Abstract
Abstract 122
Cancer clone development is widely regarded as an evolutionary or Darwinian process of genetic diversification and natural (or therapeutic) selection within tissue ecosystems. Emerging studies are providing strong evidence that dynamic and complex branching sub-clonal genetic architectures are a common feature of cancer (Greaves M and Maley CC Nature 2012). This complexity may underpin the intransigence of advanced cancer to therapeutic control, particularly as the critical 'driver' cells – cancer or leukaemic stem cells, also appear to be genetically diverse within individual patients (Anderson K et al Nature 2011, Notta F et al Nature 2011). Sub-clonal architecture can only be fully determined through the study of large numbers of single cells uniformly sampled from the individual cancer of interest and assessed for composite genotype. Various technologies and approaches from fluorescent in situ hybridisation (FISH) to whole-genome sequencing of single cells have been applied to cancer and leukaemic cells but each approach has limitations. We have developed a novel multiplex microfluidic Q-PCR approach that allows unbiased single cell sampling, high throughput analysis of hundreds of individual cells and simultaneous detection of multiple genetic alterations in a single cell, including fusion genes, DNA copy number alterations (CNAs) and sequence-based mutations.
As a proof of principle study we have applied this technique to REH, an acute lymphoblastic leukaemia (ALL) cell line that harbors the ETV6-RUNX1 fusion and a SNP in the EPO receptor gene, which we used as a surrogate mutation. We further determined a detailed sub-clonal genetic architecture for two ETV6-RUNX1 positive ALL patient samples with multiple point mutations and copy number alterations (determined by whole-genome sequencing) by interrogating approximately 400 flow cytometry sorted single cells with validation by FISH and standard sequencing. Briefly, single cells were lysed prior to multiplex specific (DNA) target amplification (STA) and Q-PCR using the 96.96 dynamic microfluidic array and the BioMarkï HD (Fluidigm, UK). Phylogenetic trees were constructed using maximum parsimony with PAUP analysis software.
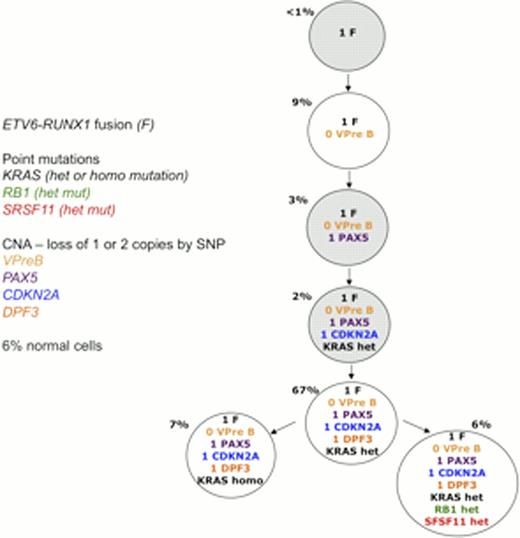
Interrogation of REH revealed that all single cells registered the ETV6-RUNX1 fusion and EPO receptor SNP, but 42% of cells gained either 1 or 2 additional copies of chromosome 21. Patient sample data revealed branching sub-clonal architectures in Case A in which all leukaemic cells harbored the fusion with additional point mutations but only sub-clones showed CNAs. In contrast, the sub-clonal architecture of Case B showed that whilst the ETV6-RUNX1 fusion was the earliest (or universal) genomic event, CNAs were relatively early events preceding the acquisition of point mutations (Figure 1). In both cases, the numerically predominant sub-clone harbored both point mutations and CNAs in addition to the presumptive initiating lesion, ETV6-RUNX1.
These detailed and complex sub-clonal architectures would be masked by other genetic techniques. Single cell genetics coupled with deep genome sequencing is now technically feasible and provides an accurate portrait of the dynamic clonal complexity in leukaemia (and other cancers). Variegated genetics and clonal complexity in individual leukaemias has important implications for our understanding of molecular pathogenesis and for therapeutic targeting.
This sub-clonal genetic architecture depicts the branching structure found for Case B, illustrating that in this case the ETV6-RUNX1 fusion was the earliest genomic event, followed by CNAs and the acquisition of point mutations. Those populations highlighted grey are within the experimental error rate but potentially true populations.
This sub-clonal genetic architecture depicts the branching structure found for Case B, illustrating that in this case the ETV6-RUNX1 fusion was the earliest genomic event, followed by CNAs and the acquisition of point mutations. Those populations highlighted grey are within the experimental error rate but potentially true populations.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal