Abstract
Abstract 1052
HIV-1 transcription is activated by HIV-1 Tat protein, which recruits CDK9/cyclin T1 and other host transcriptional co-activators to the HIV-1 promoter. Tat itself is phosphorylated by cell cycle kinase 2 (CDK2) and inhibition of CDK2 by small interfering RNA or iron chelators inhibits HIV-1 transcription. HIV-1 transcription is also activated by NF-kB that binds to HIV-1 LTR independent to Tat but can also be recruited Tat-dependently by CDK9/cyclin T1. Recently, induction of heme oxygenase-1 (HO-1) by hemin was shown to inhibit HIV-1 in vitro and in vivo. Here, we analyzed the effect of novel phenyl-1-pyridin-2yl-ethanone (PPY) based iron chelators, PPYeT and PPYaT, on HIV-1. Both chelators efficiently inhibited one round of HIV-1 replication in T cells at low nanomolar IC50s without exhibiting cytotoxicity at 24 hrs incubation. The iron chelators efficiently bound intracellular labile iron as it was determined in calcein binding assays. Because we previously showed that iron chelators inhibited the activity of CDK9, we analyzed expression of several cellular genes dependent on CDK9. Unexpectedly, chelators were found to induce the expression of IkBα, an inhibitor of NF-kB (Fig1). Treatment with the iron chelators retained NF-kB in cytoplasm of the treated cells suggesting reduction in NF-kB in nucleus (Fig2). The chelators were also shown to induce HO-1 expression in cultured monocytes, likely to do a decrease of intracellular iron pool. This effect of iron chelators mimicked the effect of hemin treatment which also induced HO-1 and inhibited HIV-1 infection in our experimental conditions. Low nanomolar IC50s for the PPY-based iron chelators and lack of toxicity suggest their potential usefulness as future anti-retroviral therapeutics. Further studies are needed to investigate additional targets for iron chelators in HIV-1 life cycle that may include reverse transcription and capsid assembly. Therefore iron chelators need to be carefully assessed not only to understand the mechanism but also as a therapeutic strategy.
This work was supported NIH Research Grants SC1GM082325, R25 HL003679, 2G12RR003048, 8G12MD007597, K25GM097501 and 1P30HL107253.
Iron chelators induce expression of IkB gene. 293T cells were treated with vehicle (DMSO), and iron chelators, PpYeT, PpYaT. After 24 treatment, RNA was extracted, reverse transcribed and analyzed by real-time PCR on Roche 4800 using primers for GAPDH,CDK2,CyclinA CyclinE and IkB. Primers for GAPDH were used for reference. The ΔΔCt analysis was performed using DMSO-treated, sample as a reference target and reference control. Both tested iron chelators induced expression of IkBα, the inhibitor of NFKB, suggesting potential mechanism for cyclin E deregulation. CyclinA is upregulated in PPYaT treated cells.
Iron chelators induce expression of IkB gene. 293T cells were treated with vehicle (DMSO), and iron chelators, PpYeT, PpYaT. After 24 treatment, RNA was extracted, reverse transcribed and analyzed by real-time PCR on Roche 4800 using primers for GAPDH,CDK2,CyclinA CyclinE and IkB. Primers for GAPDH were used for reference. The ΔΔCt analysis was performed using DMSO-treated, sample as a reference target and reference control. Both tested iron chelators induced expression of IkBα, the inhibitor of NFKB, suggesting potential mechanism for cyclin E deregulation. CyclinA is upregulated in PPYaT treated cells.
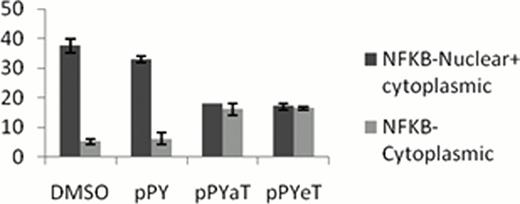
Distribution of NFKB in cytoplasm or in Nucleus and cytoplasm in cell treated and untreated with Iron Chelator. A: Number of cell having cytoplasmic and ubiquitious NFKB distribution in iron chelator treated cell and DMSO/pPY treated control.
B: Representative cells having uniform versus cytoplasmic distribution of NF-kB.
Distribution of NFKB in cytoplasm or in Nucleus and cytoplasm in cell treated and untreated with Iron Chelator. A: Number of cell having cytoplasmic and ubiquitious NFKB distribution in iron chelator treated cell and DMSO/pPY treated control.
B: Representative cells having uniform versus cytoplasmic distribution of NF-kB.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal