Abstract
Abstract 1697
Abnormal and clonal hematopoiesis resulting in peripheral blood cytopenias, and risk of progression to acute myeloid leukemia (AML) are the main characteristics of myelodysplastic syndromes (MDS) which present a high diversity of somatically mutated genes. Recently, mutations targeting genes whose products participate to the early steps of RNA splicing (SF3B1, SRSF2, ZRSR2, and U2AF35) have been reported. Mutations in ASXL1, DNMT3, EZH2, IDH1/2 and TET2, suggest deregulation of the epigenetic control of transcription. Additional genes known to be mutated in MDS include RUNX1, TEL/ETV6, TP53 and NRAS The alteration of some of these genes may carry prognostic value. Next-generation sequencing analyses of AML and related disorders recently identified mutations in two chromosome X genes, BCOR and BCORL1, that code for related transcriptional co-repressors interacting with histone deacetylases and presenting specific properties. BCOR interacts with BCL6 and constitutional inactivating mutations have been described in the Oculo-Facial-Cardio-Dental syndrome. BCOR is also affected by mutations or translocations in retinoblastoma and sarcoma. BCORL1 has been implicated in chromosomal rearrangements in hepatocellular carcinoma. In this study, we have investigated BCOR and BCORL1 gene by Sanger sequencing in a cohort of 221 MDS samples.
The 221 MDS samples were collected at diagnosis in multicenter clinical trials in France between 1999 and 2011. The coding sequences of BCOR (ENST00000378444) and BCORL1 (ENST00000218147) were analyzed by Sanger sequencing. Mutational analyses of ASXL1, CBL, DNMT3A, ETV6, EZH2 IDH1/2, JAK2, NRAS, RUNX1, SF3B1, SRSF2, TET2, TP53, U2AF35, and ZRSR2 were previously reported. Non tumoral material (buccal swab or CD3+ cells) was analyzed for the presence of the identified variations of BCOR or BCORL1 when available (n=9). The prognostic impact of BCOR mutations was evaluated in MDS patients with available follow-up information (n=203).
BCOR and BCORL1 coding sequences were analyzed in the cohort of 221 MDS patients and we found mutations in 8.6% (n=19) and 2.3% (n=5) respectively. Alterations were distributed all over the coding regions. Strikingly, two patients presented concomitant inactivating mutations of BCOR and BCORL1. Among the 19 BCOR alterations, 9 were missense, 3 nonsense, 5 frameshift and 2 splice site mutations.
No significant difference in age, sex, karyotype, blood counts or bone marrow blasts between BCORmut and BCORwt patients was observed. Mutations were found in patients of all IPSS risk-groups and WHO subtypes. Comparison of cytological bone marrow reports revealed a trend for a higher rate of dysgranulopoiesis in BCORmut patients (P=0.06).
Because only truncating mutations (splice, frameshift and nonsense mutations) are unambiguously expected to affect BCOR function, statistical analyses were restricted to truncating BCOR mutations (n=10/221, 4.5%). Truncating BCOR mutations were frequent in RUNX1mut patients (19% versus 3% in RUNX1wt patients; P=0.027). They were by trend associated with SRSF2 and DNMT3A mutations (P=0.09) and were exclusive with IDH1/2, JAK2, NRAS, TP53, and ZRSR2 mutations.
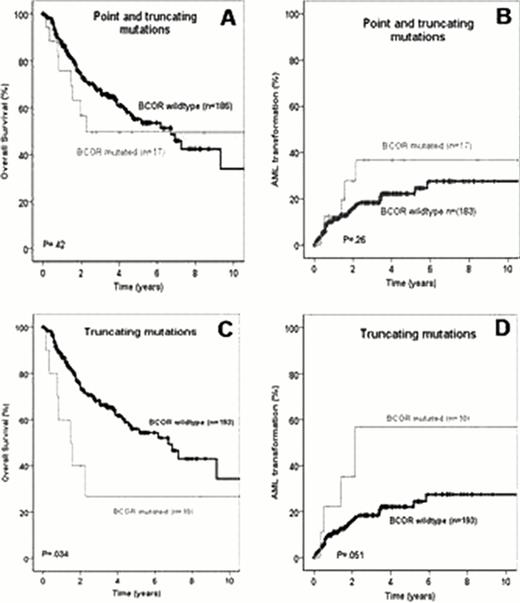
In univariate analysis, Overall Survival (OS) and AML transformation rate did not differ between patients with either missense or truncating BCOR mutations and BCORwt patients (Figure A&B). However, inferior OS (P=0.034) and higher AML transformation rate by trend (P=0.051) were observed for patients with truncating BCOR mutations (Figure C&D).
Multivariate analysis demonstrated that a truncating BCOR mutation was an independent unfavourable prognostic factor for OS (HR 3.3; 95%CI 1.4 – 8.1; P=.008). The low number of BCORL1 mutated patients precludes any statistical analyses but clinical follow-up was available for 4 out of 5 patients. Three patients died 7, 17 and 27 months after MDS diagnosis. The fourth patient received allogeneic bone marrow transplantation after AML transformation and is in complete remission 5 years after transplantation.
In summary, truncating mutations of BCOR were independently associated with a worse OS in MDS. These data support the idea that BCOR mutations appear as associated with RUNX1 and DNMT3A mutations as reported in AML. Moreover, and despite a relatively low mutation frequency in MDS, BCOR might be considered as a key gene in risk stratification.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal