Abstract
Abstract 679
Peripheral T-cell lymphoma (PTCL) represents approximately 10–12% of all non-Hodgkin lymphoma (NHL) in the Western world, with a higher incidence in Asian populations. The World Health Organization classification recognizes a number of distinctive subtypes of PTCL including angioimmunoblastic T-cell lymphoma (AITL), anaplastic large cell lymphoma (ALCL), adult T-cell leukemia/lymphoma (ATLL), extranodal NK/T-cell lymphoma of nasal type (ENKTCL), and many other rare entities that present mainly as extranodal PTCL. However, with current immunophenotypic and molecular markers, about 30–50% of PTCL cases are not classifiable and are categorized as PTCL-not otherwise specified (PTCL-NOS). With the exception of ALK(+)ALCL, the PTCLs generally have a poor outcome and, thus a better understanding of the biology of these diseases is greatly needed to improve the long-term survival of these patients.
In the current study, we performed gene expression profiling analysis on a large and well- characterized series of PTCL and ENKTCL cases (n=372) from the Lymphoma Leukemia Molecular Profiling Project (LLMPP), the International Peripheral T-cell Lymphoma Project (IPTCL) and other major institutions to define robust molecular classifiers, oncogenic pathways and prognosticators for the more common PTCL entities, as well as unique molecular and prognostic subgroups within PTCL-NOS. Molecular signatures for diagnosis and prognosis were generated in training data sets and validated in separate cohorts.
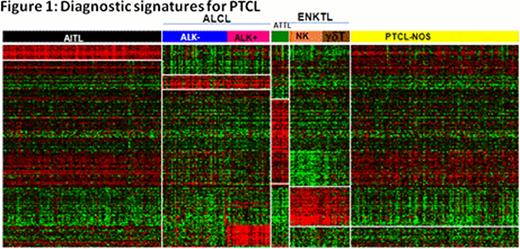
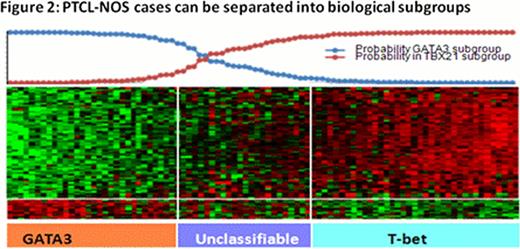
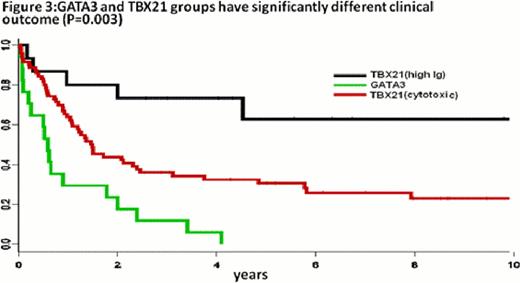
Robust molecular classifiers for AITL, two types of systemic ALCL (ALK(+) and ALK(-)), ATLL and ENKTCL were identified (Figure 1). These classifiers reflect the pathobiology of the tumor cells, as well as their microenvironment, and represent a refinement of what we reported previously (Iqbal et.al Blood, 2010; Iqbal et.al Leukemia. 2011). Importantly, ALK(-)ALCL can be differentiated from ALK(+)ALCL and PTCL-NOS with a unique gene expression signature. Approximately 14% of PTCL-NOS were re-classified as ALK(-)ALCL and showed expression of CD30 protein, TIA-1 or granzyme B by immunohistochemistry. ENKTCL can be separated molecularly into NK-cell lymphoma and gd-PTCL, the latter of which was also identified in 9% of PTCL-NOS. The remaining PTCL-NOS cases could be separated into two major subgroups related to T-cell differentiation and characterized by either high expression of GATA3 (30%) or TBX21(T-BET) (45%) and many of the corresponding target genes (Figure 2). Cases with high expression of GATA3 had poor overall survival and showed enriched Wnt and mTOR pathways, but no prominent microenvironment signature. The high TBX21 subgroup had a remarkably good outcome for patients with a high plasma cell-like gene expression signature, but poor overall survival when expressing a high cytotoxic signature (Figure 3). The molecular prognosticator for AITL largely reflected the role of the tumor microenvironment, with the presence of a high B-cell signature correlating with favorable outcome, whereas high dendritic cell/monocyte signatures were associated with inferior survival.
We have organized the most comprehensive molecular profiling study of PTCL, and have not only refined the molecular diagnostic and prognostic signatures for the common subtypes of PTCL, but also segregated PTCL-NOS in meaningful biological and prognostic subtypes. Molecular diagnostic and prognostic signatures of PTCL frequently include components of the tumor-host interactions, highlighting the importance of the microenvironment in PTCL biology. This study provides an important framework for additional analysis to identify novel therapeutic targets to improve the outcome of patients with PTCL.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal