Abstract
In patients with chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL), deletion of 17p13 (17p-) resulting in loss of one allele of TP53 and 11q22 (11q-) resulting in loss of one allele of ATM predicts a significantly poorer prognosis. These lesions are nearly always monoallelic in patients with CLL and their consequences are considerably influenced by the integrity of the remaining allele. A dysfunctional mutation in the remaining allele, resulting in more aggressive disease, ultimately occurs in most patients with 17p- (> 80%) but fewer patients with 11q- (30%). Because ATM and TP53 are integral components of the p53 pathway, we hypothesize that patients with clonal 17p- and 11q- in the same CLL cell would have a worse prognosis than patients with only one of these deletions. To test this hypothesis, we compared the clinical outcomes (overall survival (OS) and time to treatment (TTT)) of patients with 17p- and 11q- in the same cell to patients with either 17p- or 11q- or neither defect.
We used the Mayo Clinic CLL Database and the Cytogenetics laboratory database to identify all CLL patients seen in the Division of Hematology from 1/1/1995-3/29/2012 who had FISH analysis. Data was extracted on demographics, date of first (sentinel) occurrence of 11q- and 17p-, treatment, and vital status. FISH studies were performed on uncultured blood or bone marrow by standard methods using the Mayo Clinic CLL FISH panel of centromere specific control probes and probes for 6q (MYB), 11q (ATM), 13q14, and 17p (TP53) deletions, trisomy 12 and IGH. Patients with both 11q- and 17p- were further tested with the Abbott Molecular (Des Plaines, IL) ATM/TP53 combination probe set, to determine if the 11q and 17p deletions were present in the same cells. Patients were grouped into 4 cohorts: 1) clonal 17p- and 11q-; 2) 17p- only; 3) 11q- only; and 4) neither 17p- nor 11q-. OS and TTT were calculated and Kaplan-Meier curves were drawn from sentinel FISH date until their last known alive date or death date for OS and until treatment or last known untreated date for TTT. We censored all data on 3/29/2012 and analyzed using log-rank tests from date of sentinel FISH. Sanger sequencing of exons 4–9 of TP53 was done in samples from 15 patients with clonal 17p- and 11q- with an available sample collected within 1 year of the sentinel FISH test and without treatment in that interval. All mutations were confirmed by a repeat assay and analyzed for their predicted effect using the UMD p53 database.
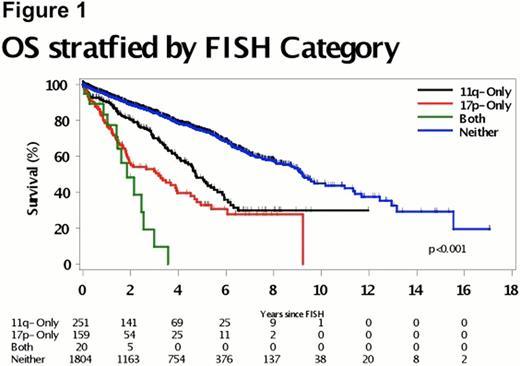
We studied 2234 CLL patients with a median follow-up of 4.8 years (range 0 – 29.6 years). Twenty (1%) patients had clonal 17p- and 11q-, 159 (7%) had 17p-, 251 (11%) had 11q-, and 1804 (81%) had neither 17p- nor 11q-. Median OS from sentinel FISH study (Figure 1) was significantly shorter in the clonal 17p- and 11q- group (1.9 years), 17p- group (3.2 y), and 11q- group (4.7 y) compared to the patients with neither 17p- nor 11q- (9.2 y) (p < 0.0001). OS was significantly shorter in the patients with clonal 17p- and 11q- cells vs those with 17p- (p=0.048). TTT was also significantly different among the 4 groups (p<0.0001) with median time to treatment of 0.07 y in the clonal 17p- and 11q-, 0.2 y in 17p-, 0.7 y in 11q-, and 5.6 y in the neither 17p- nor 11q- group. TP53 analysis performed in samples from 15 patients with clonal 17p- and 11q- showed that 10 patients had mutations in the remaining TP3 allele predicted to be deleterious to normal p53 protein function. Median OS was 2.1 years for those with TP53 mutations and 0.9 years for those without mutations (p = 0.03) and median TTT was 0.25 years for those with TP53 mutations and 0.05 years for those without mutations (p = 0.049).
“Double hit” clonal deletions of 17p and 11q occurred in 1% of this large cohort of CLL patients and were associated with a significantly poorer prognosis. We recommend that testing for this “double hit” phenomenon, by use of an ATM/TP53 combination probe set, should thus be performed in all patients with 17p- and 11q- on FISH analysis because these patients have a poorer prognosis than those with either 17p- or 11q- and could be candidates for novel therapies. The finding that dysfunctional TP53 mutations in the remaining allele did not worsen prognosis in patients with clonal 17p- and 11q- suggests that monoallelic deletion of both ATM and TP53 in a clone of CLL cells is sufficient to disable p53 pathway function in these patients. Future study of clonal 17p- and 11q- CLL could thus be informative in this regard.
Kay:Celgene: Research Funding. Shanafelt:Genentech: Research Funding; Glaxo-Smith-Kline: Research Funding; Cephalon: Research Funding; Hospira: Research Funding; Celgene: Research Funding; Polyphenon E International: Research Funding. Zent:Biothera: Research Funding; Genzyme: Research Funding; Genentech: Research Funding; Novartis: Research Funding; GlaxoSmithKline: Research Funding.

This icon denotes a clinically relevant abstract
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal