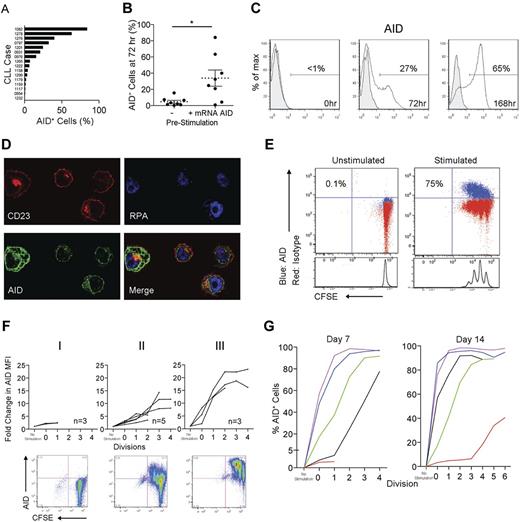
AID protein is variably induced in peripheral blood (PB) CLL cells and its expression is related to proliferation. Although circulating CLL cells variably express AID mRNA20-22 (Figure 1), protein expression by such cells has not been documented.23 Furthermore, although all clones can be induced to express mRNA,10,17,21,22 it is not known if this mRNA is translated to significant protein expression and, if so, in which cells. We therefore investigated whether AID protein becomes detectable in blood CLL cells after stimulation in the CD40 + IL-4 system that mimics the T-cell help likely present in the CLL tissue microenvironment.28,29,31 Seventy-two hours after stimulation, all cases became AID mRNA+ (not shown), in keeping with previously reported results.21,22 AID protein was simultaneously detected, with notable variability in the percentage of AID+ cells between different patient samples (0.3%-84.2% of the total CD5+CD19+ population; mean = 19.3%, median = 6.9%; Figure 3A). Interestingly, detection of AID mRNA before stimulation predicted significantly higher percentages of AID+CD19+CD5+ cells at 72 hours (Figure 3B). Extending the culture to 168 hours resulted in further AID protein up-regulation (Figure 3C). As in the LNs, confocal microscopy localized AID principally to the cytoplasm (Figure 3D), as is the case for normal B cells.32,33
In vitro–activated CLL PBMCs express AID protein. (A) Percentage of AID+ cells within the total CD5+CD19+ population derived from 16 cocultures of CLL PBMCs and CD32-transfected fibroblasts stimulated with CD40 + IL-4 for 72 hours. (B) Comparison of the presence or absence of AID mRNA prestimulation with the percentage CD5+CD19+ cells expressing AID protein 72 hours poststimulation. Means ± SEM of 16 samples are shown. *P = .01 by unpaired t test. (C) Detection of AID protein (white fill) compared with rat IgG2b isotype control mAb (gray fill) by FACS on CD5+CD19+ cells stimulated in the CD40 + IL-4 system at 0, 72, and 168 hours. (D) Confocal photomicrograph of CD23+ cells (red) from the CD40 + IL-4 system visualized at 168 hours demonstrating AID protein (green) localized only in the cytoplasm (replicating protein A-blue (RPA) is used as a nuclear stain). Original magnification was 600×. (E) Representative FACS plots of AID staining on CD5+CD19+ cells derived from unstimulated and CD40 + IL-4–stimulated, CFSE-labeled CLL PBMCs after 7 days. Percentages of AID+ cells in the total CD5+CD19+ population are shown. (F) Three patterns of AID up-regulation were observed after CD5+CD19+ cells were cultured for 7 days in the CD40 + IL-4 system: (I) no up-regulation (M-CLL922, M-CLL1227, and M-CLL1232), (II) up-regulation with each division cycle (M-CLL1082, M-CLL1201, U-CLL1238, M-CLL1252, and M-CLL1299), and (III) up-regulation after ≤ 2 cycles (M-CLL797, U-CLL976, and U-CLL1278). (G) Graphs comparing the percentage AID+CD5+CD19+ cells from 5 cultures at 7 and 14 days. Colors denote individual patient samples: purple: U-CLL1278, blue: M-CLL1082, green: M-CLL1299, black: M-CLL1252, and red: M-CLL922).
In vitro–activated CLL PBMCs express AID protein. (A) Percentage of AID+ cells within the total CD5+CD19+ population derived from 16 cocultures of CLL PBMCs and CD32-transfected fibroblasts stimulated with CD40 + IL-4 for 72 hours. (B) Comparison of the presence or absence of AID mRNA prestimulation with the percentage CD5+CD19+ cells expressing AID protein 72 hours poststimulation. Means ± SEM of 16 samples are shown. *P = .01 by unpaired t test. (C) Detection of AID protein (white fill) compared with rat IgG2b isotype control mAb (gray fill) by FACS on CD5+CD19+ cells stimulated in the CD40 + IL-4 system at 0, 72, and 168 hours. (D) Confocal photomicrograph of CD23+ cells (red) from the CD40 + IL-4 system visualized at 168 hours demonstrating AID protein (green) localized only in the cytoplasm (replicating protein A-blue (RPA) is used as a nuclear stain). Original magnification was 600×. (E) Representative FACS plots of AID staining on CD5+CD19+ cells derived from unstimulated and CD40 + IL-4–stimulated, CFSE-labeled CLL PBMCs after 7 days. Percentages of AID+ cells in the total CD5+CD19+ population are shown. (F) Three patterns of AID up-regulation were observed after CD5+CD19+ cells were cultured for 7 days in the CD40 + IL-4 system: (I) no up-regulation (M-CLL922, M-CLL1227, and M-CLL1232), (II) up-regulation with each division cycle (M-CLL1082, M-CLL1201, U-CLL1238, M-CLL1252, and M-CLL1299), and (III) up-regulation after ≤ 2 cycles (M-CLL797, U-CLL976, and U-CLL1278). (G) Graphs comparing the percentage AID+CD5+CD19+ cells from 5 cultures at 7 and 14 days. Colors denote individual patient samples: purple: U-CLL1278, blue: M-CLL1082, green: M-CLL1299, black: M-CLL1252, and red: M-CLL922).
We also analyzed AID protein induction in relation to cell division using CFSE dilution. Unstimulated CD5+CD19+cells did not divide, whereas multiple divisions could be induced in many of the stimulated cultures. In these cultures, increasing numbers of CD5+CD19+AID+ cells were apparent with each division cycle (Figure 3E). At 7 days, 3 patterns of AID protein expression were apparent: group I clones, with no significant division, exhibited no or minimal AID protein; group II clones showed increasing AID protein levels with each division cycle; and group III clones showed rapid AID protein up-regulation, even in cells undergoing no or only 1 or 2 divisions (Figure 3F).
When a subset of group II and III samples was cultured for up to 14 days, AID up-regulation occurred in each case, although in some group II cases, this was only seen after multiple divisions (eg, M-CLL922; Figure 3G). These patterns of AID protein induction were not correlated with IGHV usage, mutation status, CD38 or ZAP-70 levels, or preexisting cytogenetic aberrations (supplemental Table 1).
Multiply divided cells from both U-CLL and M-CLL clones can carry out the full range of AID functional activities. Having demonstrated that PB CLL cells can express AID protein, we wished to determine whether such cells exhibited no, some, or all of the typical AID documented activities, namely formation of dsDNA breaks, CSR, and SHM. First, we compared CFSE-labeled cells from 3 patients that were cultured for 14 days with and without CD40 + IL-4 system stimulation for the presence of phospho-histone H2A.X (pH2A.X), which localizes to dsDNA breaks.34 At day 14, increased anti-pH2A.X fluorescence was observed in stimulated CD23+ cells and not in unstimulated cells (Figure 4A). Moreover, we found significantly increased anti-pH2A.X fluorescence in stimulated cells with diminished CFSE intensity (more rounds of division) compared with cells with higher CFSE intensity (less/undivided cells; Figure 4B). AID protein expression for the same cases was on average 2.7-fold higher in the most-divided cells compared with the least-divided cells (Figure 4C).
CLL cells that divided and up-regulated AID protein exhibited more dsDNA breaks. (A) Confocal photomicrographs comparing CLL PBMCs stimulated in the CD40 + IL-4 system with unstimulated cells cocultured only with CD32-transfected fibroblasts. Original magnification was 630×. (B) Quantitative colocalization of CFSE intensity (x axis) and anti-pH2A.X staining (y-axis) on CD23+ cells derived from stimulated cultures. The shaded area (gray) represents the range of pH2A.X intensity derived from unstimulated cells, all of which had a CFSE intensity of at least 256 pixels; numbers denote the quantity of cells present in each of the 4 quadrants. ***P < .0001 by Fisher exact test. (C) Graph showing the change in AID mean fluorescence intensity (MFI) identified in CD5+CD19+ cells of the same 3 samples in panel B as determined by flow cytometry.
CLL cells that divided and up-regulated AID protein exhibited more dsDNA breaks. (A) Confocal photomicrographs comparing CLL PBMCs stimulated in the CD40 + IL-4 system with unstimulated cells cocultured only with CD32-transfected fibroblasts. Original magnification was 630×. (B) Quantitative colocalization of CFSE intensity (x axis) and anti-pH2A.X staining (y-axis) on CD23+ cells derived from stimulated cultures. The shaded area (gray) represents the range of pH2A.X intensity derived from unstimulated cells, all of which had a CFSE intensity of at least 256 pixels; numbers denote the quantity of cells present in each of the 4 quadrants. ***P < .0001 by Fisher exact test. (C) Graph showing the change in AID mean fluorescence intensity (MFI) identified in CD5+CD19+ cells of the same 3 samples in panel B as determined by flow cytometry.
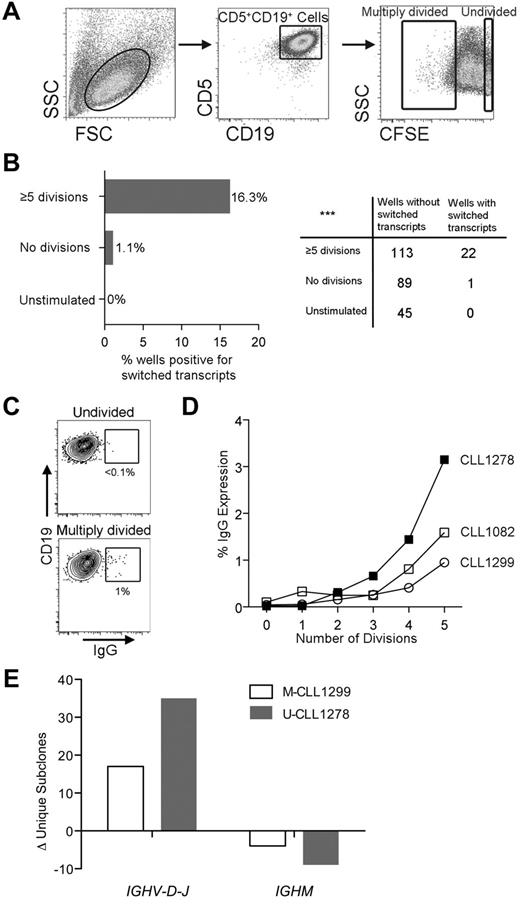
To assess for CSR, we sorted 20-cell aliquots of CFSE-labeled unstimulated cells and of cells stimulated for 14 days with CD40 + IL-4; these fractions contained pure populations of cells demonstrating 0 or 5-6 divisions, respectively (Figure 5A). A total of 16.3% of wells from divided cells contained switched (α and/or γ) and unswitched m+ transcripts with the same IGHV-D-J rearrangement as the leukemic clone; few or no switched transcripts were found in undivided or unstimulated cells (1.1% and 0%, respectively; Figure 5B). The presence of switched Ig protein was confirmed by detecting more surface IgG+ cells among multiply divided than undivided CD5+CD19+ cells (Figure 5C-D).
CLL cells that divided and up-regulated AID protein exhibited CSR and de novo SHM. (A) Sort strategy to obtain 20-cell and 1-cell aliquots of CD5+CD19+ cells from CLL PBMCs after up to 14 days of culture. (B) Comparison of the percentages and numbers of 20-cell/well aliquots that yielded α and γ (switched) CLL patient–specific IGHV-D-J transcripts from divided, undivided, and unstimulated CD5+CD19+ populations after 14 days of culture. ***P = .0002 by χ2 test. (C) Representative FACS plots showing minimal surface IgG expression by CD5+CD19+ cells that had not divided after 14 days of stimulation compared with at least a 10-fold higher expression by multiply divided cells. (D) Comparison of the division number with the percentage of surface IgG expression by CD5+CD19+ cells derived from stimulated CLL PBMCs after 14 days of culture. (E) Change in number of unique mutated subclones identified by NGS from cells stimulated in the CD40 + IL-4 system compared with prestimulated samples. The change in unique subclone count is subdivided into mutations in IGHVDJ and IGHM; a negative number was generated for IGHM in both cases because less unique subclones were found in this region after stimulation compared with prestimulated cells.
CLL cells that divided and up-regulated AID protein exhibited CSR and de novo SHM. (A) Sort strategy to obtain 20-cell and 1-cell aliquots of CD5+CD19+ cells from CLL PBMCs after up to 14 days of culture. (B) Comparison of the percentages and numbers of 20-cell/well aliquots that yielded α and γ (switched) CLL patient–specific IGHV-D-J transcripts from divided, undivided, and unstimulated CD5+CD19+ populations after 14 days of culture. ***P = .0002 by χ2 test. (C) Representative FACS plots showing minimal surface IgG expression by CD5+CD19+ cells that had not divided after 14 days of stimulation compared with at least a 10-fold higher expression by multiply divided cells. (D) Comparison of the division number with the percentage of surface IgG expression by CD5+CD19+ cells derived from stimulated CLL PBMCs after 14 days of culture. (E) Change in number of unique mutated subclones identified by NGS from cells stimulated in the CD40 + IL-4 system compared with prestimulated samples. The change in unique subclone count is subdivided into mutations in IGHVDJ and IGHM; a negative number was generated for IGHM in both cases because less unique subclones were found in this region after stimulation compared with prestimulated cells.
Finally, we examined 1 U-CLL and 2 M-CLL cases for de novo IGHV mutations, initially using single-cell RT-PCR and Sanger sequencing of IGHV-D-J rearrangements derived from cells cultured with the CD40 + IL-4 system. For U-CLL1278, we found no sequences with new mutations after 7 days of culture (supplemental Table 5). However, after 14 days of culture, a total of 5 mutations were present in 111 sequences from multiply divided cells, representing a 39-fold increase over our experimental error rate (supplemental Table 5). We therefore next examined undivided and multiply divided cells from M-CLL1082 and M-CLL1299 after 14 days stimulation. In multiply divided cells, there were 3 and 4 mutations, respectively, a 30- and 26-fold increase over the error rate derived from a total of 114 and 159 sequences (supplemental Table 5). In the undivided cells, M-CLL 1082 demonstrated only 1 new mutation among 96 sequences (12-fold increase over error) with no new mutations in 159 sequences in M-CLL1299. All mutations were observed in the IGHV-D-J region with no mutations in the IGHM constant region.
Although these data were consistent with cell stimulation resulting in de novo mutations, because the overall number of sequences with mutations was low, we could not exclude that some of the variants were not preexisting subclones present at very low frequencies in the original patient samples that would be detected using highly sensitive techniques.11,35 We therefore selected 2 of the same 3 cases (U-CLL1278 and M-CLL1299) for in-depth NGS of IGHV-D-J-μ cDNA before (ie, before stimulation) or after (ie, after stimulation) in vitro stimulation with the CD40 + IL-4 system. We obtained 58 000-84 000 sequence reads from each population with the dominant CLL clone sequence accounting for 96.7%-99.0% of the reads (supplemental Table 6). To assess for numbers of new unique IGHV-mutated subclones generated by stimulation, we analyzed the total number of individual unique and shared sequences present regardless of frequency (supplemental Tables 6 and 7). Strikingly, after stimulation, we found increased numbers of unique subclones for both U-CLL1278 and M-CLL1299 (Figure 5E and supplemental Table 6). Specifically, there were 34 and 16 subclones with unique mutations in the IGHV-D-J of U-CLL1278 and M-CLL1299, respectively, with no increase in unique subclones with mutations in the IGHM of either case (Figure 5E). Because virtually all of the new DNA mutations were in IGHV-D-J and not in IGHM, a targeted (not a nonspecific) mutational process was likely responsible. Moreover, because the number of unique subclones was much higher in the U-CLL case, which showed a type III AID up-regulation pattern compared with the M-CLL case (which showed a type II pattern; Figure 3F), the extent of new DNA mutations appeared to be correlated with the rate ± the amount of AID protein induction by CLL cells. Finally, NGS indicated that certain M-CLL1299 subclones originally identified by our single-cell approach as new mutants (supplemental Table 5 sequence E7_D) were present both before and after stimulation (supplemental Table 7 sequence 5FH2O2/6HAL9T) and therefore represented minor undetected in vivo mutants.
AID expression is associated with the presence of unfavorable genomic aberrations and poor clinical outcomes. If AID function has pathobiological significance in CLL, then its expression should be correlated with worse clinical outcomes. Because > 8 years have elapsed since our original study describing variable AID mRNA expression in circulating CD5+CD19+ cells of CLL patients,22 we compared AID mRNA expression with time to first treatment (TFT) and overall survival (OS) in those same patients (supplemental Table 2). AID mRNA+ CLL patients had a significantly shorter median TFT compared with AID mRNA− patients (58 vs 124 months; P = .0023; Figure 6A). Furthermore, AID+ patients had significantly shorter median OS than AID− CLL patients (132 vs 228 months; P < .0001; Figure 6B); this finding was not because of earlier onset of disease, because both AID+ and AID− groups were diagnosed at similar median ages (58 vs 61.5 years; P = n/s; Figure 6C). Indeed, AID+ patients died at a significantly younger median age than AID− patients (73 vs 83 years; P < .0001; Figure 6D). These data associate AID function with worse CLL disease.
AID+ CLL is correlated with an increased number of cytogenetic aberrations and worse clinical outcomes. Clinical data and prognostic factors for AID+ and AID− CLL patients were compared using the Kaplan-Meier method. (A) TFT curves comparing AID+ and AID− CLL patients (n = 58 and n = 55, respectively) were significantly different. **P = .0023. (B) OS curve of AID+ CLL patients (n = 57) differed significantly from that of AID− patients (n = 55) ***P < .0001. (C) Age of CLL diagnosis curves of AID+ and AID− CLL patients were not significantly different (n/s). (D) Age of death curve for AID+ CLL patients differed significantly from that of AID− patients. ***P < .0001. (E) U-CLL patients were more frequently AID+ (38/55), whereas M-CLL patients were more frequently AID− (54 of 66). ***P = .0001. (F) OS curve of AID+ M-CLL patients differed significantly from that of AID− M-CLL patients. *P = .0264. (G) OS curves of AID+ and AID− U-CLL patients were not significantly different (n/s). (H) AID+ patients had increased numbers of aberrations at 7 commonly tested cytogenetic loci (17p13.1, 11q22.3, 12-CEN, 13q34, 11q13, 14q32, and 6q23.3) compared with AID− patients (median = 1 vs 0). Distribution of the numbers of aberrations was significantly different between the AID+ and AID− groups. ***P = .0009.
AID+ CLL is correlated with an increased number of cytogenetic aberrations and worse clinical outcomes. Clinical data and prognostic factors for AID+ and AID− CLL patients were compared using the Kaplan-Meier method. (A) TFT curves comparing AID+ and AID− CLL patients (n = 58 and n = 55, respectively) were significantly different. **P = .0023. (B) OS curve of AID+ CLL patients (n = 57) differed significantly from that of AID− patients (n = 55) ***P < .0001. (C) Age of CLL diagnosis curves of AID+ and AID− CLL patients were not significantly different (n/s). (D) Age of death curve for AID+ CLL patients differed significantly from that of AID− patients. ***P < .0001. (E) U-CLL patients were more frequently AID+ (38/55), whereas M-CLL patients were more frequently AID− (54 of 66). ***P = .0001. (F) OS curve of AID+ M-CLL patients differed significantly from that of AID− M-CLL patients. *P = .0264. (G) OS curves of AID+ and AID− U-CLL patients were not significantly different (n/s). (H) AID+ patients had increased numbers of aberrations at 7 commonly tested cytogenetic loci (17p13.1, 11q22.3, 12-CEN, 13q34, 11q13, 14q32, and 6q23.3) compared with AID− patients (median = 1 vs 0). Distribution of the numbers of aberrations was significantly different between the AID+ and AID− groups. ***P = .0009.
In this patient cohort, AID expression was correlated with the lack of IGHV mutations (P = .0001; Figure 6E). However, because approximately one-third of patients showed discordance between these parameters, we investigated whether AID mRNA expression further segregated clinical outcomes for these subgroups. AID expression in M-CLL was significantly correlated with worse median OS (183 vs 260 months; P = .0264; Figure 6F), suggesting that AID associates with and identifies M-CLL patients with uncharacteristic poor outcomes. Furthermore, AID+ U-CLL patients tended to have a shorter median OS than AID− U-CLL patients, although this was not significant (99 vs 151 months; Figure 6G). Finally, to support the association of AID with poor clinical outcome, we also compared it with the presence of more genomic aberrations that might suggest off-target activity of AID. For these loci, AID+ CLL patients demonstrated significantly more cytogenetic aberrations than AID− CLL patients (P = .0009; Figure 6H).
Multivariate analysis could not be performed with our original prospective CLL cohort (n = 126) because not all patients had been studied for all prognostic markers (supplemental Table 2). Therefore, to determine whether AID expression is an independent risk factor in CLL, we measured AID expression in a second cohort of 83 patients for whom we had complete information on clinical outcome, IGHV mutations, CD38 and ZAP-70 levels, and high-risk genomic aberrations (supplemental Table 3). This cohort had similar characteristics as the original cohort. Multivariate effects of the univariate factors were examined for TFT when a sufficient number of events had accumulated. We found that AID expression, ZAP-70 levels, and IGHV mutations remained as independent risk factors (Table 1), with AID expression and IGHV mutation being statistically significant (P = .0244 and P = .0191, respectively).
Correlation of CLL risk factors and AID expression
| Parameter . | Cox regression proportional hazard . | |||||
|---|---|---|---|---|---|---|
| Univariate . | Multivariate . | |||||
| P . | HR . | 95% CI . | P . | HR . | 95% CI . | |
| Unmutated IGHV | .0058 | 3.991 | 1.494-10.661 | .0191 | 3.260 | 1.213-8.760 |
| CD38+ | .0855 | 2.034 | 0.906-4.567 | |||
| ZAP-70+ | .0046 | 3.267 | 1.440-7.413 | .0653 | 2.192 | 0.951-5.049 |
| High-risk aberrations | .0612 | 2.172 | 0.964-4.892 | |||
| AID+ | .0055 | 3.080 | 1.392-6.817 | .0244 | 2.524 | 1.127-5.651 |
| Parameter . | Cox regression proportional hazard . | |||||
|---|---|---|---|---|---|---|
| Univariate . | Multivariate . | |||||
| P . | HR . | 95% CI . | P . | HR . | 95% CI . | |
| Unmutated IGHV | .0058 | 3.991 | 1.494-10.661 | .0191 | 3.260 | 1.213-8.760 |
| CD38+ | .0855 | 2.034 | 0.906-4.567 | |||
| ZAP-70+ | .0046 | 3.267 | 1.440-7.413 | .0653 | 2.192 | 0.951-5.049 |
| High-risk aberrations | .0612 | 2.172 | 0.964-4.892 | |||
| AID+ | .0055 | 3.080 | 1.392-6.817 | .0244 | 2.524 | 1.127-5.651 |
Individual Cox regressions were used to examine the association between each factor and TFT on 83 CLL patients with data for all parameters listed. Factors were then included in a multivariable Cox proportional hazards model to examine the joint effects of those factors on TFT. Best subsets selection was used to select the final multivariate model. Factors that did not fit the multivariate best model are indicated by “-”. High-risk aberrations (+), that is, patient cells having at least 1 of the 3 aberrations 11q22.3, 12-CEN, or 17p12.1, were compared against those with 0 of 3.
HR indicates hazard ratio; and CI, confidence interval.
Discussion
The present study addresses and provides novel information that answers several questions about AID protein and its actions in CLL. First, we documented that a small number of CLL cells in the blood and secondary lymphoid tissue can and do express AID protein, and that these cells are those that recently divided (Figures 1,Figure 2–3). Such an AID protein is likely functional because of the presence of more dsDNA breaks (Figure 4), Ig class switching (Figure 5), and new IGHV-D-J mutations (Figure 5) in AID+ divided cells. Of major significance, we document conclusively for the first time that U-CLL patients can develop IGHV mutations in the context of AID protein up-regulation. Finally, AID expression is associated with increased numbers of genomic aberrations and with clinical courses, as documented in 2 distinct patient cohorts, one of which was analyzed prospectively (Figure 6).
It is the recently divided CLL fraction30,36 that is enriched for AID mRNA and protein expression (Figure 1B), and these cells produce clonally related switched Ig isotypes (Figure 1C). Furthermore, the AID+Ki67+ cells in LNs (Figure 2B) had the same phenotype as recently divided cells of the PB (Figure 2D). We therefore linked our observations on AID protein with those of Palacios et al,18 who found that AID mRNA expression in blood U-CLL cells was mainly limited to cells with proliferative potential, some of which were also undergoing active CSR.
For AID mRNA expression to have biologic significance, the leukemic cells must synthesize AID protein. Our novel description of the induction of AID protein in the majority of PB CLL samples after in vitro stimulation is therefore a significant finding. Although the induction rate and protein amount varied (Figure 3A,F), cell division and the presence of AID protein appeared to be linked: undivided clones did not up-regulate protein, whereas clones that had divided exhibited at least some protein up-regulation even if this was only after several cell cycles (Figure 3F-G). Most cases showed a progressive increase in AID protein levels with each division (Figure 3F), which is reminiscent of normal B cells,37,38 and consistent with the idea that, within lymphoid sections, AID+ cells were principally Ki-67+ (Figure 2 and Leuenberger et al39 ). However, because most Ki-67+ cells were AID− (Figure 2B), either only a small subset of cells are competent to express AID, which seems unlikely based on our in vitro stimulation data (Figure 3), or many of the Ki-67+ CLL cells had not divided a sufficient number of times at the time of analysis to produce AID protein.
Given the link between proliferation and AID expression, and using cases for which the divided cells clearly produced AID protein, we compared divided versus undivided cells to demonstrate AID functionality in CLL. We showed that multiply divided cells from both U-CLL and M-CLL patients exhibited the 3 well-established actions of AID: dsDNA breaks (Figure 4), isotype switching (Figure 5), and IGHV mutations (Figure 5 and supplemental Tables 5-7). Based on both single-cell Sanger (supplemental Table 5) and NGS (Figure 5E and supplemental Tables 6 and 7) DNA sequencing, we demonstrated induction of SHM in U-CLL as well as M-CLL clones, which differs from previous studies that detected mutations in pre-switch regions of U-CLL clones after in vitro stimulation, but not new IGHV mutations.21 This discrepancy may be because of our larger number of clonal reads using NGS (Figure 5E and supplemental Tables 6 and 7). Therefore, under appropriate experimental conditions, U-CLL clones not only transcribe AID mRNA,20-22 but also produce AID protein (Figure 3), which can induce both SHM (Figure 5) and CSR (Figure 4, Palacios et al,18 and Oppezzo et al21 ) in these cells, addressing the apparent paradox that U-CLL cases with no or minimal IGHV mutations can transcribe AID mRNA and undergo CSR but do not carry out SHM. It is clear from our data that new IGHV mutations can be induced in U-CLL clones in vitro. In addition, because we have shown such variants existing in CLL transcripts before in vitro stimulation (supplemental Tables 5 and 7), this process likely also occurs in vivo at a yet-to-be-defined rate.
Given that CLL cells synthesize AID protein with the potential for the full range of biologic activities, we next linked AID expression to patient outcome, which could imply a relationship between AID off-target activity and progression of CLL. Because we had studied the presence of AID mRNA in a large cohort of patients > 8 years ago,22 we compared this expression with TFT and OS, performing in effect a prospective, natural history analysis of the association of AID with clinical course. We found that both TFT and OS were significantly shorter in AID+ disease (Figure 6A-B). Previous studies have associated AID expression with the presence of adverse prognostic markers18,20,21,24-26 and one study retrospectively associated PB AID mRNA expression with progression-free survival, not OS, possibly because of the median follow-up of only 48 months.26 Therefore, ours is the first prospective analysis with a long median follow-up (144 months) and a sufficient number of events to demonstrate that AID predicts OS very powerfully, with a 10-year difference in survival between patients with AID+ and AID− disease (Figure 6G). A complementary study examining AID protein in tissues suggested such an association, albeit in a much smaller cohort.39
Of special clinical importance is our finding that AID expression pinpoints M-CLL patients with shorter TFT who are not identified by IGHV mutations (Figure 6F). This supports AID expression as a valuable negative prognostic indicator that can be applied to all CLL patient groups, and not just in differentiating cases of AID+ and AID− U-CLL with and without switched IGH transcripts.18 Indeed, our multivariate analysis of AID mRNA expression, IGHV mutations, CD38 or ZAP-70 levels, and cytogenetic aberrations indicated that quantitative RT-PCR–documented AID levels are a significant independent prognostic factor, along with IGHV mutation status (Table 1). These findings suggest that AID is epistatic with those markers that were not statistically significant in the best subsets selection multivariate model (ie, cytogenetic aberration and CD38 and ZAP-70 levels) and therefore further support the idea that AID is involved in the development of aggressive disease and is not just a passive correlate. Given that both CD38 and ZAP-70 are up-regulated on activation of normal human B lymphocytes,40,41 this multivariate analysis suggests that AID expression and higher levels of CD38 and/or ZAP-70 are correlated with worse outcome in the disease, possibly because the latter reflect cellular activation.
Furthermore, the correlation of genomic aberrations with AID expression (Figure 6H, Heintel et al,24 and Leuenberger et al39 ) also may reflect a common process consistent with the new genomic aberrations that occur in CLL during clonal evolution being produced by off-target AID activity and why AID+ disease has a worse prognosis. More specifically, although our study only examined in-depth AID actions on the IG locus, recent reports indicate that AID can interact with numerous non-IG loci,19 and therefore AID activity on off-target, non-IG genes is tenable. It is, however, important to acknowledge that the coexistence of AID protein and dsDNA breaks in the same cell populations does not indicate cause and effect. Indeed, other biochemical processes such as reactive oxygen species can lead to mutations. In addition, although both naturally occurring on-target11-13 and off-target3,42 mutations and those that we could induce in vitro by the CD40 + IL-4 system (Figure 5 and supplemental Tables 5 and 7) often occur at sites characteristic for AID activity, in some instances mutations are detected in the CLL genome at sites not typical of AID action; for example, NOTCH1 mutations that are found at a great distance downstream of the transcription start site.42,43 However, this latter finding does not exclude such mutations as being AID mediated because the NOTCH1 gene contains AID-binding sites19 and normal CSR, which is an accepted action of AID, occurs even farther away from the IG transcription start site than the NOTCH1 mutation is from its transcription start site. Therefore, detailed studies specifically examining off-target AID activity in CLL are warranted.
Finally, our present findings complement those of others18 regarding 2 significant puzzles concerning AID in CLL: (1) the paradox that U-CLL cases express more AID and exhibit greater levels of IGH switched subclones but minimal SHM and (2) why there is no major increase in the numbers of mutations in the IGHV locus and/or of isotype class switched cells during the course of the disease. Because both U-CLL and M-CLL clones can exhibit the full range of AID activity in vitro, the in vivo lack of SHM in U-CLL clones cannot be because of an inability of such clones to make AID protein or for the protein to carry out SHM in the appropriate setting. Given that U-CLL B cells produce Abs that are highly polyreactive, binding a variety of (auto)antigens,44-46 and that (auto)antigen-mediated drive appears to be important in disease pathogenesis and tumor cell survival,47 “on-target” AID activity in U-CLL may alter the antigen-binding properties of the BCR, resulting in a selective disadvantage and loss of those U-CLL clones undergoing SHM. This counterselection would only apply to replacement mutations that could change the BCR amino acid sequence and thereby possibly structure. Alternatively, AID activity in U-CLL may be actively inhibited from targeting or acting on IGV genes because in vivo the accessibility of the V regions to AID has been blocked (eg, by changes in chromatin structure) or is not achieved because of the lack of production of a targeting molecule by U-CLL clones. Finally, AID activity could be inhibited by the AID splice variants produced26,48 or the tissue microenvironments of U-CLL patients.
As to why clones do not exhibit increased IGHV mutations or increased CSR during the course of the disease, AID could promote apoptosis of isotype switched cells directly, as has been inferred from an AICDAnull murine model.49 Alternatively, or in addition, antigen-binding capacity might be altered on switching from IgM to non-IgM,50 thereby diminishing the trophic influences of BCR signaling and leading indirectly to cell death. Clearly, many questions remain to be answered.
In conclusion, because AID can be fully functional in both U-CLL and M-CLL clones, it may have an important role in the progression of CLL by contributing to clonal evolution. This is consistent with the expression of AID being associated with and predicting disease outcome. Further studies on the role of this enzyme in CLL are merited, because targeting those cells that express more AID or the enzyme itself and its associated molecular pathways might block clinical progression and could be therapeutically valuable.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Nina Kohn and Martin Lesser (Biostatistics Unit, The Feinstein Institute for Medical Research) for statistical assistance; Shahina Maqbool (Albert Einstein College of Medicine) and Agnes Viale and Dr Maryam Hassimi (Memorial Sloan-Kettering Cancer Center) for next-generation sequencing assistance; and Linda Johnson and Bill Kennedy for assistance with clinical samples and clinical data collection.
This study was supported in part by grants from the National Institutes of Health (RO1 CA81554 to N.C. and R01 CA72649 and R01 CA102705 to M.D.S.). The Karches Foundation, The Prince Family Foundation, The Nash Family Foundation, The Mildred and Frank Feinberg Foundation, The Marks Foundation, The Jerome Levy Foundation, The Leon Levy Foundation, and the Joseph Eletto Leukemia Research Fund also provided support. P.P. is funded by Leukaemia and Lymphoma Research, United Kingdom.
National Institutes of Health
Authorship
Contribution: P.E.M.P. and C.C.C. designed and performed the research, analyzed and interpreted the data, and wrote the manuscript; E.A., D.K., L.Z., and A.R.M. performed the research; R.N.D., X.Y., J.B., J.E.K., S.L.A., and K.R.R. revised the manuscript; S.R., P.K.M., T.M., and M.D.S. analyzed and interpreted the data and revised the manuscript; and N.C. conceived the study, designed the research, analyzed and interpreted the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.R. is Oncology Division, Center for Applied Medical Research, University of Navarra, Pamplona, Spain.
Correspondence: Nicholas Chiorazzi, MD, The Feinstein Institute for Medical Research, 350 Community Dr, Manhasset, NY 11030; e-mail: nchizzi@nshs.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal