Abstract
Abstract 4792
Chromosomal abnormalities associated with unfavorable prognosis in multiple myeloma patients include involvement of the TP53, FGRFR3 and MAF genes. It is important to accurately detect these chromosomal abnormalities to aid clinicians in the decision of therapeutic plans. Chromosomal abnormalities are not detected by traditional karyotyping due to low proliferative rate of myeloma cells. Conventional Fluorescence In-situ Hybridization (FISH) enhances the sensitivity but lacks the specificity as it does not distinguish plasma cells (PC) from the other hematopoetic cells. This can be overcome by identification of PCs by cytoplasmic immunoglobulin staining followed by FISH (cIg-FISH).
Currently, cIg-FISH is done by two methods – lysing red cells in the bone marrow aspirates and spinning the pellet onto charged slides using the Cytospin machine (Ahmann et al Cancer Genet Cytogenet 1998). The second technique uses cultured and fixed cells stored in Carnoy's fixative (Filkova et al 2006 http://www.myeloma.cz/res/file/archiv/2007-cytogen-sbornik-workshop.pdf). This second technique is more easily incorporated into the routine cytogenetic protocols used for chromosomal analysis. However, clumping and the small size of plasma cells seemed to be a major setback with the protocol. We have made minor modifications to this technique, with a different approach to fixing and dropping the cells on to the slides to give nicely separated plasma cells. These when subjected to immunostaining with kappa or/and lambda antibodies, followed by FISH, result in easily identifiable plasma cells with good bright signals under the fluorescence microscope. Twenty samples from patients with multiple myeloma were subjected to routine FISH, cIg-FISH, chromosomal karyotyping along with flow cytometry and the results were compared. Three FISH probes from Vysis for t(4;14), t(14;16) and deletion of TP53 were used. To test the utility of the technique for stored cell pellets, 4 fixed pellets stored from 2005–2008 were also tested with good results.
Comparative analysis of 20 patients with multiple myeloma by three different methods
| No . | % Plasma cells . | Conventional Karyotyping . | Conventional FISH . | Conventional cIg FISH . |
|---|---|---|---|---|
| 1 | 47 | Normal | Normal | Normal |
| 2 | 55 | Monosomy 14 | Monosomy 14 | Monosomy 14 |
| 3 | 71 | der(1;16) | Monosomy 16 | Monosomy 16 |
| 4 | 6 | Normal | Normal | Normal |
| 5 | 2 | Normal | Normal | Normal |
| 6 | 72 | der(12;16) | Monosomy 16 | Monosomy 16 |
| 7 | 13 | Normal | Normal | Trisomy 14 |
| 8 | 52 | Normal | Normal | Normal |
| 9 | 8 | Monosomy 17 | t(4;14),del17p | t(4;14),monosomy 16, del17p |
| 10 | 25 | Normal | Normal | Normal |
| 11 | 1 | Normal | Normal | Normal |
| 12 | 31 | Normal | Trisomy 4 | Trisomy 4 |
| 13 | Not done | Not done | Normal | Normal |
| 14 | 5 | Normal | Trisomy 17 | Monosomy 14, Trisomy 17 |
| 15 | 19.5 | Monosomy 14 | t(4;14), monosomy 16 | t(4;14), monosomy 16 |
| 16 | 34 | Normal | Normal | Normal |
| 17 | 50 | Normal | Normal | Normal |
| 18 | 1.8 | Normal | Normal | Normal |
| 19 | 28 | Normal | Normal | Normal |
| 20 | 47 | Normal | Normal | Normal |
| No . | % Plasma cells . | Conventional Karyotyping . | Conventional FISH . | Conventional cIg FISH . |
|---|---|---|---|---|
| 1 | 47 | Normal | Normal | Normal |
| 2 | 55 | Monosomy 14 | Monosomy 14 | Monosomy 14 |
| 3 | 71 | der(1;16) | Monosomy 16 | Monosomy 16 |
| 4 | 6 | Normal | Normal | Normal |
| 5 | 2 | Normal | Normal | Normal |
| 6 | 72 | der(12;16) | Monosomy 16 | Monosomy 16 |
| 7 | 13 | Normal | Normal | Trisomy 14 |
| 8 | 52 | Normal | Normal | Normal |
| 9 | 8 | Monosomy 17 | t(4;14),del17p | t(4;14),monosomy 16, del17p |
| 10 | 25 | Normal | Normal | Normal |
| 11 | 1 | Normal | Normal | Normal |
| 12 | 31 | Normal | Trisomy 4 | Trisomy 4 |
| 13 | Not done | Not done | Normal | Normal |
| 14 | 5 | Normal | Trisomy 17 | Monosomy 14, Trisomy 17 |
| 15 | 19.5 | Monosomy 14 | t(4;14), monosomy 16 | t(4;14), monosomy 16 |
| 16 | 34 | Normal | Normal | Normal |
| 17 | 50 | Normal | Normal | Normal |
| 18 | 1.8 | Normal | Normal | Normal |
| 19 | 28 | Normal | Normal | Normal |
| 20 | 47 | Normal | Normal | Normal |
The percent of plasma cells seen in bone marrow aspirates ranged from 1.8–71%. Of 20 samples, 3 samples showed a positive cIg-FISH but a normal conventional FISH result. All samples showed a uniformly higher percent of abnormality with the cIg-FISH protocol due to selection of plasma cells as expected.
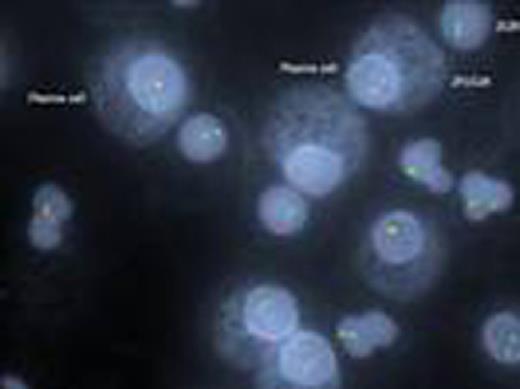
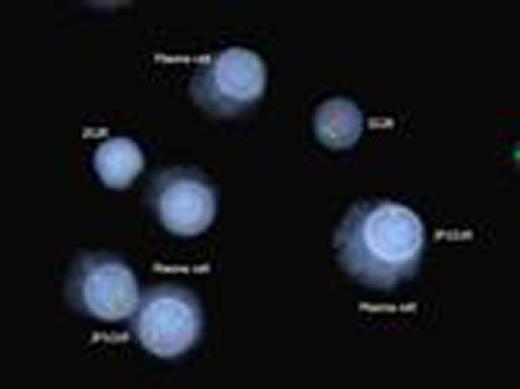
Kappa/lambda positively stained plasma cells showing FGFR3/IGH translocation
In conclusion, this technique using fixed cells will be a major asset to all cytogenetic laboratories as
It saves sample as the entire quantity of bone marrow aspirate can be used to set up cultures for chromosomal karyotyping and part of the fixed pellet can be used for cIg-FISH.
There is no necessity of making Cytospin slides - saving time and effort.
Stored pellets, especially if cells are well stored and preserved in fixative can be used for retrospective analysis.
The technique can be easily incorporated as a routine targeted FISH test for MM samples, which is not practiced in most clinical cytogenetics laboratories at present.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal