Abstract
Epistatic interactions between killer cell immunoglobulin-like receptors (KIRs) and their cognate HLA class I ligands have important implications for reproductive success, antiviral immunity, susceptibility to autoimmune conditions and cancer, as well as for graft-versus-leukemia reactions in settings of allogeneic stem cell transplantation. Although CD8 T cells are known to acquire KIRs when maturing from naive to terminally differentiated cells, little information is available about the constitution of KIR repertoires on human CD8 T cells. Here, we have performed a high-resolution analysis of KIR expression on CD8 T cells. The results show that most CD8 T cells possess a restricted KIR expression pattern, often dominated by a single activating or inhibitory KIR. Furthermore, the expression of KIR, and its modulation of CD8 T-cell function, was independent of expression of self-HLA class I ligands. Finally, despite similarities in the stochastic regulation of KIRs by the bidirectional proximal promoter, the specificity of inhibitory KIRs on CD8 T cells was often distinct from that of natural killer cells in the same individual. The results provide new insight into the formation of KIR repertoires on human T cells.
Introduction
Epidemiologic studies have found that combinations of killer cell immunoglobulin-like receptors (KIRs) and their cognate HLA class I ligands influence the outcome of human pregnancy, resistance to infections, susceptibility to autoimmune diseases and cancer, as well as the effects of hematopoietic stem cell transplantation.1-5 Although it is commonly perceived that such correlations indicate a role for natural killer (NK) cells in these settings, the scarcity of mechanistic insight at a cellular level raises the possibility that other KIR-expressing cell types might also contribute to the effects observed. Indeed, as recently shown, the presence of a specific inhibitory KIR, on the genetic level, enhanced HLA class I–restricted antiviral CD8 T-cell immunity, suggesting that KIR-expressing CD8 T cells might influence the outcome of chronic viral infections.6
KIRs represent a polygenic and polymorphic family of receptors.7 Inhibitory and activating KIRs signal via immunoreceptor tyrosine-based inhibition/activation motifs (ITIM/ITAM), respectively.8 NK cells are the main KIR-expressing immune cells.9,10 They express inhibitory KIRs in a variegated fashion to ensure a broad specificity and capacity to sense the presence of single HLA class I alleles.11 Although there is a minor influence of HLA class I on the expression of specific KIRs on NK cells,12,13 that seems more prominent in settings of viral infections,14,15 KIR expression is primarily determined by KIR gene and promoter polymorphisms as well the production of antisense RNAs in a random process that is not subject to any positive or negative selection.16,17 To preserve self-tolerance in the absence of repertoire selection, NK cells are functionally tuned in an yet not fully understood educational process in which the strength of the interactions between inhibitory KIRs and their cognate HLA class I ligands is one important factor.18
Other than NK cells, CD4 and CD8 T cells as well as γδ T cells also express KIRs.19-24 With respect to CD8 T cells, KIR expression starts to appear on effector memory CD8 cells, and a substantial fraction of terminally differentiated effector CD8 T cells are KIR+.20,22,25 The function of KIRs on CD8 T cells has been studied to some extent. Although most studies have been performed with KIR+ CD8 T-cell clones, it is clear that inhibitory KIRs can modulate T-cell receptor (TCR) signaling and dampen CD8 T-cell responses, whereas activating KIRs can enhance functional T-cell responses.26,27 However, little is known with respect to the role of cognate HLA class I molecules in shaping the KIR repertoire and function of CD8 T cells. Specifically, it is not known whether HLA class I molecules have a KIR-dependent educational effect on human CD8 T-cell function as it has on NK cells.
In the present study, we performed a high-resolution analysis of KIR expression on human CD8 T cells with the use of newly developed FACS panels, allowing also for the assessment of activating KIRs. We show that KIR expression on CD8 T cells is restricted to clonally expanded terminally differentiated CD8 T cells. These cells display a narrow KIR repertoire dominated by a single inhibitory or activating KIR. We show that the specificity of KIR expressed on CD8 T cells is random and often distinct compared with that expressed on NK cells within the same individual. Furthermore, we have attempted to dissect potential underlying mechanisms behind the diverse KIR repertoires found on CD8 T cells and NK cells by evaluating the level of selection conferred by cognate HLA class I molecules, as well as transcriptional regulation of KIRs on CD8 T cells and NK cells. Finally, we show that KIR expression down-modulates the functional responses of CD8 T cells in an HLA-independent manner, suggesting that human CD8 T cells are not subject to functional education by HLA class I molecules. Taken together, our data provide new insights into the composition of inhibitory and activating KIRs on CD8 T cells at the population and single cell levels.
Methods
Human subjects and cells
This study, conducted in accordance with the Declaration of Helsinki, was approved by the regional ethics committee in Stockholm, Sweden. Peripheral blood mononuclear cells (PBMCs) were separated from buffy coats by density gravity centrifugation (Ficoll-Hypaque; GE Healthcare). PBMCs were either cryopreserved in liquid nitrogen in 10% DMSO and 90% heat-inactivated FBS for later usage or used directly after separation. KIR and HLA genotyping were performed on genomic DNA as previously described.9 Cryopreserved samples from a total of 44 healthy subjects homozygous for the group A KIR haplotype were used for the in-depth analysis (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Directly isolated cells from an additional 199 healthy subjects, with mixed KIR haplotypes, were used to examine the expression of activating KIRs. In these 2 cohorts, age, sex, or ethnicity did not significantly influence total CD8 T-cell expression or expression of individual KIRs. For assessment of CMV-specific CD8 T cells, cryopreserved samples from 6 HLA-typed CMV-seropositive healthy individuals were used.
Monoclonal antibodies and flow cytometry
Commercially available monoclonal antibodies (mAbs) against the following proteins were used: KIR2DL1 (clone 143211), KIR2DL3 (180701), KIR3DL1 (Dx9), KIR3DL2 (Dx31), panKIR2D (NKVFS1), KIR2DS4 (179315), KIR2DL1/S1 (EB6), KIR2DL2/S2/L3 (GL183), CCR7 (3D12), CD8 (SK1), CD45RA (MEM-56), CD3 (UCHT1), CD57 (HCD57), NKG2A (z199), CD56 (NCAM16.2), CD28 (CD28.2), CD27 (O323), and CD127 (HIL-7R-M21). Anti-KIR3DL2 mAb (Dx31) was kindly provided by Dr J. Philips (DNAX Research Institute). The following tetrameric peptide/HLA class I tetramer complexes were used in this study: CMV pp65 NLVPMVATV(NV9)/HLA A*0201 and CMV pp65 TPRVTGGGAM(TM10)/HLA B*0702. FACS stainings were performed as previously described.28 Briefly, thawed PBMC samples were stained for 20 minutes on ice with saturating amounts of desired mAb combinations. To exclude dead cells from the analysis, samples were stained with the LIVE/DEAD Fixable Aqua Dead Cell Stain kit (Invitrogen). After staining, samples were washed twice and fixed in PBS containing 1% paraformaldehyde. Samples were acquired either on a CyAn ADP LX 9-color flow cytometer or on a BD LSRFortessa 18-color flow cytometer. The CyAn ADP LX instrument was equipped with a 20-mW 405-nm laser, a 20-mW 488-nm laser, and a 25-mW 635-nm laser. The LSRFortessa instrument was equipped with a 100-mW 405-nm laser, a 100-mW 488-nm laser, a 50-mW 561-nm laser, and a 40-mW 639-nm laser. Acquired data were subject to unsupervised software-based compensation with the use of FlowJo Version 9 (TreeStar) and subsequently analyzed with FlowJo Version 9.
TCR Vβ chain analysis
The TCR Vβ chain repertoire of KIR+ CD8 T cells was analyzed with the TCR Vβ kit FITC/PE (Beckman Coulter) in 20 randomly selected healthy subjects homozygous for the group A KIR haplotype. Samples were costained with mAbs against the following proteins: CD3 (UCHT1), CD8 (SK1), CD14 (MoP9), CD19 (SJ25C1), CD45RA (MEM-56), CD57 (HCD57), CD27 (O323), KIR2DL1 (EB6), KIR2DL3 (GL183), KIR3DL1 (Dx9), and KIR3DL2 (Dx31) as previously described.28
Luciferase reporter assays
Methods for the analysis of bidirectional KIR promoter activity have been reported previously.29 Briefly, fragments containing the promoter region 230 bases upstream of the transcriptional start site for each KIR were generated by PCR and cloned into the pGL3 vector (Promega) to generate constructs in both forward and reverse orientations. YT-Indy or Jurkat cells were transfected by electroporation with a BTX ECM 830 (Gentronics). A total of 5 × 106 cells were transfected with 10 μg of the specific reporter construct and 0.1 μg of the Renilla luciferase pRL-SV40 vector for normalization of transfection efficiency. Luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega) 48 hours after transfection.
Quantitative RT-PCR analysis of the KIR3DL1 proximal promotor
First, CD8 T cells and NK cells were purified by negative magnetic bead–based separation from fresh PBMCs. Purified cells underwent sorting with the use of a BD FACSAria flow cytometer for isolation of KIR3DL1+ and KIR3DL1− subsets within panKIR2D+KIR3DL2+ CD56dim NK and panKIR2D+KIR3DL2+CD45RA+CD57+ CD8 T cells. Total RNA was extracted with RNeasy columns (Qiagen). For the analysis of KIR3DL1 coding transcripts, cDNA was generated with random hexamers (Invitrogen). The primer and probe sequences for quantitative RT-PCR of the KIR3DL1 coding transcripts have been previously published.30 For the analysis of KIR3DL1 antisense transcripts, cDNA was generated at 55°C with the use of a gene-specific primer. The primer and probe sequences for quantitative RT-PCR of KIR3DL1 antisense transcripts have been published.31 All quantitative RT-PCR was performed with a 7500 Real-Time PCR System (Applied Biosystems).
CD8 T-cell functional assay
To study the functionality of KIR+ CD8 T cells, 0.5 × 106 PBMCs from donors homozygous for the group A KIR haplotype were incubated in 96-well round-bottom plates together with 5 × 104 P815 target cells with or without the addition of anti-CD3 (1:100 dilution, clone UCHT1) as well as anti-CD107a mAb. After 1 hour of incubation, brefeldin A (1:1000 dilution; Becton Dickinson) was added to each well. After 6 hours of incubation, cells were harvested and stained with antibodies against the following proteins: CD3 (UCHT1), CD8 (3B5), CD45RA (HI100), CD27 (M-T271), CD57 (TB01), KIR2DL1 (143211), and KIR2DL2/L3/S2 (GL183). Nonviable cells were excluded with the LIVE/DEAD Fixable Aqua Dead Cell Stain kit. Cells were subsequently fixed, permeabilized, and stained for intracellular IFN-γ (B27) and TNF-α (Mab11) and analyzed by flow cytometry.
Statistical analysis
The following statistical tests were used unless otherwise indicated in the figure legends. For multiple group comparisons, 1-way ANOVA or Kruskal-Wallis nonparametric tests were applied. For comparisons of independent groups, Student t test or the Mann-Whitney test was performed. For comparisons of matched groups, paired Student t test or Wilcoxon matched test was performed. In figures, n.s. indicates not significant; ***P < .001, **P < .01, and *P < .05. Statistical analyses were performed with Prism Version 5.0 software (GraphPad Software Inc).
Results
KIR+ CD8 T cells are terminally differentiated effector cells
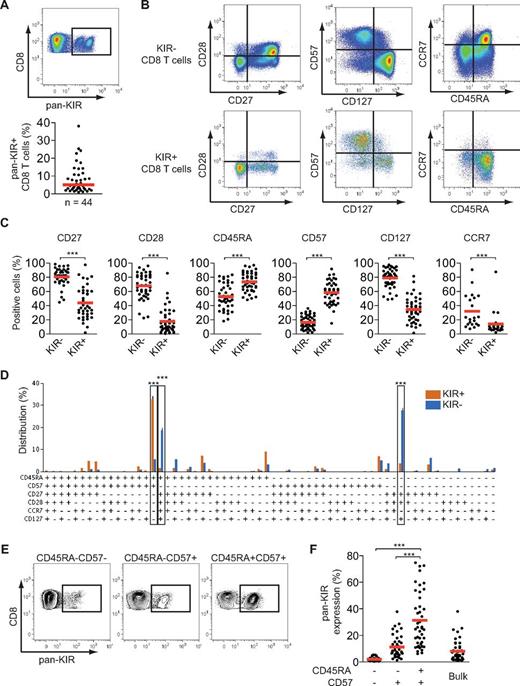
To identify and quantify the human CD8 T-cell subset that most frequently expressed KIRs, we analyzed inhibitory KIR-expressing cells within subsets of naive and differentiated CD8 T cells in peripheral blood obtained from 44 healthy subjects. On average, 5% (range, 1%-38%) of total CD8 T cells expressed KIRs (Figure 1A). Combined assessment of 6 molecules commonly used to study T-cell differentiation found that the major KIR-expressing subset was terminally differentiated (CD45RA+CD57+CCR7−CD27−CD28−CD127−) CD8 T cells, whereas the major KIR− subsets were effector memory (CD45RA−CD57−CCR7−CD27+CD28+CD127+) and naive (CD45RA+CD57−CCR7+CD27+CD28+CD127+) CD8 T cells (Figure 1B-D). Cells exhibiting a naive phenotype were sparsely represented within KIR+ CD8 T cells (Figure 1D). This analysis found that the isolated usage of CD45RA and CD57 was sufficient to identify the major KIR-expressing CD8 T-cell subset in humans (Figure 1D). Indeed, when evaluating total inhibitory KIR expression on CD45RA+CD57+ CD8 T cells, on average 30% of such cells expressed KIRs compared with 11% and 2%, respectively, for CD45RA−CD57+ and CD45RA−CD57− CD8 T cells (Figure 1E-F). These data prompted us to subsequently focus our studies on a high-resolution analysis of KIR expression on CD45RA+CD57+ CD8 T cells.
KIR+ CD8 T cells are terminally differentiated cells. (A) Staining for total inhibitory KIR (hereafter termed pan-KIR) made with 4 mAbs specific for KIR2DL1, KIR2DL3, KIR3DL1, and KIR3DL2 on CD8 T cells from 1 representative individual (top) and frequency of pan-KIR+ CD8 T cells (bottom; n = 44; median). (B) Staining for CD27, CD28, CD57, CD127, CD45RA, and CCR7 on pan-KIR− and pan-KIR+ CD8 T cells from 1 representative individual. (C) Expression of CD27, CD28, CD45RA, CD57, CD127, and CCR7 on pan-KIR− and pan-KIR+ CD8 T cells (n = 39, 44, 43, 44, 44, and 21, respectively; ***P < .001, paired Student t test; mean). (D) Distribution of pan-KIR+ and pan-KIR− CD8 T cells for expression of CD45RA, CD57, CD27, CD28, CCR7, and CD127 as defined by a Boolean gating scheme (n = 19; ***P < .001, 1-way ANOVA with Bonferroni multiple comparison test; mean ± SEM). (E) Staining for pan-KIR on CD45RA−CD57−, CD45RA−CD57+, and CD45RA+CD57+ CD8 T cells from 1 representative individual. (F) Pan-KIR expression on CD45RA−CD57−, CD45RA−CD57+, CD45RA+CD57+, and total CD8 T cells (n = 43; ***P < .001, 1-way ANOVA with Bonferroni multiple comparison test; mean).
KIR+ CD8 T cells are terminally differentiated cells. (A) Staining for total inhibitory KIR (hereafter termed pan-KIR) made with 4 mAbs specific for KIR2DL1, KIR2DL3, KIR3DL1, and KIR3DL2 on CD8 T cells from 1 representative individual (top) and frequency of pan-KIR+ CD8 T cells (bottom; n = 44; median). (B) Staining for CD27, CD28, CD57, CD127, CD45RA, and CCR7 on pan-KIR− and pan-KIR+ CD8 T cells from 1 representative individual. (C) Expression of CD27, CD28, CD45RA, CD57, CD127, and CCR7 on pan-KIR− and pan-KIR+ CD8 T cells (n = 39, 44, 43, 44, 44, and 21, respectively; ***P < .001, paired Student t test; mean). (D) Distribution of pan-KIR+ and pan-KIR− CD8 T cells for expression of CD45RA, CD57, CD27, CD28, CCR7, and CD127 as defined by a Boolean gating scheme (n = 19; ***P < .001, 1-way ANOVA with Bonferroni multiple comparison test; mean ± SEM). (E) Staining for pan-KIR on CD45RA−CD57−, CD45RA−CD57+, and CD45RA+CD57+ CD8 T cells from 1 representative individual. (F) Pan-KIR expression on CD45RA−CD57−, CD45RA−CD57+, CD45RA+CD57+, and total CD8 T cells (n = 43; ***P < .001, 1-way ANOVA with Bonferroni multiple comparison test; mean).
KIR+ CD8 T cells have a narrow TCR Vβ chain expression
TCR Vβ chain analysis is a well-established way to assess TCR diversity. Normally, more differentiated CD8 T cells have an oligoclonal, rather than polyclonal, distribution of TCR Vβ chains. To determine the TCR diversity of KIR+ CD8 T cells we analyzed expression of 24 distinct TCR Vβ chains on naive, KIR− terminally differentiated, pan-KIR+ terminally differentiated CD8 T cells as well as on the 2 largest KIR-expressing subsets within terminally differentiated CD8 T cells in 20 randomly selected healthy individuals homozygous for the group A KIR haplotype (Figure 2A-B). As expected, a larger fraction of terminally differentiated CD8 T cells than naive CD8 T cells expressed one prominent TCR Vβ chain (Figure 2A). Strikingly, however, the TCR diversity was even more restricted within KIR+ terminally differentiated CD8 T cells (Figure 2A).
KIR+ CD8 T cells display a restricted TCR Vβ chain repertoire. The distribution of 24 specific TCR Vβ chains were analyzed in distinct CD8 T-cell subsets in samples from 20 individuals homozygous for the group A KIR haplotype. (A) TCR Vβ chain distribution in KIR− CD45RA+CD27+CD57− (Naïve), KIR− CD45RA+CD27−CD57+ (EMRA), and panKIR+ CD45RA+CD27−CD57+ (EMRA) CD8 T cells summarized for all analyzed individuals. (B) TCR Vβ chain distribution in panKIR+ CD45RA+CD27−CD57+ (EMRA) CD8 T cells and within the largest (KIR+ 1) and second largest (KIR+ 2) KIR-expressing subsets of CD45RA+CD27−CD57+ (EMRA) CD8 T cells summarized for all analyzed individuals. (A-B) ***P < .001, **P < .01, paired t test. (C) TCR Vβ chain repertoires of the 2 largest KIR-expressing subsets of CD45RA+CD27−CD57+ CD8 T cells shown for 4 representative individuals. The dominant TCR Vβ chains are indicated in the respective bar graphs.
KIR+ CD8 T cells display a restricted TCR Vβ chain repertoire. The distribution of 24 specific TCR Vβ chains were analyzed in distinct CD8 T-cell subsets in samples from 20 individuals homozygous for the group A KIR haplotype. (A) TCR Vβ chain distribution in KIR− CD45RA+CD27+CD57− (Naïve), KIR− CD45RA+CD27−CD57+ (EMRA), and panKIR+ CD45RA+CD27−CD57+ (EMRA) CD8 T cells summarized for all analyzed individuals. (B) TCR Vβ chain distribution in panKIR+ CD45RA+CD27−CD57+ (EMRA) CD8 T cells and within the largest (KIR+ 1) and second largest (KIR+ 2) KIR-expressing subsets of CD45RA+CD27−CD57+ (EMRA) CD8 T cells summarized for all analyzed individuals. (A-B) ***P < .001, **P < .01, paired t test. (C) TCR Vβ chain repertoires of the 2 largest KIR-expressing subsets of CD45RA+CD27−CD57+ CD8 T cells shown for 4 representative individuals. The dominant TCR Vβ chains are indicated in the respective bar graphs.
T-cell clones with the same rearranged TCR can have distinct KIRs, an observation that is in line with late acquisition of KIRs on differentiating T cells.23,32,33 To further examine the KIR repertoire in distinct TCR Vβ chain populations ex vivo, we analyzed the diversity of the TCR Vβ chains within the largest and second largest KIR-expressing CD8 T-cell subsets in these 20 healthy individuals. Interestingly, the largest KIR-expressing CD8 T-cell subset displayed the narrowest TCR Vβ chain repertoire (Figure 2B), suggesting that such cells have undergone a phase of expansion after acquisition of the inhibitory KIR. Furthermore, we confirmed that cells expressing 2 distinct KIRs either shared TCR Vβ chains or constituted 2 distinct CD8 T-cell clones (Figure 2C). Neither of these 2 scenarios was more frequent, suggesting a random acquisition of KIRs in terminally differentiated CD8 T cells. Importantly, across the donors, no single TCR Vβ chain was dominant on KIR-expressing CD8 T cells (Figure 2C; supplemental Table 2).
The CD8 T-cell KIR repertoire is restricted toward expression of 1 dominant KIR
For an overall view of the constitution of the inhibitory KIR repertoire on CD8 T cells, coexpression patterns of KIR2DL1, KIR2DL3, and KIR3DL1 were analyzed on CD45RA+CD57+ CD8 T cells. Close to 90% of the KIR-expressing CD8 T cells had a restricted KIR repertoire and expressed only 1 inhibitory KIR (Figure 3A). Consequently, coexpression of 2-3 KIRs was less common in CD8 T cells than NK cells and less frequent than expected from a random and independent association of receptors on the cell surface as determined by the product rule (Figure 3A-B). Because the KIR repertoire of CD8 T cells was skewed toward single KIR+ cells, we determined the contribution of individual KIRs to the single KIR+ CD8 T-cell compartment. If one assumes that the 3 KIRs investigated were evenly expressed within the single KIR+CD45RA+CD57+ CD8 T-cell compartment, any one KIR would represent approximately 33% (Figure 3C, dashed line). However, no such even usage was observed; instead one specific KIR dominated on the CD8 T cells representing, on average, 68% of the total KIR+ CD8 T-cell compartment (Figure 3C). Interestingly, this deviation toward a single dominant KIR was more pronounced for CD8 T cells than for NK cells (Figure 3C). Similar results were observed when KIR expression patterns were assessed on CD45RA−CD57+ CD8 T cells (supplemental Figure 1). Furthermore, having a single, unique, dominant KIR predicted a high frequency of total KIR expression among the CD8 T cells (Figure 3D). Together, these results indicate significant differences in the composition of KIR repertoires on terminally differentiated CD8 T cells compared with NK cells and suggest a more restricted usage of 1 dominant KIR in the terminally differentiated effector CD8 T-cell compartment.
CD8 T cells express a single dominant KIR (1 KIR+ cells) often distinct from that of NK cells in the same individual. (A) Distribution of KIR2DL1-, KIR2DL3-, and KIR3DL1-expressing NK cells and CD45RA+CD57+ CD8 T cells within cells expressing 1-3 KIRs (n = 43; ***P < .001, 1-way ANOVA with Bonferroni multiple comparison test; mean ± SEM). The gating strategy used to identify NK cells included gating on CD56dim+CD3−CD14− lymphocytes after having excluded dead cells and double events. (B) The observed frequencies of CD45RA+CD57+ CD8 T cells coexpressing 2 KIRs (KIR2DL1 + KIR2DL3, KIR2DL1 + KIR3DL1, KIR2DL3 + KIR3DL1) is plotted against those given by the product of the expression frequencies for the respective KIRs. Results are shown as frequency out of total KIR-expressing cells. A perfect fit to the product rule appears as a gray line, illustrating a 1:1 relation between observed and expected frequencies. K values were derived from the slope of the linear regression of observed data relative to a perfect fit with the product rule. P values were derived from a linear regression analysis. (C) Frequency of the most expressed KIR, out of KIR2DL1, KIR2DL3, and KIR3DL1, within 1-KIR+ cells as the frequency out of total 1 KIR+ cell for NK cells and CD45RA+CD57+ CD8 T cells (n = 43; ***P < .001, paired Student t test; median). (D) Correlation between single KIR dominance within 1 KIR+ CD45RA+CD57+ CD8 T cell and pan-KIR expression of CD45RA+CD57+ CD8 T cells. (E) Expression of KIR2DL1, KIR2DL3, and KIR3DL1 on NK cells and CD45RA+CD57+ CD8 T cells (n = 43; n.s. indicates not significant; 1-way ANOVA with Bonferroni multiple comparison test; median). (F) Correlation of pan-KIR expression on CD45RA+CD57+ CD8 T cells and CD56dim NK cells. (G) Schematic pie charts of KIR2DL1, KIR2DL3, and KIR3DL1 expression within 1 KIR+ NK cell and CD45RA+CD57+ CD8 T cells for 3 representative individuals.
CD8 T cells express a single dominant KIR (1 KIR+ cells) often distinct from that of NK cells in the same individual. (A) Distribution of KIR2DL1-, KIR2DL3-, and KIR3DL1-expressing NK cells and CD45RA+CD57+ CD8 T cells within cells expressing 1-3 KIRs (n = 43; ***P < .001, 1-way ANOVA with Bonferroni multiple comparison test; mean ± SEM). The gating strategy used to identify NK cells included gating on CD56dim+CD3−CD14− lymphocytes after having excluded dead cells and double events. (B) The observed frequencies of CD45RA+CD57+ CD8 T cells coexpressing 2 KIRs (KIR2DL1 + KIR2DL3, KIR2DL1 + KIR3DL1, KIR2DL3 + KIR3DL1) is plotted against those given by the product of the expression frequencies for the respective KIRs. Results are shown as frequency out of total KIR-expressing cells. A perfect fit to the product rule appears as a gray line, illustrating a 1:1 relation between observed and expected frequencies. K values were derived from the slope of the linear regression of observed data relative to a perfect fit with the product rule. P values were derived from a linear regression analysis. (C) Frequency of the most expressed KIR, out of KIR2DL1, KIR2DL3, and KIR3DL1, within 1-KIR+ cells as the frequency out of total 1 KIR+ cell for NK cells and CD45RA+CD57+ CD8 T cells (n = 43; ***P < .001, paired Student t test; median). (D) Correlation between single KIR dominance within 1 KIR+ CD45RA+CD57+ CD8 T cell and pan-KIR expression of CD45RA+CD57+ CD8 T cells. (E) Expression of KIR2DL1, KIR2DL3, and KIR3DL1 on NK cells and CD45RA+CD57+ CD8 T cells (n = 43; n.s. indicates not significant; 1-way ANOVA with Bonferroni multiple comparison test; median). (F) Correlation of pan-KIR expression on CD45RA+CD57+ CD8 T cells and CD56dim NK cells. (G) Schematic pie charts of KIR2DL1, KIR2DL3, and KIR3DL1 expression within 1 KIR+ NK cell and CD45RA+CD57+ CD8 T cells for 3 representative individuals.
KIR specificity of CD8 T cells is random and distinct from that of NK cells in the same individual
Next, we examined the specificity of inhibitory KIRs on CD8 T cells and compared it with NK cells in the same individual. When evaluating expression of KIR2DL1, KIR2DL3, and KIR3DL1 on CD45RA+CD57+ CD8 T cells, no significant differences could be found in expression frequencies among these 3 receptors (Figure 3E). Thus, despite the biased usage of 1 KIR with a particular specificity in each individual (Figure 3A), the aggregated CD8 T-cell KIR repertoires in these healthy individuals showed that all assessed KIRs were almost equally represented. This resembled the expression pattern of distinct KIRs in NK cells (Figure 3E). However, strikingly, comparing total KIR expression on CD8 T cells with NK cells within the same individuals, no correlation was found (Figure 3F). Thus, an individual could possess CD8 T cells expressing a high frequency of KIRs, yet have NK cells with low frequencies of total KIRs, and vice versa. Thirty-six of the 44 blood donors displayed 1 dominant KIR (representing more than 50% of all KIR-expressing CD8 T cells) within their single-KIR+ CD8 T-cell compartment. Intriguingly, in these donors, the dominant KIR within CD8 T cells was often unrelated to that dominating in single-KIR+ NK cells (Figure 3G). For example, in donor 2, single-KIR+ CD8 T cells were mostly KIR2DL3+, whereas singleKIR+ NK cells were KIR2DL1+ (Figure 3G). In the cohort of 44 blood donors, 18 had a clear dominance of 1 single KIR in both CD8 T cells and NK cells. Of these, 9 donors shared the same KIR specificity, whereas 9 displayed distinct specificities. An additional 18 donors had single KIR dominance in the CD8 T-cell compartment but a variegated KIR usage in the NK-cell compartment. In the remaining donors, KIRs were more evenly distributed on both CD8 T cells and NK cells, precluding a direct comparison of their repertoires. Thus, the KIR specificity in CD8 T cells often differs from that of NK cells in the same individual.
CD8 T cells express single activating KIRs
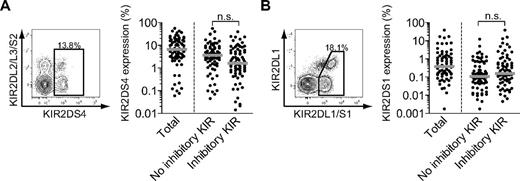
Having concluded that the composition of inhibitory KIR repertoires differs between NK cells and CD8 T cells, both at the population level and within a given individual, we next set out to study the expression pattern of activating KIRs on CD8 T cells. Expression of KIR2DS1 and KIR2DS4 was examined on CD57+ CD8 T cells in a second cohort that consisted of 199 healthy individuals with mixed KIR haplotypes (KIR haplotype group A and B). To be able to costain for activating KIRs in the group B KIR haplotype individuals, it was unfortunately not possible to include CD45RA in the analysis. Eighty-one of the 199 donors had the KIR2DS4 gene and expressed KIR2DS4. On average, 11% of total CD57+ CD8 T cells expressed KIR2DS4 in these individuals (Figure 4A). Interestingly, more than 50% of the KIR2DS4+ cells did not coexpress inhibitory KIR2DL1, KIR2DL3, or KIR3DL1 (Figure 4A; supplemental Figure 2). Furthermore, KIR2DS1 was expressed in 73 of 126 investigated group B KIR haplotype individuals (Figure 4B). As for KIR2DS4, a substantial proportion of KIR2DS1+ CD8 T cells lacked coexpression of inhibitory KIRs (Figure 4B; supplemental Figure 2). These data disclose that the expression of activating KIRs is a common event on CD8 T cells from healthy individuals and that such cells do not necessarily coexpress inhibitory KIRs to dampen their reactivity.
Expression of activating KIRs on CD8 T cells. (A) Staining for KIR2DS4 (left) on CD57+ CD8 T cells and expression of KIR2DS4 (right) on total CD57+ CD8 T cells, KIR2DL1−KIR2DL3−KIR3DL1−CD57+ CD8 T cells (no inhibitory KIR), and KIR2DL1+/−KIR2DL3+/−KIR3DL1+/−CD57+ CD8 T cells (inhibitory KIR; n = 81; median). (B) Staining for KIR2DS1 (left) on CD57+ CD8 T cells and expression of KIR2DS1 (right) on total CD57+ CD8 T cells, KIR2DL1−KIR2DL3−KIR3DL1−CD57+ CD8 T cells (no inhibitory KIR) and KIR2DL1+/−KIR2DL3+/−KIR3DL1+/−CD57+ CD8 T cells (inhibitory KIR). Costaining with antibodies against KIR2DL1 and KIR2DL1/S1 was used for specific identification of KIR2DS1+ cells (n = 73; median).
Expression of activating KIRs on CD8 T cells. (A) Staining for KIR2DS4 (left) on CD57+ CD8 T cells and expression of KIR2DS4 (right) on total CD57+ CD8 T cells, KIR2DL1−KIR2DL3−KIR3DL1−CD57+ CD8 T cells (no inhibitory KIR), and KIR2DL1+/−KIR2DL3+/−KIR3DL1+/−CD57+ CD8 T cells (inhibitory KIR; n = 81; median). (B) Staining for KIR2DS1 (left) on CD57+ CD8 T cells and expression of KIR2DS1 (right) on total CD57+ CD8 T cells, KIR2DL1−KIR2DL3−KIR3DL1−CD57+ CD8 T cells (no inhibitory KIR) and KIR2DL1+/−KIR2DL3+/−KIR3DL1+/−CD57+ CD8 T cells (inhibitory KIR). Costaining with antibodies against KIR2DL1 and KIR2DL1/S1 was used for specific identification of KIR2DS1+ cells (n = 73; median).
No evidence for HLA class I–mediated skewing of CD8 T-cell KIR repertoires
To explore whether the restricted CD8 T-cell KIR repertoires resulted from a selection process conferred by cognate HLA class I molecules, we examined the influence of KIR ligands on the KIR repertoire. Individuals included in the study were genotyped for the presence of ligands to KIR2DL1 (ie, HLA-C2), KIR2DL2/L3 (ie, HLA-C1), and KIR3DL1 (ie, HLA-Bw4). We focused on 24 individuals homozygous for the group A KIR haplotype who had 2 of the 3 KIR ligands present on a genetic level and plotted observed frequencies of self- and nonself-KIRs (ie, self and nonself with respect to cognate HLA class I ligands) within CD8 T cells (Figure 5A-B). Numerical combinatorics predicted a greater chance of expressing a self-KIR in the presence of 2 of 3 possible ligands. Indeed, on average, 21% of the expressed KIRs had a self-ligand present, compared with 8% with a nonself-ligand. Hence, the observed distribution of self- and nonself-KIRs on CD8 T cells was almost identical to the theoretical values of KIR expression expected from a random distribution without any evidence for selection imposed by the presence of cognate ligands (Figure 5B). In agreement with our previous findings,16 KIR expression on NK cells in these 24 individuals was also random without any influence by cognate HLA class I molecules (Figure 5C). Hence, despite the differing KIR repertoires of NK cells and CD8 T cells (Figure 3F-H), both cell types had random distributions of self- and nonself-KIRs (Figure 5B-C). Accordingly, the data suggest that the restricted usage of 1 dominant KIR by CD8 T cells is independent of cognate HLA class I molecules.
KIR expression on CD8 T cells is independent of self-HLA class I ligands. (A) Staining for KIR2DL1, KIR2DL3, and KIR3DL1 on CD45RA+CD57+ CD8 T cells in 3 representative individuals with known HLA genotypes where C1 is the ligand of KIR2DL3, C2 is the ligand of KIR2DL1, and Bw4 is the ligand of KIR3DL1. Numbers in the figures represent frequency of indicated KIR out of total CD45RA+CD57+ CD8 T cells. (B-C) Observed frequencies of CD45RA+CD57+ CD8 T cells (B) and NK cells (C) expressing self-and nonself-KIRs compared with theoretical values of random KIR expression analyzed for 24 individuals with 2 KIR ligands present (n.s. indicates not significant; Wilcoxon matched pairs test; mean).
KIR expression on CD8 T cells is independent of self-HLA class I ligands. (A) Staining for KIR2DL1, KIR2DL3, and KIR3DL1 on CD45RA+CD57+ CD8 T cells in 3 representative individuals with known HLA genotypes where C1 is the ligand of KIR2DL3, C2 is the ligand of KIR2DL1, and Bw4 is the ligand of KIR3DL1. Numbers in the figures represent frequency of indicated KIR out of total CD45RA+CD57+ CD8 T cells. (B-C) Observed frequencies of CD45RA+CD57+ CD8 T cells (B) and NK cells (C) expressing self-and nonself-KIRs compared with theoretical values of random KIR expression analyzed for 24 individuals with 2 KIR ligands present (n.s. indicates not significant; Wilcoxon matched pairs test; mean).
KIR promoter activity in CD8 T cells and NK cells
Thus far, we have excluded differences in selection inferred by KIR ligands as a potential cause for the different usage of KIRs by CD8 T cells and NK cells in the same individual. Next, we sought to investigate whether differences in KIR gene promoter activity contributed to the different expression of KIRs in NK cells and CD8 T cells. Bidirectional proximal KIR promoters were shown to be important in controlling KIR expression through the production of 28-base PIWI-like RNA processed from antisense transcripts that mediate epigenetic silencing of KIRs.31 To test whether bidirectional proximal promoter activity differs in NK and T cells, we analyzed the forward and reverse transcriptions from the proximal promoters of KIR2DL1, KIR2DL2, KIR3DL1*001, KIR3DL1*002, KIR3DS1, KIR3DL3, and KIR3DP1 in NK cell– and T cell–derived cell lines with the use of luciferase assays. We have shown previously that the ratios of forward versus reverse promoter activities correlates with KIR expression in NK cells.29 We observed nearly identical promoter ratios in the NK-cell and T-cell lines studied (Figure 6A).
Proximal KIR promoter activities in CD8 T cells and NK cells. (A) Relative ratios between sense (coding) and antisense transcripts from bidirectional proximal KIR promoters are depicted for YT-Indy (NK cell–derived) and Jurkat (T cell–derived) cells transfected with vectors containing proximal promoter regions from the indicated KIRs together with luciferase. Mean ± SD of 5 experiments are shown. (B) Sense (coding) and antisense transcripts generated from the bidirectional KIR3DL1 proximal promoter are depicted as relative expression ratio for total NK cells and CD8 T cells as well as for KIR3DL1+ and KIR3DL− NK cells and CD8 T cells. One representative individual of 2 investigated is shown.
Proximal KIR promoter activities in CD8 T cells and NK cells. (A) Relative ratios between sense (coding) and antisense transcripts from bidirectional proximal KIR promoters are depicted for YT-Indy (NK cell–derived) and Jurkat (T cell–derived) cells transfected with vectors containing proximal promoter regions from the indicated KIRs together with luciferase. Mean ± SD of 5 experiments are shown. (B) Sense (coding) and antisense transcripts generated from the bidirectional KIR3DL1 proximal promoter are depicted as relative expression ratio for total NK cells and CD8 T cells as well as for KIR3DL1+ and KIR3DL− NK cells and CD8 T cells. One representative individual of 2 investigated is shown.
To compare proximal sense and antisense KIR transcript levels in primary cells, we sorted KIR3DL1+ and KIR3DL1− CD8 T cells and corresponding NK cells and then performed quantitative RT-PCR, focusing on proximal KIR3DL1 promoter activity. It was observed that KIR3DL1 promoter-coding transcripts were expressed at similar levels in KIR3DL1+ CD8 T cells and NK cells, whereas no such activity was detected in KIR3DL1− cells (Figure 6B). To the contrary, only KIR3DL1− CD8 T cells and NK cells expressed KIR3DL1 promoter antisense transcripts (Figure 6B). The fact that CD8 T cells and NK cells manifested no detectable differences in proximal KIR3DL1 promoter activity indicates that the distinct KIR patterns in these 2 cell types are not caused by differences in the regulation of proximal gene promoters.
KIR+ CD8 T cells are not educated by interactions with cognate HLA class I molecules
Numerous previous studies have shown that ligation of inhibitory KIRs by their cognate ligands abrogates proximal TCR-mediated signaling, leading to poor effector responses of both CD4 and CD8 T cells.22,25,34-36 In NK cells, the strength of the inhibitory interaction between KIRs and their cognate HLA class I molecules tunes the functional responsiveness of the cell during their development in an education process termed licensing.18 However, in mice, intraepithelial T cells were not educated by interactions with self-MHC class I molecules, suggesting that such education is an NK cell–specific process.37
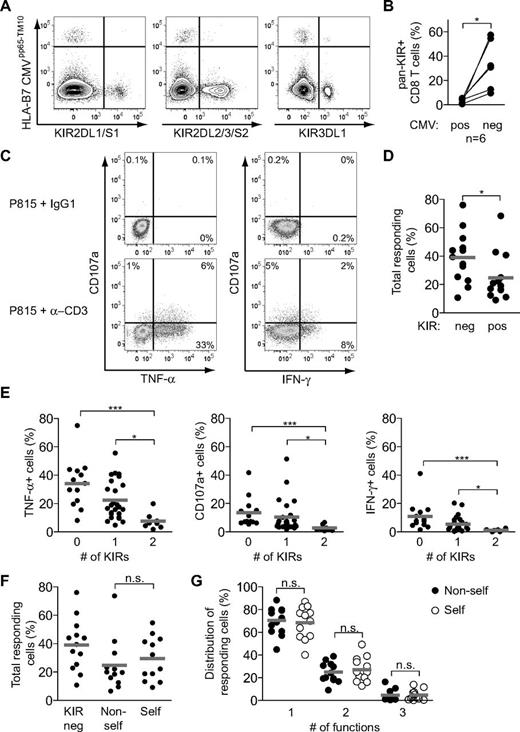
Because our studies suggested a random distribution of self- and nonself-KIRs on CD8 T cells, we set out to determine whether the specificity of the expressed KIRs had any functional consequences on CD8 T cells in humans. Ideally, such experiments should be performed by physiologic stimulation of the TCR with MHC/peptide complexes. However, in line with previous studies,20,22,25,36 we were unable to determine the natural specificity of KIR-expressing CD8 T cells. In fact, as an example, KIR expression was virtually absent on terminally differentiated CMV-specific CD8 T cells (Figure 7A-B). Therefore, overall functional responses of CD8 T cells were assessed by anti-CD3 stimulation (Figure 7C). Controlling for the stage of T-cell differentiation, KIR+ CD8 T cells responded less well than terminally differentiated KIR− CD8 T cells (Figure 7D). Strikingly, the intrinsic responsiveness to anti-CD3 stimulation correlated inversely with the number of expressed KIRs at the cell surface. Hence, CD8 T cells expressing 2 inhibitory KIRs were almost unresponsive to stimulation (Figure 7E). Finally, the hyporesponsiveness of KIR+ CD8 T cells was independent of the HLA class I ligands of the donor (Figure 7F-G), indicating that a KIR-dependent educational process (ie, licensing) probably does not take place in human CD8 T cells.
KIR+ CD8 T cells are not educated by cognate HLA class I ligands. (A) Staining for KIR2DL1/S1, KIR2DL2/3/S2, KIR3DL1, and tetramer-defined CMV-specific cells within CD45RA+CD57+ CD8 T cells in 1 CMV-seropositive healthy individual. (B) Frequency of pan-KIR+ cells within CMV tetramer–positive and –negative CD45RA+CD57+ CD8 T cells summarized for 6 CMV seropositive healthy individuals (*P < .05, paired t test). (C) Representative stainings for CD107a, IFN-γ, and TNF-α on CD45RA+CD57+ CD8 T cells from 1 healthy individual after the indicated stimulations for 6 hours. (D) Frequency of total responding (TNF-α, CD107a, or IFN-γ) cells within KIR− and KIR+ CD45RA+CD57+ CD8 T cells (n = 13; *P < .05, Wilcoxon matched pairs test; mean). (E) Frequency of cells responding with TNF-α, CD107a, and IFN-γ within KIR−, single-KIR+ or double-KIR+ CD45RA+CD57+ CD8 T cells (n = 13, 25, and 8, respectively; ***P < .01, *P < .05, Kruskal-Wallis test with Dunn posttest; mean). (F) Frequency of total responding cells in KIR− and single KIR+ cells with or without a self-ligand present in the host for the respective KIRs (n = 13, 12, and 12, respectively; n.s. indicates not significant, Wilcoxon matched pairs test; mean). (G) Distribution of responding cells into single, double, and triple function for self and nonself single KIR+ cells (n = 12; n.s. indicates not significant, Wilcoxon matched pairs test; mean). (C-G) KIR2DL1 and KIR2DL3 were analyzed.
KIR+ CD8 T cells are not educated by cognate HLA class I ligands. (A) Staining for KIR2DL1/S1, KIR2DL2/3/S2, KIR3DL1, and tetramer-defined CMV-specific cells within CD45RA+CD57+ CD8 T cells in 1 CMV-seropositive healthy individual. (B) Frequency of pan-KIR+ cells within CMV tetramer–positive and –negative CD45RA+CD57+ CD8 T cells summarized for 6 CMV seropositive healthy individuals (*P < .05, paired t test). (C) Representative stainings for CD107a, IFN-γ, and TNF-α on CD45RA+CD57+ CD8 T cells from 1 healthy individual after the indicated stimulations for 6 hours. (D) Frequency of total responding (TNF-α, CD107a, or IFN-γ) cells within KIR− and KIR+ CD45RA+CD57+ CD8 T cells (n = 13; *P < .05, Wilcoxon matched pairs test; mean). (E) Frequency of cells responding with TNF-α, CD107a, and IFN-γ within KIR−, single-KIR+ or double-KIR+ CD45RA+CD57+ CD8 T cells (n = 13, 25, and 8, respectively; ***P < .01, *P < .05, Kruskal-Wallis test with Dunn posttest; mean). (F) Frequency of total responding cells in KIR− and single KIR+ cells with or without a self-ligand present in the host for the respective KIRs (n = 13, 12, and 12, respectively; n.s. indicates not significant, Wilcoxon matched pairs test; mean). (G) Distribution of responding cells into single, double, and triple function for self and nonself single KIR+ cells (n = 12; n.s. indicates not significant, Wilcoxon matched pairs test; mean). (C-G) KIR2DL1 and KIR2DL3 were analyzed.
Discussion
Here, we examined the composition of KIR expression on human CD8 T cells. Terminally differentiated CD8 T cells had a restricted KIR repertoire dominated by a single KIR with a specificity often distinct from that of NK cells within the same individual and expressed independently of self-HLA class I molecules. CD8 T cells also often expressed activating KIRs in the absence of self-specific inhibitory KIRs. Further understanding of KIR repertoire formation in human NK and T cells is important and may provide insights into the cellular mechanisms behind genetic disease association studies in which interactions between KIR and HLA class I molecules have been implicated.
We showed further that a substantial fraction (on average 30%) of terminally differentiated CD8 T cells express KIRs and that the repertoire in most individuals is restricted to expression of a single dominant KIR. By examining the TCR Vβ chain diversity in subsets of CD8 T cells that express distinct KIRs, we concluded that the largest KIR-expressing CD8 T-cell subset displayed the narrowest TCR repertoire. Hence, although acquisition of KIRs is a late event during T-cell differentiation, these results indicate that KIR-expressing T cells are not completely exhausted but may continue to proliferate. Previous work that used mice transgenic for KIR2DL3 and its cognate HLA class I ligand suggested that engagement of inhibitory KIRs on CD8 T cells drives an accumulation of KIR+ memory CD8 T cells because of reduced levels of activation-induced cell death (AICD) within this subset.38 Similar results have been obtained from studies with human KIR+ T-cell clones in which KIR+ CD8 T cells expressed higher levels of the antiapoptotic molecule Bcl-2 in vitro and were relatively resistant to AICD.20 In the present study, we compared observed distributions of self- and nonself-KIR+ CD8 T cells with theoretical distributions of random KIR repertoires and showed that the ex vivo KIR repertoires in CD8 T cells were generated in the absence of selection by cognate HLA class I ligands. Hence, if protection from AICD is a major mechanism for the gradual appearance of KIR expression during late stages of T-cell differentiation, this process seems to be independent of cognate ligands. Indeed, transfection of KIRs into Jurkat T cells led to constitutive and HLA-independent interference with protein kinase C activation and Fas-ligand up-regulation after T-cell activation.39 In line with such constitutive inhibition of proximal T-cell activation, our data indicated that expression of KIRs in terminally differentiated CD8 T cells abrogated several functional modalities regardless of whether the donor expressed the cognate KIR ligands. Moreover, this effect was dose dependent, and cells with 2 KIRs were completely unresponsive to stimulation through the TCR. Additional support for this notion comes from studies of KIR+ CD8 T cells in patients with chronic HIV-1 infection, whereby inhibitory KIRs affected TCR-mediated stimulation of CD8 T cells in a ligand-independent fashion.40 Thus, it is possible that both self- as well as nonself-specific KIRs may rescue CD8 T cells from AICD, thereby contributing to the gradual accumulation of KIRs during CD8 T-cell differentiation. Furthermore, the general hyporesponsiveness of human CD8 T cells that express self-specific KIRs supports the finding in mice that, unlike NK cells, CD8 T cells are not functionally educated by interactions with self-MHC class I molecules.18,37
The nearly monoclonal TCR Vβ chain distribution on KIR+ CD8 T cells could indicate that KIR repertoires are shaped when CD8 T cells undergo clonal expansion in response to pathogenic challenges or to autoantigens. Previous studies have attempted to delineate the TCR specificity of KIR+ CD8 T cells. At steady state, in healthy individuals, few KIR+ CD8 T cells have a TCR directed against CMV or EBV peptides, despite the shared phenotypic features of terminal differentiation between KIR+ CD8 T cells and, especially, CMV-specific CD8 T cells.20,25,41 Although heterogeneous expression of KIRs was noted on Nef-specific CD8 T cells in 4 patients chronically infected with HIV-1,40 neither CMV-specific nor HIV-1–specific CD8 T cells during acute phases of infection as well as HCV-specific CD8 T cells from patients with chronic infection, all of which display a more immature T-cell differentiation phenotype, show signs of KIR expression.22,36,41 Thus, from these studies, performed at steady state in healthy individuals as well as during acute and chronic phases of viral infection, there is only limited information on the TCR specificity of KIR-expressing cells.
Interestingly, in addition to viral infections, KIRs have been linked to susceptibility in many autoimmune diseases.1 For example, CD8 T cells from patients with systemic lupus erythematosus expressed higher frequencies of KIR than those from healthy individuals.42 Furthermore, celiac disease has been associated with the appearance of KIR+ intraepithelial CD8 T cells in the gut lumen,43 and rheumatoid arthritis caused an accumulation of KIR+CD28− CD4 T cells.21,23 Because most available antibodies against KIRs display cross-reactivity and recognize both activating and inhibitory receptor forms, the role of activating KIRs in the context of these autoimmune conditions could not be examined in detail on primary cells in those published studies. With the use of an established flow cytometric panel that includes KIR2DS4 and allows discrimination of KIR2DS1 from KIR2DL1, our analysis found that CD8 T cells frequently express activating KIRs. Together, with the finding that CD8 T cells often lack inhibitory KIRs for self-HLA class I molecules, these results fit with a potential role for KIR+ CD8 T cells in causing immune disorders.26 Whether these disease conditions alter the KIR repertoire of CD8 T cells toward the expression of specific KIR combinations in peripheral blood and in affected tissues remains to be determined.
We found that the KIR expression patterns on CD8 T cells often was distinct from that of NK cells in the same individual. One obvious basis for such intraindividual discrepancies in KIR repertoires between the 2 cell types could be that one of them has been subject to an active selection process conferred by interactions with cognate HLA class I. However, we found no support for this possibility. Instead, our data suggest that KIR expression, both on NK cells and CD8 T cells, occurs independently of expressed HLA class I molecules. Another conceivable explanation for intraindividual discrepancies in KIR expression is cell type–specific differences in the transcriptional machinery that regulate KIR expression. Most T cells and NK cells have the potential to support KIR expression.44 Xu et al have shown, using cell lines, that the transcriptional control of KIR2DL2 differs between T cells and NK cells.44 In the present study, we focused on the bidirectional proximal KIR3DL1 promoter and compared activity in KIR3DL1+ and KIR3DL1− CD8 T cells and NK cells. Interestingly, we detected similar patterns of promoter antisense RNA expression, as well as equivalent levels of promoter activity, in KIR3DL1+ and KIR3DL1− CD8 T cells and NK cells. In addition, an in vitro comparison of KIR promoter activities in YT-Indy (NK cell–derived) versus Jurkat (T cell–derived) cells indicated that the ratio of forward to reverse promoter activity is similar, predicting a similar probability of KIR expression in T and NK cells. Taken together, these results suggest that differences in transcriptional regulation are not the underlying mechanism for intraindividual KIR repertoire differences between CD8 T cells and NK cells.
Epigenetic control mechanisms are important for regulating KIR expression in NK cells.45,46 Although NK cells are considered to retain stable KIR repertoires over time,11,47 KIR expression accumulates on CD8 T cells in individuals as they reach older age.48 Explanations for the latter observation include the accumulation of terminally differentiated CD8 T cells,49 as well as progressive KIR gene promoter demethylation in the terminally differentiated CD8 T cells,48 both occurring with increasing age. Thus, it is plausible that differences in the epigenetic control of KIR gene expression between NK cells and CD8 T cells, with a patchy, stochastic, and nonspecific promoter demethylation occurring in CD8 T cells, might have contributed to the intraindividual differences in KIR specificity that we observed in the present study.
In conclusion, we have performed a high-resolution analysis of the KIR repertoire of CD8 T cells and found that these cells have a highly restricted KIR repertoire, often dominated by a single inhibitory KIR, and formed independently of selection by cognate HLA class I molecules. Our findings challenge the notion that inhibitory self-KIRs are involved in fine-tuning of functional responses and being essential for survival of CD8 T cells during clonal expansion. Further studies of complete KIR repertoires of both NK cells and CD8 T cells should clarify factors underlying the relation of KIRs and HLA class I genes to disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the Swedish Research Council, the Swedish Cancer Society, the Royal Swedish Academy of Sciences, the Tobias Foundation, the Wenner-Gren Foundation, and the Karolinska Institutet.
Authorship
Contribution: N.K.B. designed and performed research, analyzed data, and wrote the paper; V.B., F.C., L.L.L., J.L., R.A.K., and S.K.A. designed and/or performed research and analyzed data; S.L. coordinated collection of clinical material; H.-G.L. analyzed data and contributed to writing the paper; and K.-J.M. designed research, analyzed data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Niklas K. Björkström, Center for Infectious Medicine, F59, Department of Medicine, Karolinska Institutet, Karolinska University Hospital Huddinge, S-141 86 Stockholm, Sweden; e-mail: niklas.bjorkstrom@ki.se; and Karl-Johan Malmberg, Center for Infectious Medicine, F59, Department of Medicine, Karolinska Institutet, Karolinska University Hospital Huddinge, S-141 86 Stockholm, Sweden; e-mail: kalle.malmberg@ki.se.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal