In this issue of Blood, Schuh et al report the detection of heterogeneous patterns of clonal evolution in 3 patients with IgHV unmutated CLL over the lifetime of their disease using whole-genome sequencing (WGS).1
One of the clinical hallmarks of chronic lymphocytic leukemia (CLL) is the high degree of variability in its disease course. Whereas some patients experience an indolent disease course, others succumb to the disease rapidly despite intensive treatment. Over time, most patients develop refractory disease that does not respond to chemotherapy. The advent of next-generation sequencing (NGS), with its unprecedented ability to systematically discover key genetic alterations that underlie cancer, offers the tantalizing possibility of unraveling the genetic basis of this heterogeneity. Indeed, the high degree of molecular variation existing between individuals with the same hematologic malignancy (ie, intertumoral genetic heterogeneity) has been revealed over the past few years by applying this transformative technology across various blood malignancies, including CLL.2 Thus, to a certain degree, clinical variability can be explained by the existence of patient subsets bearing distinct molecular genotypes. However, because NGS has the potential for sensitive genome-wide detection of mutations harbored in only a fraction of the total cell population,3 an even more striking recent discovery has been the detection of genetic heterogeneity within malignant cells of the same individual (ie intratumoral heterogeneity4 ), including blood malignancies such as acute myelogenous leukemia (AML)5 and multiple myeloma (MM).6
Schuh et al demonstrate that this additional layer of complexity is also a feature of CLL. While the existence of subclonal populations in CLL has been long appreciated on the basis of FISH cytogenetic and SNP data,7,8 the inherent limitations of these technologies restrict the range of genetic alterations that can be used for sensitively detecting and tracking subpopulations over time. In contrast, WGS offers the detection of approximately 2000 mutations per CLL sample. Through the clustering of genetic alterations of similar allelic frequency, subclonal architecture can be resolved with unparalleled granularity.
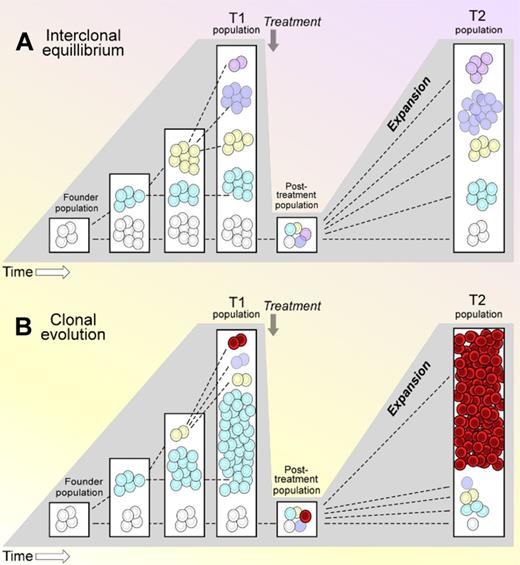
Using this approach, Schuh and colleagues followed 3 patients of a particular prognostic category—unmutated IGHV—and for each, CLL cells were sampled at 5 time points for up to 7 years. All 3 patients received 3 therapies; initially alkylator or fludarabine-based chemotherapy, followed by antibody-based therapy. Across all time points, subclonal populations were consistently observed, with each sample harboring up to 5 distinct subpopulations. Some mutations were detected in all cells and across all time points and were interpreted to represent events in the founder subclone, while other events were considered secondary events because they were common to only a subset of leukemia cells and demonstrated varying allelic frequency after treatment. Notably, although a several log reduction in white blood cell counts was observed after therapy in each case, 3 very different temporal patterns of repopulation were observed. One patient revealed stable equilibrium of 5 subpopulations over the course of years, despite therapy (see figure panel A). In contrast, a separate patient demonstrated the near complete replacement of the tumor after cytotoxic therapy by a subclone that was originally only a small subpopulation at an earlier time point (panel B). Finally, the third patient demonstrated a hybrid pattern—an initial stable balance between subclones that over time led to expansion of 1 subclone that became the dominant subclone at the time of the patient's death.
Schematic representation of 2 diverse dynamic patterns observed in whole-genome sequenced CLL samples, sampled at multiple times. T1 indicates time point 1; and T2, time point 2. One pattern is of interclonal equilibrium (A), while the other is of clonal evolution (B).
Schematic representation of 2 diverse dynamic patterns observed in whole-genome sequenced CLL samples, sampled at multiple times. T1 indicates time point 1; and T2, time point 2. One pattern is of interclonal equilibrium (A), while the other is of clonal evolution (B).
What do these patterns tell us? Although only 3 cases were analyzed, the results of sampling multiple serial time points per patient suggest the presence of dynamic forces that can impact the stability of CLL genetic composition over time. For example, the scenario provided in panel A suggests the presence of clonal interference9 —that co-existing subclonal populations may limit each others' growth by mutual competition. In contrast, a separate insight is suggested by the scenario illustrated in panel B, in which a more aggressive clone emerges after therapy that was present in the pretreatment sample at low frequency (detectable by deep sequencing). On one hand, this could be explained by the active selection of particular genetic alterations by therapy. Alternately, harkening to paradigms formulated in evolutionary biology, cytotoxic therapy could act as a classic evolutionary restriction point and reset interclonal dynamics in favor of a more aggressive clone by markedly reducing disease bulk and by removing the incumbent clone.10
These data invite one to speculate that the composition of CLL of any particular individual results from personal “ecology”—a result of the interplay between both intrinsic (eg, inherent “driverness” among subclones) and extrinsic/environmental factors (eg, time since transformation, exposure to cytotoxic therapy). The work by Schuh et al thus serves as an important starting point, and inspires numerous questions. How frequently are the patterns of interclonal equilibrium or of clonal evolution observed in patients and how to do they relate to known prognostic risk factors in CLL? Do these patterns hold prognostic or predictive significance? What are the underlying genetic factors that determine whether subclones recede or expand? Can identification of subclonal mutations before therapy anticipate the composition of the relapsing tumor? Answering each of these questions will require tumor sequencing of larger and preferably uniformly treated patient sample sets along with an analysis pipeline that can accommodate these large datasets. In turn, addressing these questions has the potential to reveal whether the presence of subclonal populations actually impacts CLL disease tempo or responsiveness to therapy.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal