Abstract
Understanding the process of myeloid differentiation offers important insights into both normal and abnormal developmental processes but is limited by the dearth of experimental models. Here we show that myeloid progenitors can be derived from embryonic stem cells, immortalized, and applied to the study of the mechanisms underlying myeloid differentiation. The embryonic stem cell–derived myeloid progenitors, when immortalized with estrogen-regulated Hoxb8 protein, demonstrate normal karyotyping, are genetically tractable, and can be differentiated into functional neutrophils. Using this model, we identified mammalian target of rapamycin complex 1 as a critical regulator of myeloid differentiation. Together, our studies led to a convenient, karyotypically normal, and genetically manipulatable cellular system, which can be used to shed new light on the mechanisms for myeloid differentiation.
Introduction
Myeloid progenitors derived from multipotential hematopoietic stem cells can be differentiated into myeloid cells, including neutrophils, monocytes, and macrophages, which act as key mediators of innate immunity and play a central role in host defense against infections and to tissue damage.1-3 Conversely, defective regulation of myeloid differentiation has devastating consequences, leading to myeloid diseases and disorders, such as myeloid aplasia, dysplasia, and leukemia. Therefore, an improved understanding of the molecular mechanisms that control myeloid differentiation will not only provide new insights into fundamental developmental processes but also improve our abilities to treat leukemia and other myeloid disorders.
Two in vitro experimental models (primary normal myeloid precursors and leukemic cells arrested at various developmental stages) have been used for the studies of myeloid differentiation. These models have their limitations and drawbacks. Primary myeloid progenitors isolated from bone marrows are physiologic, but they are generally of limited quantities, hard to purify to homogeneity, refractory to genetic manipulations, and not suited for long-term culture,4 thus limiting their applications. Leukemia cell lines that can be induced to myeloid cells in the presence of chemical inducers, such as DMSO and retinoid acid, are karyotypically abnormal and thus may not recapitulate the normal myeloid cells. Therefore, there are imperative needs to establish new physiologic and yet genetically tractable models for analyzing myeloid differentiation and functions.
To develop such models, we turned to embryonic stem cells (ESCs), which self-renew almost indefinitely in vitro while maintaining stable karyotypes, are genetically tractable and can be differentiated into nearly all cell types, including hematopoietic precursor cells and functional myeloid cells.5-13 We also took advantage of a recently developed method, which is based on induced ectopic expression of β-estradiol–regulated-Hoxb8 protein (Hoxb8-ER),14 to immortalize ESC-derived myeloid progenitors. The ESC-derived immortalized progenitor cells demonstrate normal karyotyping, are genetically manipulatable, and can be differentiated into functional neutrophils. Using this model, we screened a collection of kinase inhibitors and identified mammalian target of rapamycin complex 1 (mTORC1) as a critical regulator of myeloid differentiation.
Methods
Cell culture
W4/129S6 mESCs (Taconic) were plated on γ-irradiated mouse embryonic fibroblasts or 0.1% gelatin-coated 6-well plates and maintained in DMEM (high glucose, Invitrogen) with 15% FBS, 1000 U/mL leukemia inhibitory factor (Chemicon), 0.1mM nonessential amino acids, 2mM l-glutamine, 1mM sodium pyruvate, 10−6M 2-mercaptoethanol, 100 U/mL penicillin, and 100 U/mL streptomycin. Medium was changed every other day. HEK293T cells and OP9 bone marrow stromal cells were purchased from ATCC and were cultured following ATCC's recommendations.
Inhibitor and antibodies
All inhibitors were purchased from Calbiochem. Antibodies against mTOR, Raptor, Rictor, or S6K1 were from Cell Signaling Technology. Antibodies against Gr-1, CD11b, CD16, CD80, CD45, CD41, TER119, B220, c-Kit, and Sca-1 were from BD Biosciences.
Isolation of murine bone marrow progenitors
Per the protocol of Animal Care and Use Committee approval, mouse bone marrow progenitor cells were isolated from femurs and tibias of C57Bl/6 mice, cultured and expanded in medium containing 10 ng/mL IL-3, 20 ng/mL IL-6, and 25 ng/mL stem cell factor (SCF), as described previously.14
EB induction and differentiation of myeloid progenitors and neutrophils
Embryoid body (EB) induction from ESCs, isolation of myeloid progenitors, and subsequent neutrophil differentiation were as described previously.5 Briefly, EBs were induced from ESC and cultivated for 8 days, trypsinized to single cells, and coated onto semiconfluent OP9 cells in medium containing 25 ng/mL oncostatin M, 10 ng/mL basic fibroblast growth factor, 5 ng/mL IL-6, 20 ng/mL SCF, 5 ng/mL IL-11, and 1 ng/mL recombinant mouse leukemia inhibitory factor. After 3-day expansion, the progenitor cells were transferred onto fresh semiconfluent OP9 cells and cultured in neutrophil-differentiation medium containing 60 ng/mL G-CSF, 3 ng/mL GM-CSF, and 5 ng/mL IL-6. After 6-10 days, cells were harvested for further analysis.
To induce neutrophil differentiation of the mEB8-ER and the mBB8-ER cells, cells were washed 3 times with PBS to remove β-estradiol, as described earlier,14 and cultured in OptiMem medium containing 10% FBS, 1% glutamine, 30μM β-mercaptoethanol, 2 ng/mL G-CSF or GM-CSF, 100 U/mL penicillin, and 100 U/mL streptomycin. Kinase inhibitors were added to the differentiation medium 1 hour before addition of G-CSF. For induction of neutrophil differentiation of bone marrow progenitor cells, the same medium was used, except that 100 ng/mL G-CSF was provided.
Retrovirus production and infection
To immortalize progenitors derived from ESCs or the mouse bone marrow, cells were infected with retrovirus containing the Hoxb8-ER construct as described previously.14 After infection, cells were cultured and maintained in OptiMem containing 10% FBS, 1% Pen/Stre/Glutamine, 10 ng/mL SCF, 30μM β-mercaptoethanol, and 1μM β-estradiol, in the absence of OP9 feeder cells, which were no longer needed after immortalization, as described previously.14
For small hairpin RNA (shRNA)–mediated knockdown of mTOR, Raptor, and Rictor, LMP-puro-shRNA containing retroviruses were used. The shRNA retroviral constructs and sequences were as described previously.15 For S6K1 knockdown, the S6K1 shRNA construct, described earlier16 and kindly provided by Dr Rafi Ahmed (Emory University), was used. Retroviruses were generated as previously described,14 added with polybrene (6 μg/mL, Sigma-Aldrich) to the Hoxb8-ER cells and incubated for 20 hours, after which fresh growth medium was provided. Cells were selected with puromycin (2 μg/mL) 2 days after infection and used for subsequent analyses 3 days after selection.
CFU assay
MethoCult GF M3434 medium (StemCell Technologies) was used for the colony formation assay. The procedures were conducted according to the manufacturer's instructions. Briefly, a concentrated (10×) EB8-ER cell suspension (5 × 105/mL cells with or without 10μM β-estradiol) was prepared, and 0.3 mL cell suspension was added to 3 mL of MethoCult. The cell MethoCult mixture (1.1 mL) was transferred to 35-mm dishes, which were gently tilted and rotated to evenly distribute methylcellulose. Cells were then cultivated at 37°C for 10 days.
Transwell assay and calcium flux
Transwell assays were performed as described previously.17 The procedure for measuring the intracellular calcium release after formyl-methionyl-leucyl-phenylalanine (fMLP) stimulation was performed as reported previously.18 EB8-ER cells (5 × 106) were suspended in 1 mL of RPMI medium with 1% BSA. Cells were incubated with Fura-2 AM (5μM) at 37°C for 20 minutes. The cells were washed twice with PBS and suspended in 1 mL of calcium flux buffer (145mM NaCl, 4mM KCl, 1mM NaHPO4, 1.8mM CaCl2, 25mM HEPES, 0.8mM MgCl2, and 22mM glucose). Fluorescence readings were recorded at 37°C in a fluorimeter (Molecular Device M5) before and after the addition of fMLP. Intracellular calcium concentrations were presented as the relative ratio of excitation fluorescence intensity emitted at 510 nm in response to sequential excitation at 340 nm and 380 nm.
Effects of rapamycin in vivo
C57BL/6 mice (8-12 weeks old) were used. Mice were injected intraperitoneally with vehicle or rapamycin (4 mg/kg) per day for 7 days as described earlier.19 Mice were killed on day 8, and cells from the peripheral blood and bone marrow were collected for further analysis. Blood routine test was conducted to analyze lymphocytes in the periphery blood. To collect cells from the bone marrow, femurs and tibias were flushed, washed with PBS, and resuspended in PBSE at 107 cells/mL. Cells for various lineages were labeled using lineage cell detection cocktail (Miltenyi Biotec), as reported previously.20 Briefly, neutrophils from the bone marrow were labeled with antibodies against Gr-1 and CD11b, and granulocyte-macrophage progenitors (GMPs) were labeled as Lin−Sca-1−c-Kit+CD127−CD34+CD16/32+ cells. The flow cytometric analysis was conducted using LSRII (BD Biosciences).
Western blotting
Cells were harvested and lysed directly with Laemmli sample buffer (Bio-Rad). A total of 20 μL of each sample was separated by SDS-PAGE and analyzed by Western blot following the protocol as described.21 The blots were developed using SuperSignal West Pico Chemiluminescent Substrate (Pierce Chemical).
FACS analysis of myeloid progenitors or neutrophils
To determine expression of c-Kit, Sca-1, TER119, B220, CD41, CD45, CD16, CD80, Gr-1, and CD11b, cells were collected, fixed with paraformaldehyde in PBS for 10 minutes, washed twice with 0.1% BSA, and incubated for 30 minutes at room temperature with the FITC- or PE-conjugated antibodies (BD Biosciences) in buffer containing 1% BSA. Cells were analyzed on a FACSDiva machine (BD Biosciences).
To obtain c-Kit–positive and Sca-1–negative progenitors from the mouse bone marrow, cells were stained with FITC- and PE-conjugated Sca-1 and c-Kit antibodies (BD Biosciences) and sorted using flow cytometry.
Apoptosis assay
Neutrophils or progenitors were harvested, stained with FITC-conjugated annexin V and propidium iodide using an annexin V–FITC Apoptosis Detection Kit (BD Biosciences), and analyzed by flow cytometry.21 Cells negative for both annexin V and propidium iodide were scored as nonapoptotic cells.
Wright-Giemsa staining
For morphologic analysis of immortalized progenitor cells, differentiated neutrophils, or other cells, Wright-Giemsa (Sigma-Aldrich) staining was used. Images were collected using a Zeiss EC Plan-Neofluar 40×/0.3 Ph1 objective on a Zeiss Axiovert 200M microscope and processed using Adobe Photoshop (Adobe).
Statistical analyses
Results are shown as mean ± SEM. Statistical significance was determined using t test. Results were considered significant when P < .05.
Results
Directed differentiation of ESCs into myeloid progenitors
In an effort to establish a simple, physiologic, and genetically tractable system for myeloid differentiation, we attempted to derive myeloid progenitor cells from ESCs. We dissociated ESCs into single cells and induced them to form EBs, as described previously.5 Hematopoietic progenitors were isolated from EBs and subsequently expanded by culture on the bone marrow–derived mouse stromal cell line OP9, in the presence of cytokines.5 Using differentiation medium containing G-CSF or GM-CSF, we further induced directed differentiation of the progenitor cells to neutrophils or macrophages (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). This method used ESCs and thus bypassed the use of mouse tissues. However, the procedure is tedious, produces a low cell yield, and requires one to generate progenitors from ESCs freshly for each analysis. As such, this method is in need of further improvement.
Immortalization of ESC-derived myeloid progenitors
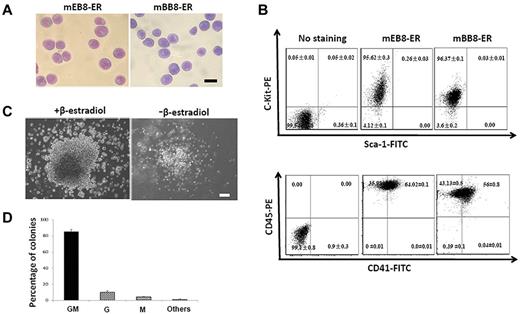
It was recently reported that bone marrow–derived myeloid progenitors can be immortalized by the use of ectopic expression of β-estradiol–regulated Hoxb8-ER.14,22 The Hoxb8-ER–immortalized progenitor cells maintained stable karyotyping and could be differentiated into functional neutrophils and macrophages on ER-hoxb8 inactivation.14 A major advantage of this system is tightly regulated Hoxb8 expression, leading to low expression of differentiation genes in progenitor cells (< 2.5% in differentiated cells), low expression of promyelocyte genes in differentiated cells (< 1%), and high differentiation efficiency of neutrophils.14 We found that this approach was also effective for immortalization of ESC-derived myeloid progenitors. We infected ESC-derived progenitor cells using retrovirus containing Hoxb8-ER as described previously14 (supplemental Figure 1A-B). As a positive control, we also isolated progenitor cells from the mouse bone marrow and infected them with the Hoxb8-ER virus. After 20-day culture in SCF-containing medium and stimulated with β-estradiol, the ESC-derived progenitors proliferated rapidly and overextended passages (Figure 1A; supplemental Figure 1C),14 suggesting that they became immortalized. Importantly, these cells exhibited a nearly homogeneous myeloblast-like morphology, as evidenced by large oval nuclei and relatively scant cytoplasm (Figure 1A), and expressed high levels of progenitor markers, including c-Kit, CD41, and CD45 and little Sca-1 (Figure 1B), similar to immortalized bone marrow cells (Figure 1A-B; supplemental Figure 1C). In addition, in experiments with the CFU assay, the ESC-derived progenitors formed large blast-like colonies (82% ± 6%) in the presence of β-estradiol (Figure 1C). Without β-estradiol, however, such colonies were hardly seen (Figure 1C). Instead, the cells formed more restricted colonies, with a colony-forming efficiency of 80% ± 4%, indicating high ratios of progenitor cells. Among the colonies formed, the majority (85% ± 3%) were granulocyte-macrophage (GM) colonies, with the remaining ones as granulocyte (G; 10% ± 2%) and macrophage (M; 4% ± 1%) colonies (Figure 1D). Furthermore, the ESC-derived immortalized progenitors maintained a stable karyotype, similar to ESC-derived and immortalized from the bone marrow (data not shown).14 We named the 2 types of immortalized cells mEB8-ER and mBB8-ER, respectively, based on the source of derivation (mouse ESCs or the mouse bone marrow).
Characterization of ESC-derived immortalized myeloid progenitors. (A) Wright-Giemsa staining of mEB8-ER (left) or mBB8-ER (right) cells. Bar represents 10 μm. (B) Surface expression of Sca-1, c-Kit, CD45, and CD41 in mEB8-ER and mBB8-ER cells assessed with flow cytometry. Nonspecific IgG was used as a negative control (denoted as “No staining”). (C) Analysis of EB8-ER cells using the CFU assay. Phase-contrast images of cells with (left) or without (right) β-estradiol treatment. (D) The relative percentage of GM, G, and M colonies formed from mEB8-ER cells in the absence of β-estradiol. Quantification of 4 separate experiments. Each represents the mean ± SEM (error bars). (C-D) Cells were cultivated in MethoCult GF M3434 medium for 10 days. Bar represents 50 μm.
Characterization of ESC-derived immortalized myeloid progenitors. (A) Wright-Giemsa staining of mEB8-ER (left) or mBB8-ER (right) cells. Bar represents 10 μm. (B) Surface expression of Sca-1, c-Kit, CD45, and CD41 in mEB8-ER and mBB8-ER cells assessed with flow cytometry. Nonspecific IgG was used as a negative control (denoted as “No staining”). (C) Analysis of EB8-ER cells using the CFU assay. Phase-contrast images of cells with (left) or without (right) β-estradiol treatment. (D) The relative percentage of GM, G, and M colonies formed from mEB8-ER cells in the absence of β-estradiol. Quantification of 4 separate experiments. Each represents the mean ± SEM (error bars). (C-D) Cells were cultivated in MethoCult GF M3434 medium for 10 days. Bar represents 50 μm.
In the absence of β-estradiol and with the addition of G-CSF, mEB8-ER cells can be differentiated into neutrophils within 5-6 days, with an efficiency of > 98%. In these experiments, a range of concentrations of G-CSF (2-50 ng/mL) was tested (data not shown), and 2 ng/mL G-CSF, which was found to suffice to induce mEB8-ER cells into neutrophils, was used for the rest of the study (Figure 2A). The differentiated cells showed typical morphology of mouse neutrophils with segmented multilobulated and ring-like nuclei, expressed neutrophil markers Gr-1, CD16, and myeloid differentiation markers Mac-1 (CD11b), CD80 (Figure 2A-B; supplemental Figure 2),23 and responded to chemoattractants by increasing intracellular calcium and migrating directionally (Figure 2C-D), suggesting that they are functional neutrophils.5,14,24,25 As expected, G-CSF stimulation did not increase the level of B220 (marker for B cells) and TER119 (marker for erythroid lineage; supplemental Figure 3A). Furthermore, similar to the mBB8-ER cells,14 the ESC-derived progenitor cells could be induced to differentiate to macrophages (with up to 50% efficiency) in the presence of GM-CSF (supplemental Figure 3B), in keeping with an earlier report.26 Interestingly, compared with the mEB8-ER cells, the mBB8-ER cells expressed a relative higher level of Gr-1 and lower level of CD11b when differentiated with GM-CSF, which may reflect subtle intrinsic differences between the 2 types of progenitor cells. In sum, because the mEB8-ER cells can be cultivated indefinitely, differentiated rapidly, and are karyotypically normal, they offer a convenient and physiologic experimental system for the study of myeloid differentiation.
Differentiation of ESC-derived progenitors to neutrophils. (A) Wright-Giemsa staining of mEB8-ER (top) or mBB8-ER cells (bottom) 6 days after the addition of 2 ng/mL G-CSF. (B) Surface expression of Gr-1, CD11b, CD16, and CD80 in mEB8-ER (top) and mBB8-ER cells (bottom) assessed with flow cytometry. Cells were induced to differentiate for 6 days. Cells before induction of differentiation (“0 day”) were used as a negative control (“Control”). (C) Analysis of intracellular [Ca2+] in mEB8-ER and mBB8-ER–derived neutrophils in response to fMLP stimulation. Fluorescence was recorded using SpectraMax M2 spectrometer (excitation, 340-380 nm; emission, 510 nm). Results are plotted as emission ratio versus time. Arrow indicates the time of addition of fMLP (100nM). A representative experiment from 5 separate experiments. (D) Migration in the transwell assay of mEB8-ER and mBB8-ER–derived neutrophils. Left: Representative images of cells that crossed the transwell membrane with or without the gradient of fMLP (100nM). Right: The count of cells that crossed the transwell membrane. Each bar represents the mean ± SEM (error bars). *P < .05. All values were normalized to the level (= 1) in cells without fMLP stimulation.
Differentiation of ESC-derived progenitors to neutrophils. (A) Wright-Giemsa staining of mEB8-ER (top) or mBB8-ER cells (bottom) 6 days after the addition of 2 ng/mL G-CSF. (B) Surface expression of Gr-1, CD11b, CD16, and CD80 in mEB8-ER (top) and mBB8-ER cells (bottom) assessed with flow cytometry. Cells were induced to differentiate for 6 days. Cells before induction of differentiation (“0 day”) were used as a negative control (“Control”). (C) Analysis of intracellular [Ca2+] in mEB8-ER and mBB8-ER–derived neutrophils in response to fMLP stimulation. Fluorescence was recorded using SpectraMax M2 spectrometer (excitation, 340-380 nm; emission, 510 nm). Results are plotted as emission ratio versus time. Arrow indicates the time of addition of fMLP (100nM). A representative experiment from 5 separate experiments. (D) Migration in the transwell assay of mEB8-ER and mBB8-ER–derived neutrophils. Left: Representative images of cells that crossed the transwell membrane with or without the gradient of fMLP (100nM). Right: The count of cells that crossed the transwell membrane. Each bar represents the mean ± SEM (error bars). *P < .05. All values were normalized to the level (= 1) in cells without fMLP stimulation.
Small-scale pharmacologic screening using the mEB8-ER progenitors
We next attempted to use the mEB8-ER model to dissect the signaling mechanisms that control myeloid differentiation. The function of protein kinases in mediating signal transduction during development, transcription, immune response, metabolism, apoptosis, and cell differentiation has been well established. Therefore, to identify potential key regulators of myeloid differentiation, we screened a collection of specific kinase inhibitors (∼ 30, EMD Biosciences; Table 1). In earlier studies, we applied a similar strategy and identified key regulatory networks governing pluripotency and early differentiation of human pluripotent stem cells.21,27,28 We induced mEB8-ER cells to undergo differentiation in the presence of G-CSF, treated them with inhibitors over the course of differentiation, and determined the expression of differentiation markers, such as Gr-1 and Mac-1 (CD11b; supplemental Figure 4). We found that, although most inhibitors exerted little effect, those against protein kinase C (PKC: bisindolylmaleimide I and Gö 6983), mTOR (rapamycin), and Syk (Syk inhibitor) markedly reduced expression of Gr-1 and Mac-1 (CD11b). Furthermore, the PI3Kα inhibitor and the NF-κB inhibitor induced rapid apoptosis (within 2-3 days; data not shown). The role of mTOR was further explored (the rest of the study), whereas analysis of other lead pathways awaits future experimentation.
Inhibitors, their targets and effects
| Name . | Target . | Outcome . |
|---|---|---|
| PD98059 | MEK | − |
| AG1478 | EGFR | − |
| SB203580 | P38 | − |
| JNK inhibitor | JNK | − |
| STAT3 inhibitory peptide | Stat3 | − |
| JAK3 inhibitor | JAK3 | − |
| AG1296 | PDGFR/C-kit | − |
| Bisindolylmaleimide I | PKC | + |
| G6976 | aPKC | − |
| KT5720 | PKA | − |
| PP2 | SRC | − |
| G6983 | PKC | + |
| GSK3 inhibitor IV | GSK3 | − |
| NF-κB SN50 | NF-κB | D |
| Rapamycin | mTOR | + |
| PI3K-α inhibitor IV | PI3K-α | D |
| Syk inhibitor IV | Syk kinase | + |
| Name . | Target . | Outcome . |
|---|---|---|
| PD98059 | MEK | − |
| AG1478 | EGFR | − |
| SB203580 | P38 | − |
| JNK inhibitor | JNK | − |
| STAT3 inhibitory peptide | Stat3 | − |
| JAK3 inhibitor | JAK3 | − |
| AG1296 | PDGFR/C-kit | − |
| Bisindolylmaleimide I | PKC | + |
| G6976 | aPKC | − |
| KT5720 | PKA | − |
| PP2 | SRC | − |
| G6983 | PKC | + |
| GSK3 inhibitor IV | GSK3 | − |
| NF-κB SN50 | NF-κB | D |
| Rapamycin | mTOR | + |
| PI3K-α inhibitor IV | PI3K-α | D |
| Syk inhibitor IV | Syk kinase | + |
+ indicates impairs differentiation; −, no effect; and D, cell death.
Rapamycin disrupts differentiation of the mEB8-ER progenitors
mTOR is a serine/threonine kinase and master regulator of cell growth, proliferation, and differentiation. It senses nutrient availability and energy levels.29 Inhibition of mTOR prevents proliferation and differentiation of endothelial or megakaryocyte progenitors in vitro30,31 and reduces the growth of acute myeloid leukemia cell lines.32,33 The role of mTOR in myeloid differentiation under G-CSF stimulation remains to be defined.
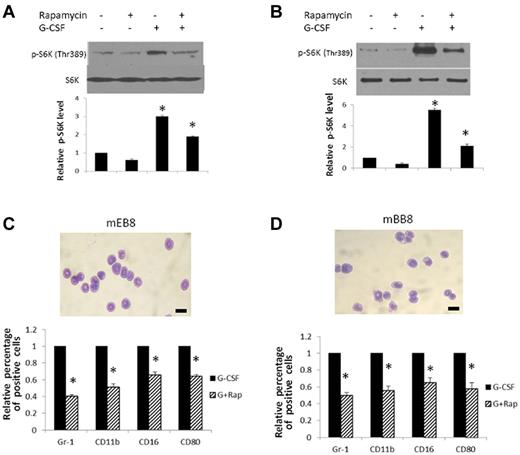
Stimulation of mEB8-ER cells with G-CSF robustly activated mTOR, as indicated by a significant increase in phosphorylation (activation) of ribosomal p70S6 kinase (p70S6K; Figure 3A), a downstream molecule of mTOR and a widely used readout for mTOR activity,28 suggesting that mTOR might serve as a key signaling intermediate downstream of G-CSF signaling during myeloid differentiation. In keeping with this notion, rapamycin treatment markedly inhibited differentiation of mEB8-ER cells induced by G-CSF, as shown by a decrease of Gr-1, CD11b, CD16, and CD80 surface expression, without significantly increasing the percentage of apoptotic cells (< 10%; Figure 3C; supplemental Figure 5A). Furthermore, rapamycin did not alter the level of TER119 and B220, suggesting that the treatment did not enhance erythroid and B-cell lineage activities (supplemental Figure 6A). Rapamycin's effects were also observed in mBB8-ER cells (Figure 3B,D). We used 100nM of rapamycin for these analyses; this is a dose commonly used for mTOR inhibition,32,33 which potently inhibited both the basal and G-CSF–induced p70S6K activation (Figure 3A-B).
Rapmycin disrupts G-CSF–induced myeloid differentiation of mEB8-ER/mBB8-ER cells. (A-B) Top panel: Western blot of threonine-phosphorylated S6K (at residue 389: p-S6K) in mEB8-ER (A) or mBB8-ER (B) cells with or without rapamycin treatment. Cells were treated or not treated with G-CSF (2 ng, 30 minutes). Total S6K was a loading control. A typical experiment of 4 independent experiments. Bottom panel: Relative protein levels of p-S6K in mEB8-ER or mBB8-ER cells with various treatments. Quantification of blots from 4 separate experiments. Each bar represents the mean ± SEM. *P < .05. (C-D) Wright-Giemsa staining (top) and relative percentage of CD11b, Gr-1, CD16, and CD80-positive cells (bottom) in mEB8-ER (C) or mBB8-ER cells (D) measured with flow cytometry. Cells were induced to differentiate in the presence of G-CSF (2 ng/mL, 6 days) with (“G + Rap”) or without (“G-CSF”) rapamycin treatment (100nM). Four separate experiments were conducted, and quantification of 3 replicates of a typical experiment. Each bar represents the mean ± SEM (error bars). All values were normalized to the level (= 1) in cells without rapamycin treatment. *P < .05. Bar represents 10 μm.
Rapmycin disrupts G-CSF–induced myeloid differentiation of mEB8-ER/mBB8-ER cells. (A-B) Top panel: Western blot of threonine-phosphorylated S6K (at residue 389: p-S6K) in mEB8-ER (A) or mBB8-ER (B) cells with or without rapamycin treatment. Cells were treated or not treated with G-CSF (2 ng, 30 minutes). Total S6K was a loading control. A typical experiment of 4 independent experiments. Bottom panel: Relative protein levels of p-S6K in mEB8-ER or mBB8-ER cells with various treatments. Quantification of blots from 4 separate experiments. Each bar represents the mean ± SEM. *P < .05. (C-D) Wright-Giemsa staining (top) and relative percentage of CD11b, Gr-1, CD16, and CD80-positive cells (bottom) in mEB8-ER (C) or mBB8-ER cells (D) measured with flow cytometry. Cells were induced to differentiate in the presence of G-CSF (2 ng/mL, 6 days) with (“G + Rap”) or without (“G-CSF”) rapamycin treatment (100nM). Four separate experiments were conducted, and quantification of 3 replicates of a typical experiment. Each bar represents the mean ± SEM (error bars). All values were normalized to the level (= 1) in cells without rapamycin treatment. *P < .05. Bar represents 10 μm.
To better understand the effect of rapamycin, we assessed whether it influenced the growth of mEB8-ER cells. Rapamycin treatment of cells cultured as monolayers moderately reduced cell growth (by 21% ± 6%, after 5-day growth; supplemental Figure 5B). In addition, in experiments with the colony formation assay, cells with rapamycin treatment showed slightly reduced colony sizes (19% ± 7%) compared with control cells (supplemental Figure 5C). A small decrease in cell number (9% ± 2%) was also observed in mEB8-ER cells undergoing myeloid differentiation in the presence of G-CSF (after 6-day differentiation; supplemental Figure 5D). Thus, mTOR inhibition moderately reduces the growth of mEB8-ER progenitors under self-renewal and differentiation conditions.
mTORC1, but not mTORC2, is essential for differentiation of mEB8-ER progenitors
The high selectivity of rapamycin toward mTOR has been documented.34 Nevertheless, to further confirm the specificity of rapamycin, we exploited RNAi-induced knockdown to deplete mTOR in mEB8-ER cells. We used retrovirus-mediated infection followed by drug selection to stably express shRNAs specifically targeting mTOR15 and other genes of interest.
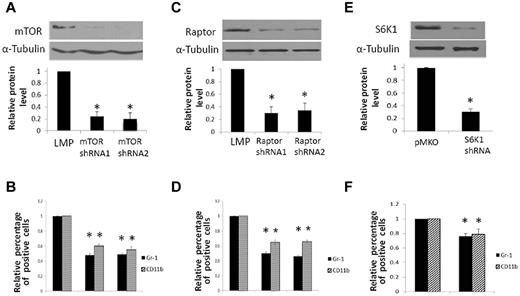
Infection of mEB8-ER cells with retroviruses containing mTOR-targeting shRNAs reduced the level of mTOR by ∼ 80% compared with control cells without the treatment (Figure 4A). Depletion of mTOR markedly reduced the level of Gr-1 and CD11b in cells induced to differentiate to neutrophils (Figure 4B), highly reminiscent of rapamycin treatment (Figure 3C).
mTORC1, but not mTORC2, is necessary for myeloid differentiation of mEB8-ER cells. (A,C,E) Top panel: Western blot of mTOR (A), Raptor (C), or S6K1 (E) in mEB8-ER cells with or without shRNAs targeting mTOR, Raptor, or S6K1. α-tubulin was a loading control. A typical blot from 4 independent experiments. Bottom panel: Quantification of blots from 4 separate experiments. Each bar represents the mean ± SEM. *P < .05. LMP/pMKO: the empty retrovector. (B,D,F) Relative percentage of CD11b and Gr-1–positive cells with or without mTOR (B), Raptor (D), and S6K1 (F) depletion, measured with flow cytometry. Four separate experiments were conducted, and quantification of 3 replicates of a typical experiment. Each bar represents the mean ± SEM (error bars). All values were normalized to the level (= 1) in cells without depletion. *P < .05.
mTORC1, but not mTORC2, is necessary for myeloid differentiation of mEB8-ER cells. (A,C,E) Top panel: Western blot of mTOR (A), Raptor (C), or S6K1 (E) in mEB8-ER cells with or without shRNAs targeting mTOR, Raptor, or S6K1. α-tubulin was a loading control. A typical blot from 4 independent experiments. Bottom panel: Quantification of blots from 4 separate experiments. Each bar represents the mean ± SEM. *P < .05. LMP/pMKO: the empty retrovector. (B,D,F) Relative percentage of CD11b and Gr-1–positive cells with or without mTOR (B), Raptor (D), and S6K1 (F) depletion, measured with flow cytometry. Four separate experiments were conducted, and quantification of 3 replicates of a typical experiment. Each bar represents the mean ± SEM (error bars). All values were normalized to the level (= 1) in cells without depletion. *P < .05.
mTOR participates in 2 signaling complexes with distinct cellular functions: mTORC1 and mTORC2.29 Furthermore, it has been shown that mTORC1, but not mTORC2, is sensitive to rapamycin.35-37 These findings and the effect of rapamycin on myeloid differentiation (Figure 3) led us to ask whether mTORC1 might be involved in the regulation of myeloid differentiation. Indeed, depletion of Raptor, a major component of mTORC1, markedly reduced expression of Gr-1 and CD11b in differentiating mEB8-ER cells (Figure 4C-D), which mimicked the effect of rapamycin treatment and mTOR depletion. As expected, Raptor depletion caused the level of p70S6K phosphorylation to reduce (data not shown). In contrast, depletion of Rictor, a major component of mTORC2,38 failed to alter the level of Gr-1 and CD11b (supplemental Figure 6B-C). Together, these results indicated that mTORC1, but not mTORC2, is essential for differentiation of mEB8-ER progenitors.
To further dissect the mechanism by which mTORC1 controls myeloid differentiation, we asked whether S6K1 might be involved in mediating the function of mTORC1 in myeloid differentiation. S6K1 is a principal downstream effector of mTORC139,40 and is required for mTOR regulation of early adipocyte differentiation41 and vascular muscle cell differentiation.42 Consistent with these earlier findings, S6K1 depletion in mEB8-ER cells reduced the level of Gr-1 and CD11b (Figure 4E-F). Interestingly, the effect of S6K1 depletion appeared weaker than that of mTOR and Raptor depletion, implying that S6K1 may partially mediate the function of mTORC1 during myeloid differentiation.
mTORC1 is required for differentiation of primary myeloid progenitors
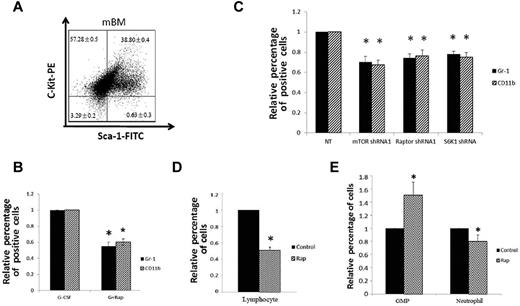
Our experiments with mEB8-ER progenitors suggested that the mTOR pathway is essential for myeloid differentiation. To further validate these results, we isolated c-Kit–positive and Sca-1–negative progenitors from the mouse bone marrow (Figure 5A)14 and assessed the effect of rapamycin on G-CSF–induced cell differentiation. Stimulation of these primary myeloid progenitors with G-CSF led to up-regulation of Gr-1 and CD11b, whereas rapamycin markedly reduced expression of these markers (by 50% and 41%, respectively; Figure 5B). Consistent with these results, depletion of mTOR, Raptor, and S6K1 in primary progenitors also induced similar down-regulation of the differentiation markers, whereas Rictor depletion had little effect (Figure 5C; and data not shown). These results provided further evidence that mTOR is an essential regulator of myeloid differentiation and again demonstrated that the mEB8-ER cells are a valid system for the study of myeloid differentiation.
Effects of mTOR inhibition in primary progenitors and in vivo. (A) Surface expression of Sca-1 and c-Kit in mouse bone marrow progenitors (mBM) assessed by flow cytometry. c-Kit–positive and Sca-1–negative cells were sorted and used for myeloid differentiation. (B) Relative percentage of CD11b and Gr-1–positive cells in mouse bone marrow progenitors with or without rapamycin treatment (100nM), measured with flow cytometry. Cells were induced to differentiate in the presence of G-CSF (100 ng/mL, 10 days). Four separate experiments were conducted, and quantification of 3 replicates of a typical experiment are shown. Each bar represents the mean ± SEM (error bars). All values were normalized to the level (= 1) in cells without rapamycin treatment. *P < .05. (C) Relative percentage of CD11b and Gr-1–positive cells in mouse bone marrow progenitors with or without mTOR, Raptor, and S6K1 depletion, measured with flow cytometry. mTORshRNA1, Raptor shRNA1, and S6K1 shRNA were used to deplete mTOR, Raptor, and S6K1, respectively. NT (nontargeting) denotes cells infected with virus containing the empty retrovector (latent membrane protein or pMKO). Each bar represents the mean ± SEM (error bars) of 4 separate experiments. All values were normalized to the level (= 1) in cells without rapamycin treatment. *P < .05. (D) Relative percentage of lymphocytes in peripheral blood after 1-week rapamycin administration in vivo. “Control” and “Rap” indicate injection of vehicle and rapamycin, respectively. (E) Relative percentage of neutrophils and GMPs in the bone marrow after 1-week rapamycin administration in vivo. “Control” and “Rap” indicate injection of vehicle and rapamycin, respectively. (D-E) Three separate experiments were conducted, and in each experiment 10 mice were used. Each bar represents the mean ± SEM (error bars). All values were normalized to the level (= 100%) in cells without rapamycin treatment. *P < .05.
Effects of mTOR inhibition in primary progenitors and in vivo. (A) Surface expression of Sca-1 and c-Kit in mouse bone marrow progenitors (mBM) assessed by flow cytometry. c-Kit–positive and Sca-1–negative cells were sorted and used for myeloid differentiation. (B) Relative percentage of CD11b and Gr-1–positive cells in mouse bone marrow progenitors with or without rapamycin treatment (100nM), measured with flow cytometry. Cells were induced to differentiate in the presence of G-CSF (100 ng/mL, 10 days). Four separate experiments were conducted, and quantification of 3 replicates of a typical experiment are shown. Each bar represents the mean ± SEM (error bars). All values were normalized to the level (= 1) in cells without rapamycin treatment. *P < .05. (C) Relative percentage of CD11b and Gr-1–positive cells in mouse bone marrow progenitors with or without mTOR, Raptor, and S6K1 depletion, measured with flow cytometry. mTORshRNA1, Raptor shRNA1, and S6K1 shRNA were used to deplete mTOR, Raptor, and S6K1, respectively. NT (nontargeting) denotes cells infected with virus containing the empty retrovector (latent membrane protein or pMKO). Each bar represents the mean ± SEM (error bars) of 4 separate experiments. All values were normalized to the level (= 1) in cells without rapamycin treatment. *P < .05. (D) Relative percentage of lymphocytes in peripheral blood after 1-week rapamycin administration in vivo. “Control” and “Rap” indicate injection of vehicle and rapamycin, respectively. (E) Relative percentage of neutrophils and GMPs in the bone marrow after 1-week rapamycin administration in vivo. “Control” and “Rap” indicate injection of vehicle and rapamycin, respectively. (D-E) Three separate experiments were conducted, and in each experiment 10 mice were used. Each bar represents the mean ± SEM (error bars). All values were normalized to the level (= 100%) in cells without rapamycin treatment. *P < .05.
The effect of rapamycin in vivo
We next asked whether inhibition of mTOR impairs neutrophil differentiation in vivo. We injected C57BL/6 mice intraperitoneally with rapamycin (4 mg/kg per day) for 7 days as described.19 Consistent with earlier studies,19 treatment of mice with rapamycin markedly reduced the number of lymphocytes (Figure 5D), indicating that the treatment was effective. Furthermore, rapamycin treatment consistently reduced the number of neutrophils in the mouse bone marrow (by 17%; Figure 5E), in keeping with its effect in vitro. The reduced neutrophil number was not the result of low abundance of progenitor cells. To the contrary, rapamycin treatment robustly increased the number of GMPs in the bone marrow (Figure 5E). Notably, the increase in GMPs after rapamycin treatment in vivo contrasted the reduced growth of mEB8-ER progenitors caused by rapamycin in vitro (see supplemental Figure 5B-C). This might be attributed to the differences between in vitro and in vivo cellular microenvironments and/or other factors that remain to be defined.
Discussion
In conclusion, we used ESC-derived myeloid progenitors and the Hoxb8-ER–based inducible expression system to establish immortalized myeloid progenitor cells. Unlike leukemia cell lines that have been extensively used in previous studies,43,44 the Hoxb8-ER–based cells demonstrate normal karyotyping and are therefore more physiologic. Furthermore, in contrast to the primary myeloid progenitors isolated from bone marrows, these cells can be maintained almost indefinitely in culture and readily manipulated genetically. Moreover, in the absence of β-estradiol and cytokines, they can be rapidly differentiated into functional myeloid cells. Therefore, our studies led to a convenient, physiologic, and genetically tractable cellular system for the study of myeloid differentiation and potentially myeloid diseases and disorders.
To test the feasibility of applying the Hoxb8-ER–based model to exploration of the molecular mechanisms that govern myeloid differentiation, we screened a small collection of kinase inhibitors, leading us to identify several potential key regulatory molecules, including PKC, Syk, and mTOR. Using RNAi-mediated knockdown, we showed that mTOR is necessary for G-CSF–induced myeloid differentiation, thus validating the results from the inhibitor studies. In addition, we isolated myeloid progenitor cells from the mouse BM and confirmed the effects of mTOR inhibition in the primary cells. Together, the results from our studies outlined an effective strategy and provided proof of concept for future larger-scale chemical and genomic screens to identify the signaling components and networks that underlie myeloid differentiation. Furthermore, our studies set the stage for future experimentation to confirm and characterize the functions of PKC and Syk in myeloid progenitor cells during differentiation.
The cellular target of the bacterial macrolide rapamycin, mTOR, belongs to the PI3K-related family of Ser/Thr kinases and functions as a master regulator of cell growth and proliferation by regulating multiple downstream effectors.40,45 Recent studies have begun to demonstrate that mTOR plays a significant role in regulating self-renewal and lineage specification of a variety of stem and progenitor cells. For instance, mTOR signaling is required for differentiation of mouse myoblasts,46,47 self-renewal and differentiation potential of human hemangioma stem cells,48 and Drosophila germline stem cells,49 self-renewal50 and early adipocyte differentiation of mESCs,41 and long-term self-renewal and pluripotency of human ESCs.28,39 In this study, we show that mTOR is also necessary for differentiation of mouse myeloid progenitor cells. In addition, our results suggest that mTORC1 and a downstream kinase S6K1, but not mTORC2, is involved in the regulation of myeloid differentiation. Therefore, we provide additional evidence for the importance of mTOR signaling in controlling the fate decisions of stem/progenitor cells. It is currently unclear how mTORC1 and S6K1 might signal in myeloid progenitors to regulate differentiation. More complete understanding of the detailed mechanisms that underlie the actions of mTORC1 awaits future experiments.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Rafi Ahmed for providing the S6K1 shRNA construct and members of F.W.'s laboratory for helpful discussions.
This work was supported by the National Institutes of Health (grant GM083812; F.W.), National Basic Research Program of China (2012CB966403; J.Z.), Tianjin Natural Science Foundation (12JCZDJC24600; Y.X.), 863 project from the Ministry of Science & Technology of China (2011AA020118, J.Z.; GM083601, F.W.; HD059002, F.W.; and HL063819, P.J.S. and T.W.), and National Science Foundation (CAREER award 0953267, F.W.; and Science and Technology Center Emergent Behaviors of Integrated Cellular Systems CBET-0939511). M.P.K. receives a small portion of licensing fees for the HoxB8-ER system, which is patented by the University of California.
National Institutes of Health
Authorship
Contribution: D.L. designed and performed research, analyzed data, and prepared the manuscript; H.Y., H.N., P.L., S.P., Q.Z., Y.X., and J.Z. performed research; R.K., M.P.K., T.W., and P.J.S. contributed crucial reagents and analytic tools; and F.W. designed research, analyzed data, and prepared the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Fei Wang, Department of Cell and Developmental Biology and Institute for Genomic Biology, University of Illinois at Urbana-Champaign, 601 S Goodwin Ave, Urbana, IL 61801; e-mail: feiwang@life.uiuc.edu; and Dong Li, Department of Cell and Developmental Biology and Institute for Genomic Biology, University of Illinois at Urbana-Champaign, 601 S Goodwin Ave, Urbana, IL 61801; e-mail: dongly@uiuc.edu.

![Figure 2. Differentiation of ESC-derived progenitors to neutrophils. (A) Wright-Giemsa staining of mEB8-ER (top) or mBB8-ER cells (bottom) 6 days after the addition of 2 ng/mL G-CSF. (B) Surface expression of Gr-1, CD11b, CD16, and CD80 in mEB8-ER (top) and mBB8-ER cells (bottom) assessed with flow cytometry. Cells were induced to differentiate for 6 days. Cells before induction of differentiation (“0 day”) were used as a negative control (“Control”). (C) Analysis of intracellular [Ca2+] in mEB8-ER and mBB8-ER–derived neutrophils in response to fMLP stimulation. Fluorescence was recorded using SpectraMax M2 spectrometer (excitation, 340-380 nm; emission, 510 nm). Results are plotted as emission ratio versus time. Arrow indicates the time of addition of fMLP (100nM). A representative experiment from 5 separate experiments. (D) Migration in the transwell assay of mEB8-ER and mBB8-ER–derived neutrophils. Left: Representative images of cells that crossed the transwell membrane with or without the gradient of fMLP (100nM). Right: The count of cells that crossed the transwell membrane. Each bar represents the mean ± SEM (error bars). *P < .05. All values were normalized to the level (= 1) in cells without fMLP stimulation.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/120/24/10.1182_blood-2012-03-414979/4/m_zh89991200110002.jpeg?Expires=1769104126&Signature=0gnGlbIxje8r~BnzE9nQthiBSPizFSbk7~DU-ZoLUCWejXv7xpydRB0nhn4GKCAgbgKZstjzPL7LRO9tx4uHOkY8BlP83p9N19OExGW6Rmh8a-xdylaqGBnVGxOWqhdoAiH1ODVsi4gyoIGgXXPr9emLIwtlOWJEBjjQIzGXdomb0CbAqIvo0ZEFq9BMDeEllY8OCizKZ930NyDV2~atHavFExZD3csdJATlaECdA4kN~atCuBqmBd6xeJiIopDP-KbrD-Cg-ThmVrwwnksUxhyoaueTaQZ9bZSPCJfXEJ5nfPR-N3z6ST72u5VElXp4lX82yX1yJ83ZjMz30Xx2bQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal