Abstract
Plasmacytoid dendritic cells (pDCs), originating from hematopoietic progenitor cells in the BM, are a unique dendritic cell subset that can produce large amounts of type I IFNs by signaling through the nucleic acid–sensing TLR7 and TLR9 (TLR7/9). The molecular mechanisms for pDC function and development remain largely unknown. In the present study, we focused on an Ets family transcription factor, Spi-B, that is highly expressed in pDCs. Spi-B could transactivate the type I IFN promoters in synergy with IFN regulatory factor 7 (IRF-7), which is an essential transcription factor for TLR7/9-induced type I IFN production in pDCs. Spi-B–deficient pDCs and mice showed defects in TLR7/9-induced type I IFN production. Furthermore, in Spi-B–deficient mice, BM pDCs were decreased and showed attenuated expression of a set of pDC-specific genes whereas peripheral pDCs were increased; this uneven distribution was likely because of defective retainment of mature nondividing pDCs in the BM. The expression pattern of cell-surface molecules in Spi-B–deficient mice indicated the involvement of Spi-B in pDC development. The developmental defects of pDCs in Spi-B–deficient mice were more prominent in the BM than in the peripheral lymphoid organs and were intrinsic to pDCs. We conclude that Spi-B plays critical roles in pDC function and development.
Introduction
Dendritic cells (DCs) are activated by pathogen sensors such as TLRs and produce various cytokines, thereby playing critical roles in linking innate and adaptive immunity.1-3 DCs are heterogeneous and DC responses depend on the subsets. Plasmacytoid DCs (pDCs) are a unique subset that is distinct from conventional DCs (cDCs).4 pDCs originate from hematopoietic progenitor cells in the BM and their development depends on several transcription factors, including E2-2.5,6 However, the molecular mechanisms for pDC development remain largely unknown.
pDCs express nucleic acid–sensing TLR7/9 and produce vast amounts of type I IFNs, including IFN-α and IFN-β, on TLR7/9 signaling, whereas TLR7/9-stimulated cDCs produce some IFN-β, but not IFN-α. Therefore, robust production of type I IFN by TLR7/9 signaling is a hallmark of pDCs.7 TLR7/9 signaling for type I IFN production depends on IRF-7, which is phosphorylated and activated by several signaling molecules, including IκB kinase α.7-12 This signaling pathway functions selectively in pDCs with constitutively high expression of IRF-7.13,14 However, IRF-7 alone is not sufficient for this pDC function because, despite rapid and abundant induction of IRF-7 after TLR7/9 stimuli, cDCs fail to produce IFN-α, indicating the involvement of an additional factor(s).
Considering that Ets family transcription factors cooperatively function with IRF family members,15 in the present study, we focused on Spi-B, an Ets family member highly expressed in pDCs among the DC subsets.16,17 There are approximately 30 Ets family members and all possess a common Ets domain that binds to the purine-rich GGAA/T consensus sequence.18 Knock-down of human SPIB expression inhibited pDC generation from CD34+ precursor cells, indicating that Spi-B is critical for pDC expansion or development.19 Spi-B–deficient mice have defects in B-cell activation and T cell–dependent humoral immune responses20 ; however, the in vivo roles of Spi-B in pDC function or development have not been well characterized.
In the present study, we found that Spi-B can transactivate the type I IFN promoters and has the best synergy with IRF-7 among the IRF family members. By analyzing Spi-B–deficient mice, we also clarified in vivo roles of Spi-B in pDC function and development.
Methods
Plasmids
The Ifna4 promoter (approximately −486 to −55)–driven luciferase reporter plasmid was described previously.10 For the Ifnb1 promoter–driven luciferase reporter plasmid, the Ifnb1 promoter region (approximately −140 to +42) was amplified by PCR using a sense primer, 5′-AGCTTGAATAAAATGAATATTAGAAGC-3′, and an antisense primer, 5′-CAAGATGAGGCAAAGGCTGTCAAAGGC-3′, and ligated into pGL3 (Promega).21 Four potential Ets-binding sites in the Ifna4 and Ifnb1 promoters and one IRF-binding site in the Ifna4 promoter were mutated using the QuickChange II Site-Directed Mutagenesis Kit (Stratagene; supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).22 For the Il12b promoter–driven luciferase reporter plasmid, the Il12b promoter region (approximately −412 to +12) was amplified by PCR using a sense primer, 5′-GGGTACCTTCTTTGGGCCTGTAACACCTACTTATTTG-3′, and an antisense primer, 5′-GAAGCTTACTGTTCCTTCTGCTGCCTTGGCTGCTC-3′, and ligated into pGL3. For the Tnf promoter–driven luciferase reporter plasmid, the Tnf promoter region (approximately −1260 to +140) was amplified by PCR using a sense primer, 5′-GGGTACCCACGAAGCTCTAAAAGCCAGCCACTG-3′, and an antisense primer, 5′-GAAGCTTGGAGATGTGGCGCCTTGGGCCAGTGAG-3′, and ligated into pGL3. Expression vectors for murine Spi-B, Spi-B mutants, IRF-1, IRF-3, IRF-4, IRF-5, IRF-7, and IRF-8 were generated as follows. The hemagglutinin (HA)–tagged murine Spi-B cDNA fragments encoding the full-length transactivation domain deletion mutant and the Ets domain deletion mutant were amplified by PCR from a Spi-B cDNA clone (msh30167) as a template and subcloned into pCSII-EF-MCS-IRES2-venus (HA-SpiB-IRES2-venus, HA-SpiBdTA-IRES2-venus, and HA-SpiBdEts-IRES2-venus). The FLAG-tagged murine IRF-1 cDNA fragment was amplified by PCR from an IRF-1 cDNA clone (msj01193) and subcloned into pEF-BOS (pEF-BOS-FLAG-mIRF-1). The FLAG-tagged murine IRF-3, IRF-4, and IRF-5 cDNA fragments were amplified by PCR from IRF-3 (3110001G18), IRF-4 (F630019B18), and IRF-5 (F830012G18) cDNA clones,23 respectively, and subcloned into pEF-BOS (pEF-BOS-FLAG-mIRF-3, pEF-BOS-FLAG-mIRF-4, and pEF-BOS-FLAG-mIRF-5). The FLAG-tagged murine IRF-8 cDNA fragment was amplified by PCR from a CpG DNA-stimulated GM-CSF–induced BM DC cDNA library and subcloned into pEF-BOS (pEF-BOS-FLAG-mIRF-8). The FLAG-tagged murine IRF-7 expression vector (pEF-BOS-FLAG-mIRF-7) and FLAG-tagged murine NF-κB p65 expression vector (pCDNA3-FLAG-p65) were described previously.10,24
Luciferase assay
293T cells were seeded in 24-well plates (7 × 104 cells/well) and cultured overnight. These cells were transiently transfected with 75 ng of a luciferase reporter plasmid together with a combination of expression plasmids for Spi-B, IRFs, or NF-κB p65 using Lipofectamine 2000 (Invitrogen). Cell lysates were prepared 18 hours after transfection and luciferase activity was measured by the dual-luciferase reporter assay system (Promega).
Immunoprecipitation and immunoblotting
293T cells were transiently transfected with a combination of plasmids and then immunoprecipitation and immunoblotting experiments were performed with anti-HA mAb (3F10; Roche) and/or anti-FLAG M2 mAb (Sigma-Aldrich), as described previously.10
Mice
C57BL/6J mice were purchased from CLEA Japan. Ly49Q transgenic mice were described previously.25 Ly49Q transgenic Spi-B–deficient mice were generated by mating Ly49Q transgenic Spib+/− mice with Spib+/− mice. Mice were maintained under the specific pathogen-free conditions in the animal facility of the RIKEN Research Center for Allergy and Immunology and Osaka University. All animal experiments were approved by the animal research committees of RIKEN Yokohama Research Institute and Osaka University.
Generation of Spi-B–deficient mice
A targeting vector was designed to replace a 1.6-kb genomic fragment with the neomycin resistance gene from pMC1-neo polyA (Stratagene). The replaced genomic fragment contains exon 4, exon 5, and a part of exon 6 encoding the Ets domain. A herpes simplex virus thymidine kinase gene (HSV-TK) was inserted at the downstream end of the long arm. A C57BL/6-derived embryonic stem cell line, Bruce4, was transfected with the targeting vector by electroporation and selected with G418 and ganciclovir. Doubly resistant clones were screened for homologous recombination by PCR and verified by Southern blot analysis.
Northern blot analysis
Total RNA was extracted with the RNeasy mini kit (QIAGEN), electrophoresed, and transferred to nylon membranes. The cDNA probes were amplified from a Spi-B cDNA clone (msh30167) by PCR and used. Hybridization was performed as described previously.26
Reagents
The following reagents were used for cell stimulation: 8-mercaptoguanosine (Sigma-Aldrich), polyuridylic acid (Sigma-Aldrich), R848 (Invivogen), and ODN1668 and D19 (Hokkaido System Science).26 The vesicular stomatitis virus (VSV) was kindly provided by Dr Hiroshi Watarai (RIKEN Research Center for Allergy and Immunology).
Flow cytometric analysis
Cells from spleen, BM, inguinal lymph nodes (iLNs), and peripheral blood (PB) were incubated with anti-CD16/32 (93; eBiosciences) to block Fc receptors and stained with the following mAbs: allophycocyanin (APC)–Cy7-anti-CD11c (N418; BioLegend), peridinin chlorophyll protein (PerCP)–Cy5.5–anti-CD45R/B220 (RA3-6B2; BioLegend), Horizon V450-anti-CD19 (1D3, BD Biosciences), APC-anti-CD317 (BST2, eBio927; eBiosciences), phycoerythrin (PE)–anti-Ly49Q (2E6; MBL), FITC–anti-Siglec-H (440c; Hycult Biotech), APC–anti-H-2Kb (AF6-88.5.5.3; eBiosciences), FITC–anti-CD199 (CCR9, CW-1.2; eBiosciences), PE–anti-CD135 (Flt3, A2F10; eBiosciences), FITC–anti-CD45.1 (A20; eBiosciences), phycoerythrin-Cy7 (PE-Cy7)-anti-CD45.2 (104; BD Biosciences), biotinylated anti-CD200 (R&D Systems), and PE–anti-CD86 (GL1; BD Biosciences). Streptavidin-PE-Cy7 (BD Biosciences) was used to develop biotinylated mAbs. Stained cells were analyzed with a FACSCalibur flow cytometer (BD Biosciences), a FACSAria II cell sorter (BD Biosciences), and FlowJo Version 8.8.7 software (TreeStar).
Cell sorting
Mononuclear cells were enriched by histopaq1083 (Sigma-Aldrich) from spleen or BM and stained with FITC–anti-BST2, PE–anti-B220, and APC–anti-CD11c. CD11c+B220+BST2+ and CD11c+B220−BST2− cells were purified by FACS sorting with a FACSVantage, FACSAria, or FACSAria II cell sorter (BD Biosciences) and used as pDCs and cDCs, respectively. In certain experiments, pDCs were stained with Alexa Fluor 647–anti-BST2 (927; eBiosciences), labeled by anti-Cy5/anti–Alexa Fluor 647 Microbeads (Miltenyi Biotec), and purified with an MACS Column (Miltenyi Biotec).
Cytokine production by DCs
DCs were plated in 96-well flat-bottom plates (1 × 105 cells/well) and stimulated with the indicated concentrations of the various stimuli. Polyuridylic acid was mixed with Lipofectamine 2000 (Invitrogen) and used. After 24 hours of cell stimulation, culture supernatants were harvested and the cytokine levels in the supernatants were measured by ELISA. IFN-α and IFN-β ELISA kits were from PBL. The IL-12p40 ELISA kit was from TECHNE. TNF-α ELISA kits were purchased from R&D Systems.
Expression analysis
Total RNA was reverse transcribed and assayed by quantitative real-time PCR using an ABI PRISM 7000 (Applied Biosystems). TaqMan probes (TaqMan Gene Expression Assay; Applied Biosystems) were used for 18S rRNA (internal control), Irf7, Ifna4, Ifna5, Ifna6, Ifnb1, Tnf, and Il12b. For the other genes, SYBR Green primers were used (supplemental Methods). The expression of all genes was normalized to that of 18S rRNA and is represented as the ratio to the indicated reference samples. All primers were validated for linear amplification.
Serum cytokine levels after injection of TLR agonists
Mice were injected with various TLR agonists. Fifty micrograms of polyuridylic acid complexed with 1.5 μmol in vivo–jetPEI (polyplus transfection) per mouse was intravenously injected. Thirty-two micrograms of ODN1668 complexed with 30 μg of DOTAP (Roche) per mouse was intravenously injected. One hundred and fifty micrograms of poly(I:C) per mouse was intraperitoneally injected. After injection, sera were taken at the indicated time points and analyzed for cytokine levels by ELISA.
Generation of BM chimeric mice
CD45.1+ C57BL/6 recipient mice were lethally irradiated (8 Gy) using an X-ray irradiator (HITACHI MBR-1520R-3) and intravenously injected with a 1:1 mixture of CD45.2+ wild-type (WT) C57BL/6 or Spi-B–deficient BM cells with CD45.1+ WT C57BL/6 BM cells. Cells were analyzed 6 to 8 weeks after BM transfer with a FACSAria II cell sorter (BD Bioscience) and FlowJo software (TreeStar).
BrdU incorporation assay
Mice were injected intraperitoneally with 1 mg of 5-bromo-2′-deoxyuridine (BrdU; Wako) and fed with continuously with light-protected drinking water containing 1 mg/mL of BrdU. At the indicated days, splenic or BM cells were harvested and analyzed for BrdU incorporation. Intracellular staining for BrdU was performed with the FITC BrdU Flow kit (BD Biosciences) according to the manufacturer's protocol.
DNA microarray analysis
A total of 300 000 CD11c+B220+BST2+ BM or splenic cells from WT or Spi-B–deficient mice were sorted and total RNA was extracted with the RNeasy micro kit (QIAGEN). The quality of the prepared RNA was assessed with a 2100 Bioanalyzer (Agilent Technologies). Probes were synthesized using 2 successive rounds of cRNA amplification according to the standard Affymetrix protocols, and hybridized to mouse 430 2.0 chips (Affymetrix). Raw data were transformed with the GCRMA algorithm. The microarray data have been deposited in the RIKEN Research Center for Allergy and Immunology RefDIC database (http://refdic.rcai.riken.jp/). Sample IDs for WT splenic, WT BM, Spib−/− splenic, and Spib−/− BM pDCs are RSM04570, RSM04571, RSM04572, and RSM04573, respectively. More than 10-fold down-regulated genes in Spi-B–deficient pDCs were subjected to GeneSpring GX11 software (Agilent technologies) analysis to draw the clustering tree and heat map.
Statistical analyses
Statistical significance was determined by a 2-tailed Student t test. P < .05 was considered statistically significant.
Results
Spi-B can activate type I IFN promoters in synergy with IRF-7
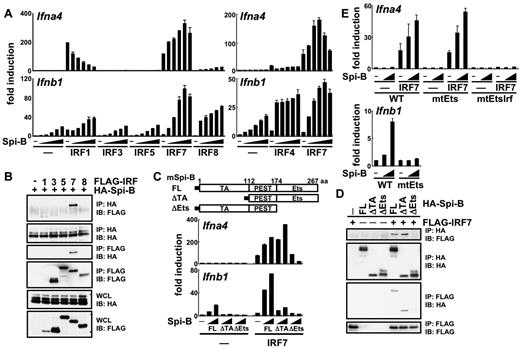
An Ets family transcription factor, Spi-B, is abundantly expressed in pDCs compared with other DC subsets.16,17 Spi-B can transactivate the Ig light chain enhancer in cooperation with IRF-8.15 In the present study, we first investigated whether Spi-B could transactivate type I IFN promoters with or without IRFs using the luciferase assay. Spi-B alone failed to activate the Ifna4 promoter (Figure 1A). However, it could synergistically enhance IRF-7–induced transactivation. Spi-B did not show such activity when coexpressed with other IRFs, although it could down-regulate IRF-1–induced activation. The Ifnb1 promoter was activated by Spi-B alone (Figure 1A). This activity was enhanced by IRF-7, although IRF-7 alone failed to activate the promoter. The synergism was also observed with IRF-4 or IRF-8, although to a lesser extent than with IRF-7. Spi-B also augmented IRF-1–induced Ifnb1 promoter activation, but this effect was additive. IRF-3 and IRF-5 did not activate the Ifna4 or Ifnb1 promoters despite coexpression of Spi-B. We conclude that Spi-B can activate type I IFN promoters in the best synergy with IRF-7.
Molecular mechanisms for Spi-B–mediated type I IFN promoter activation. (A) 293T cells were transiently transfected with an Ifna4 or Ifnb1 promoter-driven luciferase reporter plasmid alone or together with a combination of expression vectors for Spi-B (5.25, 10.5, 21, 42, or 84 ng/well) and IRF family members (8.4 ng/well). After 18 hours, cell lysates were prepared and subjected to the luciferase assay. (B) Interaction of Spi-B with IRF family members in 293T cells. IP indicates immunoprecipitation; IB, immunoblot; and WCL, whole cell lysate. (C) 293T cells were transiently transfected with an Ifna4 or Ifnb1 promoter-driven luciferase reporter plasmid alone or together with a combination of expression vectors for Spi-B mutants (21 or 84 ng/well) and IRF-7 (8.4 ng/well). The luciferase assay was carried out as indicated in panel A. Filled boxes denote HA tag. FL indicates full-length Spi-B; ΔTA, transactivation domain deletion mutant of Spi-B; ΔEts, Ets domain deletion mutant of Spi-B; and PEST, proline, glutamic acid, serine and threonine rich domain. (D) Interaction of Spi-B mutants with IRF-7 in 293T cells. (E) The following mutant promoters were used for the luciferase assay. Ifna4-am4Ets, the Ifna4 promoter carrying the mutation in all 4 Ets-binding sites; Ifna4-am4Ets9596, the Ifna4 promoter carrying the mutation in all 4 Ets-binding sites and an IRF-binding site; and Ifnb1-bm4Ets, the Ifnb1 promoter carrying the mutation in all 4 Ets-binding sites. Data are representative of 2 independent experiments (means ± SD in panels A, C, and E).
Molecular mechanisms for Spi-B–mediated type I IFN promoter activation. (A) 293T cells were transiently transfected with an Ifna4 or Ifnb1 promoter-driven luciferase reporter plasmid alone or together with a combination of expression vectors for Spi-B (5.25, 10.5, 21, 42, or 84 ng/well) and IRF family members (8.4 ng/well). After 18 hours, cell lysates were prepared and subjected to the luciferase assay. (B) Interaction of Spi-B with IRF family members in 293T cells. IP indicates immunoprecipitation; IB, immunoblot; and WCL, whole cell lysate. (C) 293T cells were transiently transfected with an Ifna4 or Ifnb1 promoter-driven luciferase reporter plasmid alone or together with a combination of expression vectors for Spi-B mutants (21 or 84 ng/well) and IRF-7 (8.4 ng/well). The luciferase assay was carried out as indicated in panel A. Filled boxes denote HA tag. FL indicates full-length Spi-B; ΔTA, transactivation domain deletion mutant of Spi-B; ΔEts, Ets domain deletion mutant of Spi-B; and PEST, proline, glutamic acid, serine and threonine rich domain. (D) Interaction of Spi-B mutants with IRF-7 in 293T cells. (E) The following mutant promoters were used for the luciferase assay. Ifna4-am4Ets, the Ifna4 promoter carrying the mutation in all 4 Ets-binding sites; Ifna4-am4Ets9596, the Ifna4 promoter carrying the mutation in all 4 Ets-binding sites and an IRF-binding site; and Ifnb1-bm4Ets, the Ifnb1 promoter carrying the mutation in all 4 Ets-binding sites. Data are representative of 2 independent experiments (means ± SD in panels A, C, and E).
To gain further insight into how Spi-B synergizes with IRF-7, we examined its association with IRF family members (Figure 1B). Spi-B was coimmunoprecipitated with IRF-7, but not with other IRFs such as IRF-3, IRF-5, and IRF-8. It was difficult to assess whether Spi-B can associate with IRF-1, because the amount of IRF-1 in the extracts was much lower than that of other IRFs. Therefore, we have demonstrated that Spi-B preferentially associates with IRF-7 among the IRF family members.
Spi-B has an N-terminal transactivation (TA) domain and C-terminal DNA-binding Ets domain. We analyzed whether these domains are required for Spi-B function. Deletion of an Ets domain, but not a TA domain, abolished the synergistic activity of Spi-B with IRF-7 on the Ifna4 promoter (Figure 1C). Association of Spi-B with IRF-7 was also eliminated by deletion of an Ets domain, whereas a TA domain deletion mutant of Spi-B retained the association with IRF-7 (Figure 1D). Deletion of an Ets domain or a TA domain abolished the transactivating effect of Spi-B on the Ifnb1 promoter. Therefore, an Ets domain of Spi-B is required for both type I IFN promoter activation and association with IRF-7. A TA domain of Spi-B, although dispensable for association with IRF-7, is involved in Ifnb1 promoter activation.
We also examined whether Spi-B acts through the Ets-binding sites in type I IFN promoters (Figure 1E and supplemental Figure 1). Spi-B could augment IRF-7–induced Ifna4 promoter activation even if 4 potential Ets-binding sites in the promoter were mutated. The IRF-binding site was required for IRF-7–induced activation, as reported previously.27 Spi-B failed to transactivate the Ifnb1 promoter when 4 potential Ets-binding sites were mutated. Therefore, the Ets-binding sites were dispensable for Spi-B to synergistically activate Ifna4 promoter with IRF-7, whereas they were required for Spi-B to activate the Ifnb1 promoter.
TLR7/9 signal–induced cytokine production in Spi-B–deficient pDCs and mice
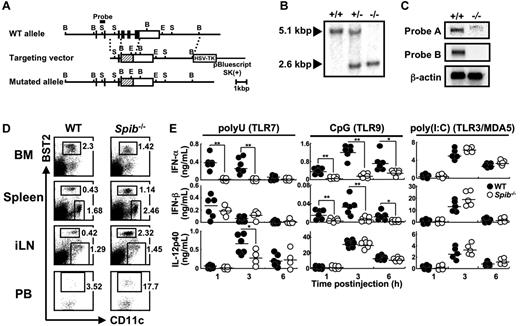
We have then generated Spi-B–deficient mice (Figure 2A). The targeting was confirmed by Southern blot analysis (Figure 2B). Northern blot analysis also showed that the expression level of the transcript from the mutated allele was faint (Figure 2C and supplemental Figure 2). Therefore, it can be assumed that the functional Spi-B protein is not generated in Spi-B–deficient mice.
Generation and cytokine responses of Spi-B–deficient mice. (A) Schematic representation of the gene targeting. Filled and open boxes represent coding and 3′-untranslated regions of Spib gene, respectively. Hatched boxes represent the coding region for neomycin resistance gene. B indicates BamH I; S, Sph I; and E, EcoR I. (B) Southern blot analysis of offspring from the heterozygote intercrosses. Genomic DNA was digested with BamH I, electrophoresed, and hybridized with the probe indicated in panel A. (C) For Northern blot analysis, Spi-B cDNA fragments corresponding to the 5′ region (nondeleted region, probe A) or to the deleted region (probe B) were used as probes. The same membrane was also hybridized with a β-actin cDNA fragment as a control. (D) FACS profiles of BM, spleen, iLN, and PB cells from WT or Spi-B–deficient mice. Dot plots of CD11c versus BST2 in BM, spleen, and iLN cells are shown. Numbers indicate percentages of gated cells among total live cells. For PB cells, dot plots of CD11c versus BST2 in CD19−CD11c+B220+ cells are shown. Numbers indicate the percentages of gated cells among CD19−CD11c+B220+ cells. Data are representative of at least 3 independent experiments. (E) Mice were injected intravenously with polyuridylic acid (polyU, a TLR7 agonist) or ODN1668 (a TLR9 agonist) or injected intraperitoneally with poly(I:C) (a TLR3/MDA5 agonist) and serum cytokine levels were measured at the indicated time points. Each symbol represents the data from 1 mouse and the bars indicate the mean. *P < .05; **P < .01
Generation and cytokine responses of Spi-B–deficient mice. (A) Schematic representation of the gene targeting. Filled and open boxes represent coding and 3′-untranslated regions of Spib gene, respectively. Hatched boxes represent the coding region for neomycin resistance gene. B indicates BamH I; S, Sph I; and E, EcoR I. (B) Southern blot analysis of offspring from the heterozygote intercrosses. Genomic DNA was digested with BamH I, electrophoresed, and hybridized with the probe indicated in panel A. (C) For Northern blot analysis, Spi-B cDNA fragments corresponding to the 5′ region (nondeleted region, probe A) or to the deleted region (probe B) were used as probes. The same membrane was also hybridized with a β-actin cDNA fragment as a control. (D) FACS profiles of BM, spleen, iLN, and PB cells from WT or Spi-B–deficient mice. Dot plots of CD11c versus BST2 in BM, spleen, and iLN cells are shown. Numbers indicate percentages of gated cells among total live cells. For PB cells, dot plots of CD11c versus BST2 in CD19−CD11c+B220+ cells are shown. Numbers indicate the percentages of gated cells among CD19−CD11c+B220+ cells. Data are representative of at least 3 independent experiments. (E) Mice were injected intravenously with polyuridylic acid (polyU, a TLR7 agonist) or ODN1668 (a TLR9 agonist) or injected intraperitoneally with poly(I:C) (a TLR3/MDA5 agonist) and serum cytokine levels were measured at the indicated time points. Each symbol represents the data from 1 mouse and the bars indicate the mean. *P < .05; **P < .01
We analyzed DC population in various tissues of Spi-B–deficient mice. pDCs are included in CD11c+B220+ cells and expresses several pDC-specific surface markers such as BST2. In Spi-B–deficient mice, the frequency and absolute number of pDCs were significantly decreased in the BM, whereas those of pDCs in peripheral lymphoid organs such as spleen, iLNs, and PB were 2- to 3-fold increased (Figure 2D and Table 1). The frequency and absolute cell numbers of CD11c+B220− cDCs in the iLNs were comparable between WT and Spi-B–deficient mice. Although those of splenic cDCs were slightly increased in Spi-B–deficient mice, splenic cDC subsets (CD4−CD8α+, CD4+CD8α−, and CD4−CD8α−) were detected at comparable levels in WT and Spi-B–deficient mice (Figure 2D, supplemental Figure 3, and Table 1). Therefore, Spi-B deficiency led to a reduction of BM pDCs concomitant with increase of peripheral pDCs.
Cell numbers and percentages in BM, spleen, iLNs, and PB
| . | WT . | Spi-B−/− . |
|---|---|---|
| BM (n = 8) | ||
| Total cells, × 10−7 | 2.4 ± 0.3 | 2.6 ± 0.5 |
| pDCs, × 10−5 | 3.7 ± 1.1 | 2.2 ± 1.1* |
| pDCs, % | 1.5 ± 0.4 | 0.9 ± 0.3† |
| B cells, × 10−6 | 3.1 ± 1.1 | 3.7 ± 2.3 |
| B cells, % | 12.7 ± 4.6 | 13.7 ± 6.8 |
| Spleen (n = 8) | ||
| Total cells, × 10−7 | 3.3 ± 0.7 | 3.2 ± 0.9 |
| pDCs, × 10−5 | 1.1 ± 0.4 | 2.3 ± 0.7† |
| pDCs, % | 0.3 ± 0.1 | 0.7 ± 0.1‡ |
| cDCs, × 10−5 | 5.6 ± 1.4 | 7.2 ± 1.5* |
| cDCs, % | 1.7 ± 0.2 | 2.3 ± 0.2‡ |
| B cells, × 10−7 | 1.6 ± 0.4 | 1.5 ± 0.5 |
| B cells, % | 47.3 ± 4.5 | 47.3 ± 5.7 |
| iLNs (n = 6) | ||
| Total cells, × 10−6 | 1.7 ± 0.9 | 0.8 ± 0.4* |
| pDCs, × 10−4 | 0.5 ± 0.2 | 1.3 ± 0.6* |
| pDCs, % | 0.3 ± 0.1 | 1.5 ± 0.4‡ |
| cDCs, × 10−3 | 8.0 ± 4.3 | 6.2 ± 4.1 |
| cDCs, % | 0.6 ± 0.2 | 0.6 ± 0.2 |
| B cells, × 10−5 | 3.9 ± 2.4 | 2.1 ± 0.9 |
| B cells, % | 22.2 ± 4.2 | 25.7 ± 3.4 |
| PB (n = 8) | ||
| Total cells, × 10−6 | 2.4 ± 0.7 | 2.5 ± 0.8 |
| pDCs, × 10−2 | 1.7 ± 1.3 | 5.9 ± 4.8* |
| pDCs, % | 0.1 ± 0.1 | 0.2 ± 0.2* |
| B cells, × 10−5 | 5.3 ± 2.3 | 5.7 ± 1.9 |
| B cells, % | 21.4 ± 3.3 | 22.9 ± 3.3 |
| . | WT . | Spi-B−/− . |
|---|---|---|
| BM (n = 8) | ||
| Total cells, × 10−7 | 2.4 ± 0.3 | 2.6 ± 0.5 |
| pDCs, × 10−5 | 3.7 ± 1.1 | 2.2 ± 1.1* |
| pDCs, % | 1.5 ± 0.4 | 0.9 ± 0.3† |
| B cells, × 10−6 | 3.1 ± 1.1 | 3.7 ± 2.3 |
| B cells, % | 12.7 ± 4.6 | 13.7 ± 6.8 |
| Spleen (n = 8) | ||
| Total cells, × 10−7 | 3.3 ± 0.7 | 3.2 ± 0.9 |
| pDCs, × 10−5 | 1.1 ± 0.4 | 2.3 ± 0.7† |
| pDCs, % | 0.3 ± 0.1 | 0.7 ± 0.1‡ |
| cDCs, × 10−5 | 5.6 ± 1.4 | 7.2 ± 1.5* |
| cDCs, % | 1.7 ± 0.2 | 2.3 ± 0.2‡ |
| B cells, × 10−7 | 1.6 ± 0.4 | 1.5 ± 0.5 |
| B cells, % | 47.3 ± 4.5 | 47.3 ± 5.7 |
| iLNs (n = 6) | ||
| Total cells, × 10−6 | 1.7 ± 0.9 | 0.8 ± 0.4* |
| pDCs, × 10−4 | 0.5 ± 0.2 | 1.3 ± 0.6* |
| pDCs, % | 0.3 ± 0.1 | 1.5 ± 0.4‡ |
| cDCs, × 10−3 | 8.0 ± 4.3 | 6.2 ± 4.1 |
| cDCs, % | 0.6 ± 0.2 | 0.6 ± 0.2 |
| B cells, × 10−5 | 3.9 ± 2.4 | 2.1 ± 0.9 |
| B cells, % | 22.2 ± 4.2 | 25.7 ± 3.4 |
| PB (n = 8) | ||
| Total cells, × 10−6 | 2.4 ± 0.7 | 2.5 ± 0.8 |
| pDCs, × 10−2 | 1.7 ± 1.3 | 5.9 ± 4.8* |
| pDCs, % | 0.1 ± 0.1 | 0.2 ± 0.2* |
| B cells, × 10−5 | 5.3 ± 2.3 | 5.7 ± 1.9 |
| B cells, % | 21.4 ± 3.3 | 22.9 ± 3.3 |
Percentages and numbers of pDCs (CD19−CD11c+B220+BST2+), cDCs (CD19−CD11c+B220−), and B cells (CD19+CD11c−) among total live cells are shown. Data represent means ± SD.
P < .05.
P < .01.
P < .001.
We then examined serum cytokine levels in vivo after TLR7/9 stimuli (Figure 2E). Serum IFN-α elevation was abolished in Spi-B–deficient mice. The mutant mice also showed defects in IFN-β and IL-12p40 responses, although the defect was not so prominent as in the IFN-α response. In response to TLR7/9 agonists, only pDCs can produce IFN-α, whereas both pDCs and cDCs can produce IFN-β and IL-12p40.4 Therefore, with a pDC-specific defect, the most pronounced effect should be on IFN-α induction, and our results indicate that Spi-B is required for TLR7/9-induced pDC responses in vivo.
Double-stranded RNA can induce production of IL-12p40 and IFN-α in a TLR3- and MDA5-dependent manner, respectively,28 and this activity does not depend on pDCs, but on cDCs or other cells such as fibroblasts. Double-stranded RNA-induced responses were normal in Spi-B–deficient mice (Figure 2E).
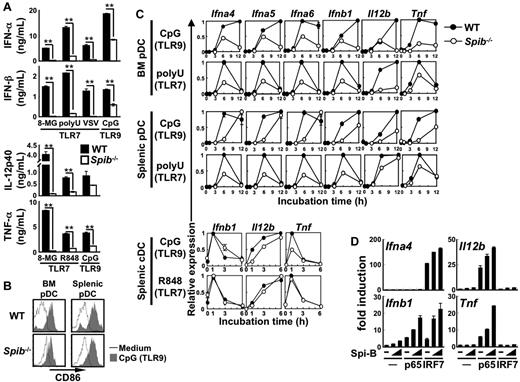
We also analyzed TLR7/9 signal-induced cytokine production by pDCs. The stimuli included synthetic agonists or VSV.10,29 Cytokine responses were impaired in Spi-B–deficient BM pDCs (Figure 3A). TLR7 and TLR9 stimuli could up-regulate CD86 expression in both WT and Spi-B–deficient pDCs, indicating that TLR activation for CD86 up-regulation is intact in Spi-B–deficient pDCs (Figure 3B and supplemental Figure 4). We also analyzed the expression of cytokine genes, including various IFN-α subtypes, in pDCs and cDCs after TLR7/9 stimulation (Figure 3C). All types of Spi-B–deficient BM and splenic pDCs showed defective induction of these genes (Figure 3C). In contrast, induction of genes such as Ifnb1, Il12b, and Tnf in Spi-B–deficient splenic cDCs was normal (Figure 3C). Therefore, Spi-B is selectively required for TLR7/9-induced cytokine production by BM and splenic pDCs.
Roles of Spi-B in in vitro pDCs responses to TLR7/9 stimuli. (A) MACS-purified BM pDCs were stimulated with 8-mercaptoguanosine (8-MG, a TLR7 agonist), polyuridylic acid (polyU, a TLR7 agonist), VSV (a TLR7 agonist), CpG DNA (a TLR9 agonist), or R848 (a TLR7 agonist). 8-MG was used at 1mM. PolyU was added at 3μg/mL as a complex with lipofectamine 2000. As CpG DNA, D19 was used at 3μM for measurement of IFN-α and IFN-β and ODN1668 was used at 1μM for measurement of IL-12p40 and TNF-α. R848 was used at 100nM. VSV was used at a multiplicity of infection of 0.5. After 20-24 hours, cytokine production was measured by ELISA. Data are representative of 3 independent experiments (means ± SD). **P < .01. (B) CD86 expression in TLR9-stimulated pDCs. MACS-purified splenic or BM pDCs were stimulated with (shaded histograms) or without (open histograms) 1μM ODN1668 for 12 hours. Surface expression of CD86 in CD19−CD11c+BST2+ cells was analyzed with a FACSCalibur. Data are representative of 2 independent experiments. (C) Gene induction in TLR7/9-stimulated pDCs and cDCs. FACS-sorted BM or splenic pDCs were stimulated with 3μM D19 or 3 μg/mL of polyU. FACS-sorted splenic cDCs were stimulated with 1μM ODN1668 or 100nM R848. Cells were harvested at the indicated time points and subjected to quantitative real-time PCR. Data are representative of 2 independent experiments (means ± SD). (D) 293T cells were transiently transfected with an Ifna4, Ifnb1, Il12b, or Tnf promoter-driven luciferase reporter plasmid alone or together with a combination of expression vectors for Spi-B (2.1 or 8.4 ng/well), NF-κB p65 (8.4 ng/well), and IRF-7 (8.4 ng/well). After 18 hours, cell lysates were prepared and subjected to the luciferase assay. Data are representative of 2 independent experiments (means ± SD).
Roles of Spi-B in in vitro pDCs responses to TLR7/9 stimuli. (A) MACS-purified BM pDCs were stimulated with 8-mercaptoguanosine (8-MG, a TLR7 agonist), polyuridylic acid (polyU, a TLR7 agonist), VSV (a TLR7 agonist), CpG DNA (a TLR9 agonist), or R848 (a TLR7 agonist). 8-MG was used at 1mM. PolyU was added at 3μg/mL as a complex with lipofectamine 2000. As CpG DNA, D19 was used at 3μM for measurement of IFN-α and IFN-β and ODN1668 was used at 1μM for measurement of IL-12p40 and TNF-α. R848 was used at 100nM. VSV was used at a multiplicity of infection of 0.5. After 20-24 hours, cytokine production was measured by ELISA. Data are representative of 3 independent experiments (means ± SD). **P < .01. (B) CD86 expression in TLR9-stimulated pDCs. MACS-purified splenic or BM pDCs were stimulated with (shaded histograms) or without (open histograms) 1μM ODN1668 for 12 hours. Surface expression of CD86 in CD19−CD11c+BST2+ cells was analyzed with a FACSCalibur. Data are representative of 2 independent experiments. (C) Gene induction in TLR7/9-stimulated pDCs and cDCs. FACS-sorted BM or splenic pDCs were stimulated with 3μM D19 or 3 μg/mL of polyU. FACS-sorted splenic cDCs were stimulated with 1μM ODN1668 or 100nM R848. Cells were harvested at the indicated time points and subjected to quantitative real-time PCR. Data are representative of 2 independent experiments (means ± SD). (D) 293T cells were transiently transfected with an Ifna4, Ifnb1, Il12b, or Tnf promoter-driven luciferase reporter plasmid alone or together with a combination of expression vectors for Spi-B (2.1 or 8.4 ng/well), NF-κB p65 (8.4 ng/well), and IRF-7 (8.4 ng/well). After 18 hours, cell lysates were prepared and subjected to the luciferase assay. Data are representative of 2 independent experiments (means ± SD).
Spi-B–deficient pDCs showed defective production of not only type I IFNs, but also proinflammatory cytokines in response to TLR7/9 signaling (Figure 3A,C). Therefore, we analyzed whether Spi-B can activate promoters of Il12b and Tnf (Figure 3D). Spi-B expression alone failed to activate these promoters. However, NF-κB p65 could activate these promoters and these activities were synergistically enhanced by coexpression of Spi-B. Spi-B synergy with NF-κB p65 was also observed in transactivating the Ifnb1, but not Ifna4, promoter (Figure 3D).
As described previously,20 anti-Ig–induced B-cell proliferation was severely impaired in Spi-B–deficient B cells (supplemental Figure 5). However, both proliferative responses and IL-6 production in response to TLR7/9 agonists were normal, indicating that Spi-B is dispensable for TLR7/9-induced B-cell responses.
We also tested antiviral responses of WT and Spi-B–deficient mice. Mice were intravenously infected with 107 median tissue culture infective dose (TCID50) per head of VSV. At 6 hours after injection, viral titer and serum IFN-α levels in WT versus Spi-B–deficient mice were 51.7 ± 20.6 × 103 versus 58.8 ± 41.1 × 103 TCID50/100 mg of spleen and 1.19 ± 0.83 versus 1.75 ± 0.59 ng/mL (n = 5), respectively. Therefore, although Spi-B–deficient mice showed defective responses against TLR7/9 agonists, the mutant mice showed comparable responses against VSV with WT mice in this condition.
pDC development in Spi-B–deficient mice
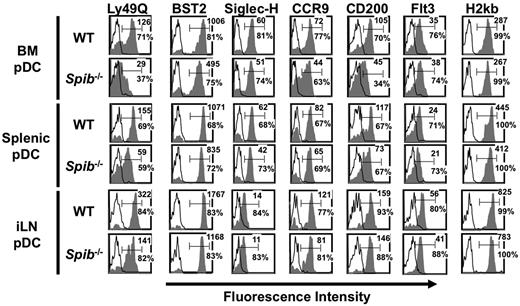
We further analyzed expression of surface markers on BM and peripheral pDCs. Expression of Ly49Q, a type II C-type lectin, increases along with pDC maturation, but is not detected on cDCs.30,31 The expression level of Ly49Q on Spi-B–deficient BM pDCs was severely impaired (Figure 4). Expression levels of BST2, Siglec-H, CCR9 or CD200, which are also expressed abundantly by pDCs but not cDCs, was also defective, although the defect was not so prominent for Ly49Q expression (Figure 4). The expression level of MHC class I or Fms-like tyrosine kinase 3 (Flt3) was not impaired. Similar defects were also observed in Spi-B–deficient splenic and iLN pDCs, although to a lesser extent than in BM pDCs. CD200 expression on Spi-B–deficient B-lineage cells was unimpaired (supplemental Figure 6A). Therefore, Spi-B deficiency leads to a selective defect in the expression of a set of surface markers on BM and peripheral pDCs.
FACS analysis of Spi-B–deficient mice. CD19−CD11c+B220+ cells were gated and analyzed for the expression of the indicated surface markers. Open histograms indicate isotype controls. Numbers indicate percentages and mean fluorescence intensity of positive cells for the indicated surface markers. Data are representative of at least 3 independent experiments.
FACS analysis of Spi-B–deficient mice. CD19−CD11c+B220+ cells were gated and analyzed for the expression of the indicated surface markers. Open histograms indicate isotype controls. Numbers indicate percentages and mean fluorescence intensity of positive cells for the indicated surface markers. Data are representative of at least 3 independent experiments.
According to Schlitzer et al,32 CCR9+ mature pDCs are generated from their immediate precursors, CCR9− pDCs, which show low expression of Sca-1 and lack I-Ab expression. In Spi-B–deficient mice, the CCR9− population was increased (supplemental Figure 6B). The CCR9− population in the mutant mice showed lower expression of Sca-1 and lacked I-Ab expression than those in WT mice (supplemental Figure 6C). The results indicate that transition of CCR9− pDCs to CCR9+ pDCs is impaired in Spi-B–deficient BM.
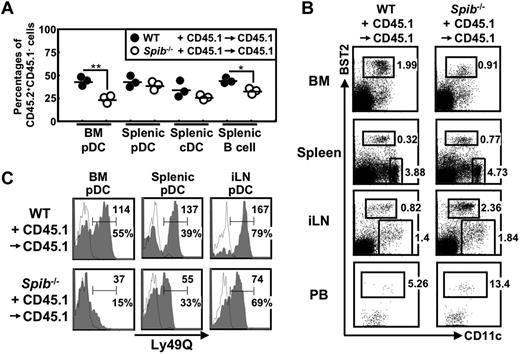
To clarify whether these defects in Spi-B–deficient mice result from a cell-autonomous effect or from the dysfunction of non-pDC populations, we generated BM chimeras by transplanting a 1:1 mixture of CD45.2+ WT or Spi-B–deficient BM cells with CD45.1+ WT BM cells into irradiated CD45.1+ WT mice (WT + CD45.1→CD45.1 or Spib−/− + CD45.1→CD45.1 chimeric mice; Figure 5). In the Spib−/− + CD45.1→CD45.1 chimeric mice, CD45.2+ Spi-B–deficient cells were predominated by CD45.1+ WT cells in BM pDCs. Such predominance was observed also in B cells, although to a lesser extent than in BM pDCs. In contrast, splenic pDCs and cDCs contained comparable percentages of Spi-B–deficient and WT cells (Figure 5A). In addition, in Spib−/− + CD45.1→CD45.1 chimeric mice, the frequency of BM pDCs among CD45.2+ cells was decreased, whereas that of splenic or iLN pDCs was increased (Figure 5B). Furthermore, decreased expression of Ly49Q in Spi-B–deficient pDCs was also observed in Spib−/− + CD45.1→CD45.1 chimeric mice (Figure 5C). Therefore, pDC defects were detected despite the coexistence of WT hematopoietic and nonhematopoietic cells, indicating that Spi-B in pDCs is required for optimal generation and Ly49Q expression of BM pDCs.
pDC defects in Spi-B–deficient mice are cell intrinsic. A 1:1 mixture of CD45.2+ WT C57BL/6 or Spi-B–deficient BM cells with CD45.1+ WT BM cells were transferred to irradiated CD45.1+ WT C57BL/6 recipients. The resulting chimeric mice, WT + CD45.1→CD45.1 and Spib−/− + CD45.1→CD45.1 mice, were analyzed 6-8 weeks after BM transfer. (A) BM pDCs, splenic pDCs, splenic cDCs, and splenic B cells were gated as CD19−CD11c+B220+, CD19−CD11c+B220+, CD19−CD11c+B220−, and CD19+CD11c− cells, respectively. The percentages of CD45.2+CD45.1− cells among these cells are shown. *P < .05; **P < .01. (B) CD45.2+CD45.1− cells were gated and dot plots of CD11c versus BST2 in BM, spleen, and iLN cells are shown. Numbers indicate percentages of gated cells among CD45.2+CD45.1− cells. For PB cells, dot plots of CD11c versus BST2 in CD19−CD11c+B220+ cells are shown. Numbers indicate the percentages of gated cells among CD45.2+CD45.1−CD19−CD11c+B220+ cells. (C) CD45.2+CD45.1−CD19−CD11c+B220+ cells were gated and analyzed for the expression of Ly49Q. Open histograms indicate isotype controls. Numbers indicate percentages and mean fluorescence intensity of Ly49Q+ cells. Experiments were performed 2 times using 6 WT + CD45.1→CD45.1 and 7 Spib−/− + CD45.1→CD45.1 mice in total. Panel A shows the data from one experiment; panels B and C show representative data.
pDC defects in Spi-B–deficient mice are cell intrinsic. A 1:1 mixture of CD45.2+ WT C57BL/6 or Spi-B–deficient BM cells with CD45.1+ WT BM cells were transferred to irradiated CD45.1+ WT C57BL/6 recipients. The resulting chimeric mice, WT + CD45.1→CD45.1 and Spib−/− + CD45.1→CD45.1 mice, were analyzed 6-8 weeks after BM transfer. (A) BM pDCs, splenic pDCs, splenic cDCs, and splenic B cells were gated as CD19−CD11c+B220+, CD19−CD11c+B220+, CD19−CD11c+B220−, and CD19+CD11c− cells, respectively. The percentages of CD45.2+CD45.1− cells among these cells are shown. *P < .05; **P < .01. (B) CD45.2+CD45.1− cells were gated and dot plots of CD11c versus BST2 in BM, spleen, and iLN cells are shown. Numbers indicate percentages of gated cells among CD45.2+CD45.1− cells. For PB cells, dot plots of CD11c versus BST2 in CD19−CD11c+B220+ cells are shown. Numbers indicate the percentages of gated cells among CD45.2+CD45.1−CD19−CD11c+B220+ cells. (C) CD45.2+CD45.1−CD19−CD11c+B220+ cells were gated and analyzed for the expression of Ly49Q. Open histograms indicate isotype controls. Numbers indicate percentages and mean fluorescence intensity of Ly49Q+ cells. Experiments were performed 2 times using 6 WT + CD45.1→CD45.1 and 7 Spib−/− + CD45.1→CD45.1 mice in total. Panel A shows the data from one experiment; panels B and C show representative data.
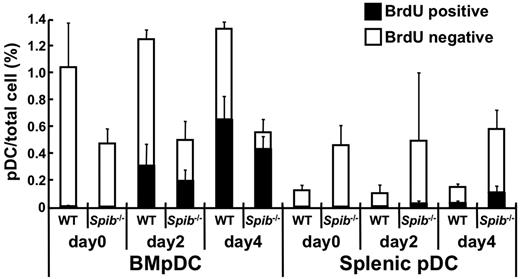
Considering that the number of pDCs in the BM was decreased in Spi-B–deficient mice, we then evaluated the proliferation status of pDCs in the BM or spleen by BrdU incorporation (Figure 6). In WT mice, the percentages of BrdU+ pDCs were higher in the BM than in the spleen, suggesting that pDCs actively proliferate in the BM. The percentages of BrdU+ BM pDCs significantly increased in both WT and Spi-B–deficient mice. In contrast, BrdU− pDCs were prominently decreased in Spi-B–deficient mice. Most of the splenic pDCs in WT and Spi-B–deficient mice were BrdU−. These results indicate that the decrease of BM pDCs in Spi-B–deficient mice is not because of defective proliferation and that nondividing pDCs are severely decreased in the BM and accumulated in the periphery in Spi-B–deficient mice.
The kinetics of BrdU labeling in WT and Spi-B–deficient pDCs. Mice were injected intraperitoneally at day 0 with BrdU and given BrdU continuously in the drinking water. At the indicated days, BM and spleen cells were analyzed for BrdU incorporation. The percentages of CD19−CD11c+B220+BST2+ cells (ie, pDCs) among total cells are shown. BrdU+ and BrdU− cells among the pDCs are indicated by black and white columns, respectively (4 mice at each time point, means ± SD). Each symbol represents the data from one mouse and the bars indicate the mean. **P < .01.
The kinetics of BrdU labeling in WT and Spi-B–deficient pDCs. Mice were injected intraperitoneally at day 0 with BrdU and given BrdU continuously in the drinking water. At the indicated days, BM and spleen cells were analyzed for BrdU incorporation. The percentages of CD19−CD11c+B220+BST2+ cells (ie, pDCs) among total cells are shown. BrdU+ and BrdU− cells among the pDCs are indicated by black and white columns, respectively (4 mice at each time point, means ± SD). Each symbol represents the data from one mouse and the bars indicate the mean. **P < .01.
Spi-B is required for expression of a set of pDC-specific genes
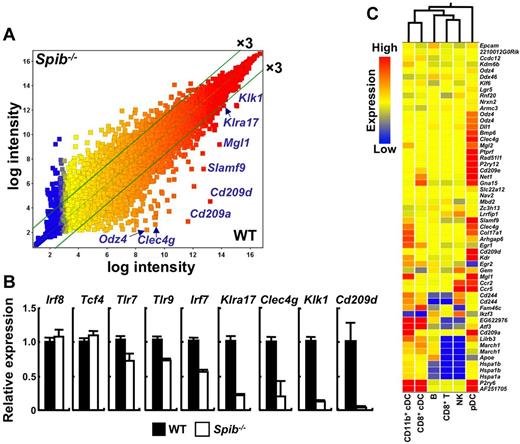
To analyze how Spi-B regulates gene-expression programs, we performed microarray analysis of WT and Spi-B–deficient BM pDCs (Figure 7). In Spi-B–deficient BM pDCs, expression of 597 and 56 genes was down-regulated more than 3- and 10-fold, respectively (Figure 7A and supplemental Table 1). Reduced expression of Klra17 (Ly49Q), Clec4g, Klk1, and Cd209d was further verified by quantitative real-time PCR (Figure 7B). The reduction in expression was not so prominent for Irf8, Tcf4 (E2-2), Tlr7, Tlr9, and Irf7, all of which are important for pDC function or development.
Gene expression in Spi-B–deficient pDCs. (A) Expression profiles of WT and Spi-B–deficient BM pDCs. CD11c+B220+BST2+ cells from WT and Spi-B–deficient mice were sorted as BM pDCs and used. The scatter plot represents normalized log intensities of individual probes. The lines indicate the 3-fold difference. (B) Expression profiles of indicated genes in sorted BM pDCs were analyzed by quantitative real-time PCR. BM pDCs were pooled from 4 mice for each experiment. Data are representative of 2 independent experiments (means ± SD). (C) Clustering of cell populations by probe sets with expression that was more than 10-fold down-regulated in Spi-B–deficient BM pDCs. The results were compared with the expression database of normal immune cell populations (Gene Expression Omnibus dataset GSE9810).
Gene expression in Spi-B–deficient pDCs. (A) Expression profiles of WT and Spi-B–deficient BM pDCs. CD11c+B220+BST2+ cells from WT and Spi-B–deficient mice were sorted as BM pDCs and used. The scatter plot represents normalized log intensities of individual probes. The lines indicate the 3-fold difference. (B) Expression profiles of indicated genes in sorted BM pDCs were analyzed by quantitative real-time PCR. BM pDCs were pooled from 4 mice for each experiment. Data are representative of 2 independent experiments (means ± SD). (C) Clustering of cell populations by probe sets with expression that was more than 10-fold down-regulated in Spi-B–deficient BM pDCs. The results were compared with the expression database of normal immune cell populations (Gene Expression Omnibus dataset GSE9810).
We also performed microarray analysis of WT and Spi-B–deficient splenic pDCs (supplemental Figure 7 and supplemental Table 2). Expression of certain sets of genes such as Clec4g and Cd209d were also down-regulated in Spi-B–deficient splenic pDCs, although the degree of down-regulation was not so prominent as in BM pDCs. The expression of Klra17 and Klk1 was also down-regulated by 1.9-fold and 2.5- to 2.7-fold, respectively. In human pDCs, an antiapoptotic gene, BCL2A1, was identified as a target gene of Spi-B.33 However, the expression levels of the Bcl2a1 in BM and splenic pDCs were low and at comparable levels between WT and Spi-B–deficient mice (data not shown). Clustering analysis of Spi-B–dependent genes against the database of gene expression in normal immune cell lineages34 distinguished pDCs from the other cell types, indicating that pDC-specific genes are enriched among Spi-B–dependent genes (Figure 7C). Deletion of a transcription factor, E2-2, a family of basic helix-loop-helix transcription factors, leads to pDC ablation.16 We compared Spi-B–dependent genes with E2-2–dependent ones (supplemental Table 1A). Among Spi-B–dependent genes, 8 were E2-2 dependent and 16 were up-regulated in Tcf4+/− pDCs. These results indicate that Spi-B regulates pDC-specific gene-expression programs in a distinct manner with E2-2.
We next analyzed how Spi-B controls expression of the Klra17 gene. Spi-B could transactivate the potential Klra17 promoter region (supplemental Figure 8A). The critical region was located between −352 and −70 bp and the essential Ets-binding site was identified (supplemental Figure 8A-B). Consistently, histone H3 acetylation, which is associated with chromatin opening, was severely decreased in the Klra17 locus of Spi-B–deficient BM pDCs (supplemental Figure 8C). Therefore, Spi-B is required for transactivating the promoter of the pDC-specific gene Klra17 through its Ets-binding site.
Ly49Q is involved in type I IFN production from TLR7/9-stimulated pDCs.35 Therefore, we investigated whether enforced expression of Ly49Q could rescue the defective responses of Spi-B–deficient mice. For this purpose, we generated Ly49Q transgenic Spi-B–deficient mice.25 The mutant mice showed increased expression of Ly49Q in both BM and splenic pDCs compared with control Spi-B–deficient mice (data not shown). However, neither TLR7- nor TLR9-mediated type I IFN induction was restored in the mutant mice (supplemental Figure 8D). This indicates that the pDC defect in Spi-B–deficient mice cannot be ascribed solely to impaired expression of Ly49Q.
Discussion
Spi-B, together with PU.1 and Spi-C, constitutes an Ets subfamily. Spi-B and PU.1 bind the target DNA cooperatively with IRF-4 and IRF-8, respectively.15 In the present study, we demonstrated the best cooperativity of Spi-B with IRF-7, which is essential for type I IFN production from TLR7/9-stimulated pDCs.36 Spi-B showed synergy with IRF-4, but IRF-4 is dispensable for pDC function.37 TLR7/9 stimuli up-regulated the expression of IRF-7, but not Spi-B, in pDCs (supplemental Figure 9). The stimuli could also activate IRF-7 by inducing phosphorylation and nuclear translocation of IRF-7. It is a reasonable assumption that the constitutive high expression of Spi-B, together with IRF-7 activation, contributes to the ability of pDCs to produce vast amounts of type I IFNs in response to TLR7/9 signaling. A C-terminal Ets domain of Spi-B is involved in the ability of Spi-B to transactivate the type I IFN promoters and to associate with IRF-7.
Spi-B–deficient pDCs showed defects in production of not only type I IFN, but also IL12p40 and TNF-α. The promoters of these proinflammatory cytokines were not activated by IRF-7, but by NF-κB p65. Spi-B could synergistically augment this NF-κB p65-induced promoter activation. Therefore, Spi-B should be involved in potentiating gene expression of proinflammatory cytokines in cooperation with NF-κB p65.
Mutant mice with defective expression of several transcription factors show impaired pDC development. For example, E2-2 is preferentially expressed in pDCs and CD11c+B220+ cells were not detected in E2-2–deficient mice.16 A zinc finger protein, Ikaros, is essential for multilineage hematopoietic cell development38 and hypomorphic Ikaros mutant mice showed selective loss of CD11c+B220+ cells.39 Therefore, E2-2 and Ikaros are required for early pDC development. BM pDC development is defined by expression of several pDC-specific cell-surface molecules. CCR9+ pDCs are mature pDCs generated from CCR9− pDCs.32 Ly49Q expression can also distinguish immature Ly49Q− pDCs from mature Ly49Q+ pDCs.31 In the present study, immature pDC fractions, which can be defined as CCR9− or Ly49Q− cells, were increased in Spi-B–deficient BM, implying that differentiation into the CCR9+ or Ly49Q+ stage is impaired. Therefore, Spi-B is required for optimal pDC development at a later stage than E2-2. PU.1 is required for the generation of pDCs and cDCs40 and Spi-C is critical for development of the red pulp macrophage.41 Therefore, Spi-B and its related molecules play specific roles in the development of various subsets of DCs or macrophages.
The Spib promoter contains an E-protein–binding site and expression of the Spib gene is impaired in E2-2–deficient mice.16,42 The expression of the Tcf4 (E2-2) gene was not affected in Spi-B–deficient pDCs. Furthermore, E2-2–induced pDC generation in humans is attenuated by Spi-B knock-down.43 Therefore, Spi-B should function downstream of E2-2 in pDC development. However, a comparison of Spi-B- and E2-2–dependent genes indicated that Spi-B has its own specific roles distinct from E2-2 in regulating pDC gene-expression programs.
In Spi-B–deficient mice, BM pDCs were decreased and peripheral pDCs were increased. Mixed BM chimera analysis showed that this defect is intrinsic to pDCs. Because peripheral pDCs in spleen, iLNs, and PB were increased, the decrease is unlikely to be due to the enhanced pDC death. Because nondividing, rather than dividing, pDCs were severely decreased in the BM and increased in the periphery, it can be assumed that Spi-B–deficient mature pDCs precociously exit from the BM to the periphery. A homing defect to the BM can also lead to a decrease of BM pDC numbers with increased peripheral pDCs, although such a homing system to the BM has not been reported. Increased peripheral pDCs cannot fully compensate for the reduced BM pDCs in terms of TLR7/9-induced type I IFN production in mice (Figure 2E). Gene expression of chemokine receptors such as Ccr2 and Ccr5 and adhesion molecules such as Epcam were decreased in Spi-B–deficient BM pDCs. The expression of an antiapoptotic gene, Bcl2, was up-regulated in Spi-B–deficient splenic pDCs (supplemental Tables 1 and 2). Further studies are necessary to determine whether and how these expression patterns lead to a decrease of BM pDCs or an increase of splenic pDCs.
VSV-induced responses are known to be dependent on TLR744 and pDC-ablated mice show defects in early responses against VSV infection.45 However, in the present study, Spi-B–deficient mice showed comparable anti-VSV responses to WT mice, although Spi-B–deficient mice did not produce any IFN-α in response to TLR7/9 agonists. The reason for this phenotype is unclear at present. In pDC-ablated mice, serum IFN-α levels were impaired, but significantly increased in response to VSV infection,45 indicating that cells other than pDCs can also produce IFN-α. Candidate cells responsible for this type I IFN production may include fibroblasts, macrophages, or cDCs. These cells can respond to virus through cytosolic nucleic acid sensors, which can induce type I IFNs in an Spi-B–independent manner. Unlike pDC-ablated mice, cDC numbers were increased in Spi-B–deficient mice. Such increased cell numbers can contribute to VSV-induced responses by themselves or indirectly through pDCs. Further studies are required to elucidate the cell sources for type I IFN production in virally infected Spi-B–deficient mice.
In the present study, we have demonstrated unique roles of Spi-B in the function and development of pDCs. The molecular mechanisms by which Spi-B is involved are overlapped with, but distinct from, E2-2. pDCs are involved not only in antiviral immunity through the detection of viral nucleic acids, but also in pathogenesis of autoimmune diseases such as systemic lupus erythematosus and psoriasis by sensing host-derived nucleic acids.46,47 Therefore, Spi-B may be a novel target molecule for modulating defense against viral infection or treating autoimmune disorders in which pDCs are critically involved.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank A. Kato, I. Ogahara, and E. Haga for technical assistance; Y. Hachiman and H. Fujimoto for cell sorting; S. Haraguchi and Y. Matsuhisa for secretarial assistance; Drs A. Hijikata and H. Kitamura for preparing the gene-profile analysis; Y. Motomura and M. Kubo for useful advice; A. Miyawaki and H. Miyoshi for providing pCSII-EF-MCS-IRES2-venus; O. Ohara for providing cDNA clones for Spi-B and IRF-1; and P. Burrows for critical review of the manuscript.
This work was supported by the Kishimoto Foundation (to I.S., K.H., H. Hemmi, M. Sugiyama, Y.F., T.O., and T.K.); Grant-in-Aid for Scientific Research (B: 21390123 to N.T.-S., 22390096 to T.N., 20390146 and 23390124 to T.K.; and C: 22590444 to K.H., 20451924 to H. Hemmi, 20590499 and 23590577 to T.T., 22590441 to K.S., 18590483 to T.K.); Grant-in-Aid for Challenging Exploratory Research (21659123 and 23659245 to T.K.), Grant-in-Aid for Young Scientists (B: 22790376 to H. Hemmi); Funding Program for Next Generation World-Leading Researchers (to N.T.-S.) from Japan Society for the Promotion of Science; Grant-in-Aid for Scientific Research on Priority Areas (19041070, 20060033 and 21022048 to T.K.); Grant-in-Aid for Scientific Research on Innovative Areas (21117003 to T.K.) from The Ministry of Education, Culture, Sports, Science and Technology; Core Research for Evolutional Science and Technology (to T.N.); Precursory Research for Embryonic Science and Technology (to K.S.) from Japan Science and Technology Agency; Health Labor Sciences Research Grant (H23-Shinko-Ippan-015 to H.H.) from The Ministry of Health, Labour and Welfare; Takeda Science Foundation (to T.T.); and the Uehara Memorial Foundation (to T.K.). I.S., C.Y., and M. Sugiyama are supported by a RIKEN Junior Research Associate Grant.
Authorship
Contribution: I.S., K.H., T. Sugiyama, C.Y., T.Y., A.I., M. Saito, M. Sugiyama, Y.F., and T.O. performed the mouse experiments and analyzed the data; I.S., K.H., T. Sugiyama, A.I., M. Sugiyama, and Y.F. performed the in vitro analysis and analyzed the data; H. Hemmi and T.T. edited the manuscript; K.S., H.K., and T.N. provided technical support; A.A., T. Suzuki, and H. Hasegawa performed the VSV infection experiments; N.T.-S. provided the Ly49Q transgenic mice; and T.K. designed and supervised the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tsuneyasu Kaisho, Laboratory for Immune Regulation, WPI Immunology Frontier Research Center, Osaka University, Yamadaoka 3-1, Suita, Osaka 565-0871, Japan; e-mail: tkaisho@ifrec.osaka-u.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal