Abstract
Endothelial cells and macrophages are known to engage in tight and specific interactions that contribute to the modulation of vascular function. Here we show that adult endothelial cells provide critical signals for the selective growth and differentiation of macrophages from several hematopoietic progenitors. The process features the formation of well-organized colonies that exhibit progressive differentiation from the center to the periphery and toward an M2-like phenotype, characterized by enhanced expression of Tie2 and CD206/Mrc1. These colonies are long-lived depending on the contact with the endothelium; removal of the endothelial monolayer results in rapid colony dissolution. We further found that Csf1 produced by the endothelium is critical for the expansion of the macrophage colonies and that blockade of Csf1 receptor impairs colony growth. Functional analyses indicate that these macrophages are capable of accelerating angiogenesis, promoting tumor growth, and effectively engaging in tight associations with endothelial cells in vivo. These findings uncover a critical role of endothelial cells in the induction of macrophage differentiation and their ability to promote further polarization toward a proangiogenic phenotype. This work also highlights some of the molecules underlying the M2-like differentiation, a process that is relevant to the progression of both developmental and pathologic angiogenesis.
Introduction
The link between the hematopoietic and the endothelial cell lineages is rooted early in development. In fact, definitive hematopoietic stem cells (HSCs) first emerge in the embryo from a specialized endothelial intermediate that holds hemogenic capacity.1-4 Although the process of hematopoietic cells (HCs) budding from hemogenic endothelium is no longer present in the adult, the interactions between HCs and the endothelium continue to be critical for the trafficking and homing of HCs, as well as for activation and recruitment of inflammatory cells to specific tissue sites.5
More recently, sinusoidal endothelial cells were shown to be essential for the self-renewal capacity of hematopoietic stem/progenitor cells (HSPCs) through the production of specific angiocrine factors.6,7 Intriguingly, bone marrow sinusoidal endothelial cells can also constitute a platform for the differentiation of HSPCs. This dual role of endothelial cells has been best exemplified by findings communicated by Kobayashi and colleagues, where the coculture of genetically modified human umbilical vein endothelial cells (HUVECs) with HSPCs supported both self-renewal and lineage-specific differentiation of HSPCs.8 Notably, the mechanisms by which endothelial cells mediate regeneration or differentiation of HCs depend largely on organ-specific determinants. Overall, mounting evidence supports the concept that the crosstalk between endothelial cells and HCs impacts the differentiation and stem cell properties of hematopoietic progenitors.
The consequences of endothelial-hematopoietic cell interactions are not unidirectional toward the latter; endothelial cells have also shown to benefit. In fact, macrophages have been demonstrated to associate tightly with capillaries and aid in the progression of angiogenesis. Specifically, during development, tissue-resident macrophages facilitate vascular morphogenesis by bridging the neighboring tip cells and mediating anastomosis of adjacent capillaries.9-11 In pathologic situations, such as carcinogenesis, Tie2-expressing macrophages (TEMs) are actively involved in promoting tumor neovascularization. Selective depletion of TEMs significantly impairs angiogenesis and tumor growth.12,13
To further dissect the impact of the crosstalk between adult endothelial cells and HCs, we established a long-term coculture system. Here we show that adult endothelial cells of diverse origins provide critical niches for the selective growth and differentiation of macrophages from hematopoietic progenitor cells. This process involves the formation of colonies that exhibit progressive differentiation toward an M2-like phenotype. The formation and maintenance of these colonies require direct contact with endothelial cells. Overall, the findings provide novel insights into the broad impact of the endothelium on HCs and further define the interactions that are critical for angiogenesis in both physiologic and patholo-gic settings.
Methods
Mice
DsRed mice, B6.Cg-Tg(CAG-DsRed*MST)1Nagy/J, were purchased from The Jackson Laboratory. Immortalized mouse endothelial cells (IMECs) were isolated from Immortomice, CBA;B10-Tg(H2Kb-tsA58)6Kio/Crl, that were purchased from Charles River. Animal protocols were conducted in accordance with University of California, Los Angeles (UCLA) Department of Laboratory Animal Medicine's Animal Research Committee guidelines.
Isolation and purification of IMECs
Mice were perfused with PBS, followed by 500 μg/mL collagenase (c0130; Sigma-Aldrich). Liver, lung, and adipose tissue were homogenized and incubated with collagenase at room temperature for 30 minutes. Samples were mixed with MCDB-131 medium (VEC Tech) containing 20% FBS (FB-11; Omega Sci), centrifuged at 200 rcf for 5 minutes, and then resuspended in MCDB-131 medium with 20% FBS. The suspension was passed through a 40-μm filter (352340; BD Biosciences), and plated onto gelatin-coated culture dishes. After 2 hours, cells were washed extensively to remove nonadherent cells, then cultured at 33°C (for induction of the oncogene). For purification, confluent endothelial cells were washed with cold DMEM (10-017-CV; Cellgro) twice then incubated with rat anti–mouse CD31 (553370; BD Biosciences) in DMEM for 15 minutes under agitation at room temperature. Cells were washed twice with cold DMEM and incubated with anti–rat-IgG magnetic beads (Invitrogen) for 15 minutes at room temperature, followed by washes, trypsinization, and magnetic purification. After several washes, purified endothelial cells were resuspended in DMEM with 10% FBS and 20 U/mL IFNgamma and cultured at 33°C. Characterization of the IMECs confirmed the expression of Vegfr2 and VE-Cadherin and their functional activation by Vegfa (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Purity of the IMECs was evaluated by the expression of CD31 via flow cytometry (supplemental Figure 1B).
Isolation of bone marrow cells
Bone marrow cells were isolated from DsRed mice. Briefly, tibia and femur were obtained then bone marrow was flushed using DPBS (21-031-CV; Cellgro). Red blood cells were lysed. Cell suspension was passed through a 40-μm filter (BD Biosciences, 352340) and seeded on the top of the IMEC monolayer. Cells were cultured in HSC media (α-MEM; 12 000-022; Gibco) with 20% fetal bovine serum (SH30070.03E; Hyclone), penicillin-streptomycin (100 U/mL; Cellgro), β-mercaptoethanol (55μM; Gibco), murine SCF (40 ng/mL), Flt3-L (20 ng/mL), IL-6 (10 ng/mL), IL-3 (10 ng/mL), thrombopoietin (TPO; 10 ng/mL), and vascular endothelial growth factor A (VEGFA; 10 ng/mL). All cytokines were purchased from PeproTech. Culture media were changed weekly. Cultures were observed under Axiovert 200M microscope (Carl Zeiss) for phase contrast and fluorescent imaging twice every week at room temperature.
Colony cell harvesting
Cocultures of indicated days were washed twice with DPBS (21-031-CV; Cellgro) then incubated with Versene for 5 minutes. IMECs were separated from colonies by incubating with Versene at 37°C until they detached from the plate as a thin layer. Attached cells were rinsed with DPBS and collected by cell lifter. The purity of colony cells being F4/80+ and Mac1+ was evaluated by flow cytometry, as shown in Figure 1D.
May-Grünwald-Giemsa staining
Cytospin slides of bone marrow and colony cells were prepared by Shandon Cytospin 4 cytocentrifuge (Thermo Electric) following standard procedures. To identify nuclear and cytoplasmic characteristics of each cell, cytospin specimens were stained with 100% May-Grünwald solution (Merck) for 5 minutes, followed by phosphate buffer wash, and then stained with 5% Giemsa solution (Merck) in deionized water for 15 minutes. All staining procedures were performed at room temperature. Staining was analyzed with Olympus BX40 microscope (Olympus).
Flow cytometry and cell sorting
Colony cells were isolated, counted, and incubated with fluorophore-conjugated antibodies at 4°C for 30 minutes, then resuspended in FACS buffer [(HBSS; Cellgro, 21-022-CV) with 2% FBS (Omega Sci; FB-11) and 10mM HEPES (Gibco; 15 630)]. Antibodies used were APC-eFluor780-F4/80 (eBio; 47-4801), eFluor450-Ly6C (eBio; 48-5932), AlexFluor700-MHCII (eBio; 56-5321), APC-Tie2 (Biolegend; 124009), PE-Gr-1 (eBio; 12-5931), PE-Cy7-Sca1 (eBio; 25-5981), FITC-cKit (eBio; 11-1171), APC-lineage cocktail (BD Biosciences; 51-9003632), APC-CD31 (eBio, 17-0311), FITC–Mac-1 (eBio; 11-0112), APC-CD206 (Biolegend; 141707), APC-CD36 (eBio; 17-0361), FITC-iNOS (BD; 610330), and PerCP-eFluor710-CD115 (eBio; 46-1152). DAPI (Invitrogen; D1306) was used as cell viability dye. LSRII Analytic Flow Cytometer (BD Biosciences) was used for acquisition. For progenitor sorting, DsRed bone marrow cells were isolated and immunofluorescently labeled with indicated markers. FACSAriaII High-Speed Cell Sorter (BD Biosciences) was used. Data were analyzed with FlowJo Version 9.5 (TreeStar). Triplicates were analyzed for quantification.
Immunofluorescent staining
For cocultured cells, colony cells were fixed with 2% PFA after removal of the IMEC layer. Immunofluorescent labeling was performed using standard procedures. Antibodies used were anti–mouse Csf1r (eBio; 14-1152), anti–mouse F4/80 (Serotech; MCA497R), anti–mouse CD206 (Santa Cruz Biotechnology; sc-34577), and anti–mouse CD31 (BD Biosciences; 553370). DAPI (Invitrogen; D1306) was used to stain nuclei. Fluorescent microscopy was performed using confocal microscope, LSM710 (Carl Zeiss) at room temperature.
BrdU assay
BrdU (Sigma-Aldrich; B5002) was added to the coculture for 6 hours before cells were fixed with Carnoys fixative. DNA was then denatured with 2M HCl (Sigma-Aldrich) and neutralized with borate buffer (pH 8.5; Sigma-Aldrich). Immunofluorescent labeling was performed using standard procedures with anti–mouse BrdU (Pierce, MA3-071) and anti–mouse F4/80 (Serotech; MCA497R) antibodies. DAPI (Invitrogen; D1306) was used to stain nuclei. Fluorescent microscopy was performed using confocal microscope, LSM710 (Carl Zeiss) at room temperature.
Dil-Ac-LDL assay
Cells were labeled with 10 μg/mL DiI-Ac-LDL (Invitrogen; L3484) for 4 hours at 37°C, then washed with PBS, trypsinized and passed through a 40-μm filter (BD; 352340) to produce single-cell suspension. LSRII Analytic Flow Cytometer (BD Biosciences) was used. APC-CD31 (eBio; 17-0311) was used to exclude IMEC contamination.
Ultrastructural analysis
For transmission electronic microscopic (TEM) analysis, cells were cultured on Thermanox Coverslips (Ted Pella; 26028) and fixed in Karnovsky glutaraldehyde. Ultrathin sections were prepared by UCLA Electron Microscopy Core Facility and examined under transmission electron microscope (JEOL 100CX) at room temperature.
Transwell assay
All transwell assays were performed with HSC media (see “Isolation of bone marrow cells”). IMECs were seeded in the cell culture inserts (BD Biosciences; 353090) and HCs were seeded into the 6-well plate. For direct contact transwell experiment, IMECs were seeded with inserts (BD Biosciences; 353090 and 353091) upside-down, incubated with medium for 2 hours, and then place into a 6-well plate as regular cultures. Cultures were observed under Axiovert 200M microscope (Carl Zeiss) for phase contrast and fluorescent imaging at room temperature.
Real-time PCR
Total RNA was extracted using RNeasy Kit (QIAGEN; 74106). RNA was reverse transcribed to cDNA using SuperScript First-strand Synthesis System (Invitrogen; 18080-051) according to the manufacturer's instructions. Quantitative real-time PCR was performed using SYBR Green system (QIAGEN; 330509) and detected on Opticon2 PCR machine (MJ Research) by 3-stage program parameters provided by the manufacturer. Each sample was tested in triplicates and samples obtained from 2 independent experiments were used. Analysis of relative gene expression data used the 2-ΔΔCT method. Primers used are provided in supplemental Table 1.
GW2580 treatment and annexin V/PI staining
GW2580 (a gift from Dr Lily Wu, UCLA) was dissolved in dimethyl sulfoxide and added to the coculture from day 1 at the concentration of 2μM. For cell-cycle analysis, GW2580 treated colony cells and control were fixed with 75% ethanol (Gold Shield Chem) for 1 hour at 4°C. Cells were then washed twice with PBS. Propidium iodide staining solution (0.5 mL; BD Biosciences, 556463) and 0.1 mg/mL RNase A (Sigma-Aldrich; R4875) were added to the cell pellet and incubated for 1 hour at room temperature. Histogram of PE (for PI detection) was obtained using LSRII Analytic Flow Cytometer (BD Biosciences) and analyzed with FlowJo Version 9.5 (TreeStar). For annexin V/PI staining, colony cells were washed twice with cold PBS and resuspended in FACS Buffer (see “Flow cytometry and cell sorting”) at a concentration of 1 × 106 cells/mL. The solution (100 μL) was added with FITC–annexin V (BD Biosciences; 556419) and PI (BD Biosciences; 556463), and then incubated for 30 minutes at room temperature in the dark and washed before flow cytometric analysis.
Microarray hybridization and data analysis
Total RNA was extracted using RNeasy Mini Kit (Invitrogen). Integrity of the RNA was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies) and purity/concentration was determined by NanoDrop 8000 Spectrophotometer (Thermo Scientific). Agilent mouse 8 × 60 k array hybridizations were performed by UCLA Clinical Microarray Core following standard Agilent Expression Analysis protocol (GSE39660). The acquisition of array image was undertaken by Agilent Scan Control and Feature Extraction 10.7 software. Raw data were analyzed using Partek genomics Suite 6.4. Data were normalized by RMA algorithm. Thresholds for selecting significant genes were set at > = 2-fold and FDR corrected P < .05. Genes met both criteria were considered as significant changes.
Tumor growth and angiogenesis model
RM1 cells (1 × 105) alone or with colony cells (2 × 104) were injected subcutaneously on the flank of C57BL/6 mice. Tumor size was determined by caliper measurements when tumors were palpable from day 6. Tumor volume was calculated by a rational ellipse formula (m1 × m1 × m2 × 0.5236, where m1 is the shorter axis and m2 is the longer axis). Thin sections (∼ 4 μm) were prepared from PFA-fixed paraffin-embedded tumor tissues. To quantify vessels, 3 sections from 2 to 3 tumors of each group were immunostained for CD31 (BD Biosciences; 553370) and scanned by a confocal microscope (Carl Zeiss; LSM710) at room temperature. Three high-power fields from each slide were examined using ImageJ Version 1.46d software (National Institutes of Health; NIH). In all studies, values were expressed as mean ± SEM.
Matrigel assay
Cocultured colony cells (5 × 104) and IMECs (1 × 104) were resuspended in DMEM (Cellgro, 10-017-CV) and mixed with Matrigel (BD Biosciences; 354248) at the ratio of 1:1 by volume. The mixture was injected subcutaneously on the flank of nude mice. As control, DMEM was mixed with Matrigel and injected on the other side of the flank. Animals were killed at day 7 after injection and Matrigel implants were dissected out. The implants were fixed and processed as regular histologic tissue. Matrigel sections were immunostained for anti–MECA-32 (Santa Cruz Biotechnology; sc-19603) and anti-DsRed (MBL; PM005) antibodies and scanned by a confocal microscope (Carl Zeiss; LSM710) at room temperature. 3D rendering of z-stack images were acquired by Volocity Version 5.5 software (Improvision).
Results
Endothelial cells instruct the differentiation of macrophages
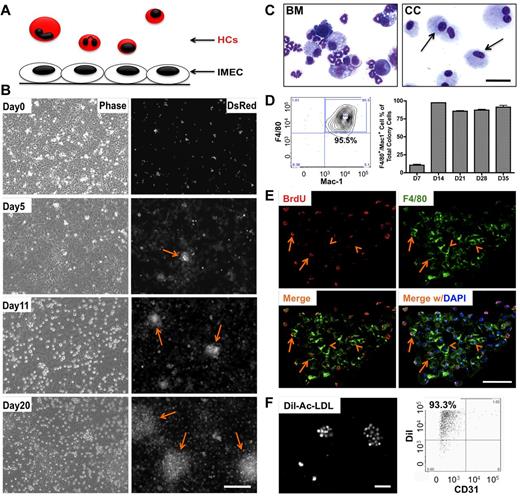
To determine the effect of heterotypic endothelial-hematopoietic cell interactions, we established an in vitro coculture system where these 2 types of cells could engage in direct contact and the impact could be investigated over time at both cellular and molecular level. Liver sinusoidal endothelial cells (IMECs) were used to support the growth of HCs isolated from the bone marrow of DsRed mice (Figure 1A). Upon coculture, a subset of HCs transmigrated across endothelium, took residence under the endothelial monolayer, and formed colonies that were visible under the fluorescent microscope 5 days after coculture. The number and size of the colonies expanded quickly (Figure 1B). Cells comprising the colonies were evaluated by cytospin. These cells were morphologically homogeneous compared with the original mixture of the bone marrow cells (Figure 1C). Further characterization revealed that the large majority of these cells were positive for F4/80 and Mac-1 (Figure 1D), markers for the macrophage lineage. Cells within the colonies were engaged in active proliferation, as shown by BrdU incorporation analysis (Figure 1E). Colony cells were also able to endocytose Dil-Ac-LDL providing further support to the macrophage identity of these cells (Figure 1F). Adult endothelial cells isolated from lung and adipose tissue were also able to induce and support macrophage differentiation (supplemental Figure 1C-D). Emergence of colonies did not occur when bone marrow was cultured alone or in the presence of OP9 stromal cells (supplemental Figure 2A-B).
Endothelial cells induce differentiation and expansion of macrophages in vitro. (A) Schematic representation of the coculture system that includes murine hematopoietic cells (HCs) and immortalized mouse endothelial cells (IMECs). All HCs express DsRed fluorescent protein. (B) Phase contrast (left) and fluorescent (right) micrographs depict the formation of DsRed+ colonies (arrow) during coculture. Note that the colonies were phase-dim or invisible under phase view. Scale bar, 200 μm. (C) Photomicrographs of isolated bone marrow (BM) cells and cells from day 27 colonies (CC, arrow) after May-Grünwald-Giemsa staining. Scale bar, 50 μm. (D) FACS profile (left) indicates that the large majority of cells in the colonies were F4/80+/Mac-1+ (n = 3). (E) Fluorescent micrographs of bromodeoxyuridine (BrdU) staining indicating active proliferation within the colony. Day 10 colonies were pulse-labeled with BrdU for 8 hours and then costained for anti-BrdU (red) and anti-F4/80 (green) antibodies. Arrow, cells positive for both F4/80 and BrdU. Arrowhead indicates F4/80+ cells not stained for BrdU. DAPI indicates staining for nucleus (blue). Scale bar, 50 μm. (F) Colony cells uptake DiI-ac-LDL. Cells were incubated with 10 μg/mL DiI-ac-LDL for 4 hours, followed by fluorescent microscopy (left). (Right) FACS profile shows the majority colony cells take up Dil-ac-LDL. CD31, to exclude possible IMEC contamination. Scale bar, 200 μm.
Endothelial cells induce differentiation and expansion of macrophages in vitro. (A) Schematic representation of the coculture system that includes murine hematopoietic cells (HCs) and immortalized mouse endothelial cells (IMECs). All HCs express DsRed fluorescent protein. (B) Phase contrast (left) and fluorescent (right) micrographs depict the formation of DsRed+ colonies (arrow) during coculture. Note that the colonies were phase-dim or invisible under phase view. Scale bar, 200 μm. (C) Photomicrographs of isolated bone marrow (BM) cells and cells from day 27 colonies (CC, arrow) after May-Grünwald-Giemsa staining. Scale bar, 50 μm. (D) FACS profile (left) indicates that the large majority of cells in the colonies were F4/80+/Mac-1+ (n = 3). (E) Fluorescent micrographs of bromodeoxyuridine (BrdU) staining indicating active proliferation within the colony. Day 10 colonies were pulse-labeled with BrdU for 8 hours and then costained for anti-BrdU (red) and anti-F4/80 (green) antibodies. Arrow, cells positive for both F4/80 and BrdU. Arrowhead indicates F4/80+ cells not stained for BrdU. DAPI indicates staining for nucleus (blue). Scale bar, 50 μm. (F) Colony cells uptake DiI-ac-LDL. Cells were incubated with 10 μg/mL DiI-ac-LDL for 4 hours, followed by fluorescent microscopy (left). (Right) FACS profile shows the majority colony cells take up Dil-ac-LDL. CD31, to exclude possible IMEC contamination. Scale bar, 200 μm.
Endothelial cell contact is essential for the emergence and maintenance of macrophage colonies
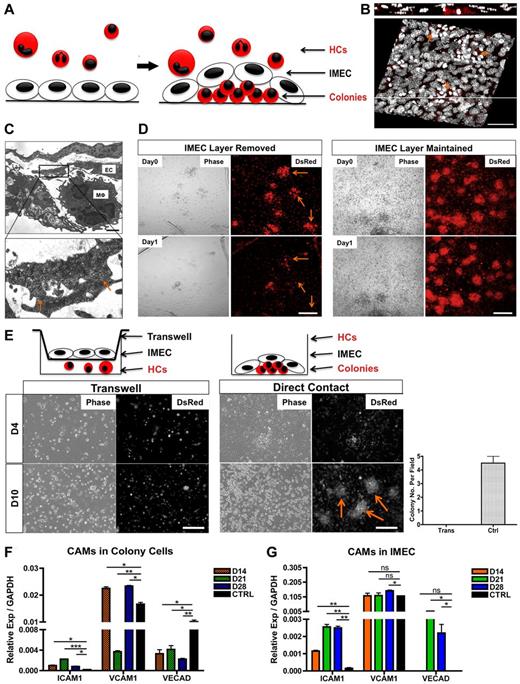
Given the intimate association between the endothelium and the colonies (Figure 2A-B), we next investigated whether the emergence and maintenance of the colonies were dependent on the physical contact with endothelium. Ultrastructural analysis confirmed the physical association between IMECs and colony cells (Figure 2C). Careful removal (peeling) of the endothelial monolayer from cultures containing colonies resulted in quick disaggregation of underlying colonies within 24 hours (Figure 2D). Furthermore, cocultures using transwells, which prevent cell-cell contact but enable exchange of secreted factors, did not support the growth of colonies, revealing that colony formation required direct contact with the endothelium (Figure 2E). Through modification of the transwell assay whereby minimal contact with the endothelium was enabled, we further determined that contact with endothelial cell processes was sufficient to elicit organization of colonies (supplemental Figure 2C). Several cell adhesion molecules were significantly expressed in both cell types. We found that VCAM1 and VE-Cadherin were present in colony macrophages with low but detectable levels of ICAM1 (Figure 2F, supplemental Figure 2D). Interestingly, contact with bone marrow cells induced an up-regulation of ICAM1 and VE-Cadherin in the endothelial monolayer (Figure 2G).
Direct contact with the endothelium is required for the formation and maintenance of macrophage colonies. (A) Schematic representation of the colonies as they take residence under the endothelial monolayer. (B) Confocal 3D reconstruction of the DsRed colonies (arrows) growing underneath the IMEC layer. Composite x-y sections and a single z-section are shown. Scale bar, 50 μm. (C) Ultrastructural analysis of cells cocultured for 28 days by transmission electron microscopy. (Bottom) Magnified micrograph reveals direct contact (arrows) between the 2 cell types. EC indicates endothelial cell; and Mφ, macrophage-like cell. Scale bar, 4 μm. (D) Colonies disaggregate (arrow) after the removal of IMEC layer for 1 day, depicted by phase contrast and fluorescent micrographs (left). Colonies with IMECs were maintained (right). Scale bar, 200 μm. (E) Colonies failed to form via transwells. (Left) Cocultures of HCs and IMECs separated by the transwell (0.4-μm pore size). (Middle) Cocultures through direct contact. Arrows indicate colonies. (Right) Quantification of colony number at day 10 of coculture (n = 3). Scale bar, 200 μm. (F) Real time PCR quantification of ICAM1, VCAM1, and VE-Cadherin in colonies. Ctrl indicates bone marrow–derived macrophages with 5 ng/mL IL-4 stimulated for 24 hours (n = 6). (G) Corresponding cell adhesion molecules (CAMs) expression in IMECs. Ctrl indicates IMECs in the absence of coculture (n = 6; *P < .05; **P < .01; ***P < .001; unpaired Student t test).
Direct contact with the endothelium is required for the formation and maintenance of macrophage colonies. (A) Schematic representation of the colonies as they take residence under the endothelial monolayer. (B) Confocal 3D reconstruction of the DsRed colonies (arrows) growing underneath the IMEC layer. Composite x-y sections and a single z-section are shown. Scale bar, 50 μm. (C) Ultrastructural analysis of cells cocultured for 28 days by transmission electron microscopy. (Bottom) Magnified micrograph reveals direct contact (arrows) between the 2 cell types. EC indicates endothelial cell; and Mφ, macrophage-like cell. Scale bar, 4 μm. (D) Colonies disaggregate (arrow) after the removal of IMEC layer for 1 day, depicted by phase contrast and fluorescent micrographs (left). Colonies with IMECs were maintained (right). Scale bar, 200 μm. (E) Colonies failed to form via transwells. (Left) Cocultures of HCs and IMECs separated by the transwell (0.4-μm pore size). (Middle) Cocultures through direct contact. Arrows indicate colonies. (Right) Quantification of colony number at day 10 of coculture (n = 3). Scale bar, 200 μm. (F) Real time PCR quantification of ICAM1, VCAM1, and VE-Cadherin in colonies. Ctrl indicates bone marrow–derived macrophages with 5 ng/mL IL-4 stimulated for 24 hours (n = 6). (G) Corresponding cell adhesion molecules (CAMs) expression in IMECs. Ctrl indicates IMECs in the absence of coculture (n = 6; *P < .05; **P < .01; ***P < .001; unpaired Student t test).
CSF1 signaling is critical for the expansion but not for the initiation of macrophage colonies
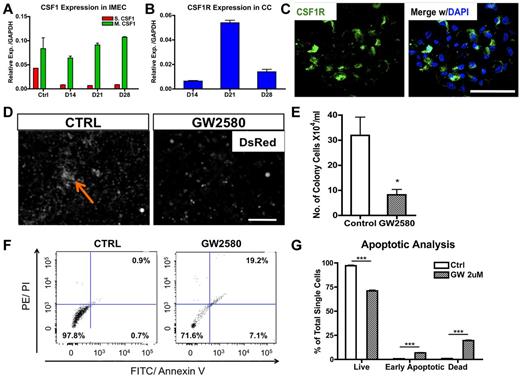
Macrophage colony-stimulating factor (MCSF; or CSF1) is a chemokine shown to stimulate proliferation, differentiation, and survival of macrophages.14 To determine whether it plays a role in endothelial-induced colony formation, we first determined transcriptional levels of both secreted and membrane-bound forms of Csf1 in cocultured IMECs. Interestingly, membrane-bound Csf1, the less dominant isoform, was prevalent in IMECs (Figure 3A). Transcripts for the Csf1 receptor (Csf1r) were highly expressed by colony macrophages with the peak at day 21, time of maximal expansion of the colonies (Figure 3B). Expression of Csf1r in the macrophage colonies was also confirmed by immunostaining (Figure 3C) and flow cytometry (supplemental Figure 3A-B).
CSF1 is essential for the expansion of macrophage colonies. (A) Membrane-bound Csf1 but not secreted Csf1 is prevalently expressed in IMECs by real-time PCR analysis. Ctrl, IMECs in the absence of coculture (n = 6). (B) Csf1 receptor (Csf1r) transcripts are highly expressed in colony cells (n = 6). (C) Immunolocalization of Csf1r (green) on a representative colony from day 10 coculture. DAPI indicates staining for nucleus (blue). Scale bar, 50 μm. (D) GW2580, a CSF1R inhibitor, significantly impairs colony emergence and growth. Coculture was treated with GW2580 (2μM) for 7 days. Ctrl indicates coculture exposed to vehicle. Arrow indicates a colony. Scale bar, 200 μm. (E) Quantification of colony cell number at day 14 of coculture in the presence or absence of GW2580 treatment (n = 3). (F) Representative dot plots showing increased apoptotic activity on GW2580 treatment. Colony cells from day 14 coculture treated with GW2580 (2μM) or vehicle were stained for propidium iodide (PI) and annexin V. (G) Apoptotic analysis of colony cells on GW2580 or vehicle treatment (n = 3; *P < .05; ***P < .001; unpaired Student t test).
CSF1 is essential for the expansion of macrophage colonies. (A) Membrane-bound Csf1 but not secreted Csf1 is prevalently expressed in IMECs by real-time PCR analysis. Ctrl, IMECs in the absence of coculture (n = 6). (B) Csf1 receptor (Csf1r) transcripts are highly expressed in colony cells (n = 6). (C) Immunolocalization of Csf1r (green) on a representative colony from day 10 coculture. DAPI indicates staining for nucleus (blue). Scale bar, 50 μm. (D) GW2580, a CSF1R inhibitor, significantly impairs colony emergence and growth. Coculture was treated with GW2580 (2μM) for 7 days. Ctrl indicates coculture exposed to vehicle. Arrow indicates a colony. Scale bar, 200 μm. (E) Quantification of colony cell number at day 14 of coculture in the presence or absence of GW2580 treatment (n = 3). (F) Representative dot plots showing increased apoptotic activity on GW2580 treatment. Colony cells from day 14 coculture treated with GW2580 (2μM) or vehicle were stained for propidium iodide (PI) and annexin V. (G) Apoptotic analysis of colony cells on GW2580 or vehicle treatment (n = 3; *P < .05; ***P < .001; unpaired Student t test).
To further ascertain the functional contribution of CSF1 in the formation and expansion of the colonies, we applied a CSF1R exclusive inhibitor, GW2580, to the cocultures. GW2580 is a selective small molecule kinase inhibitor that was shown to completely prevent CSF1R-dependent growth of macrophages in vitro and in vivo at therapeutically relevant doses.15 Exposure to GW2580 (2μM) from the onset of the coculture drastically impaired colony formation (Figure 3D-E). Small colonies were present in GW2580–treated culture, but these never expanded. Evaluation with propidium iodide indicated an increase in the early apoptotic and dead cells on treatment (Figure 3F-G) and an alteration in cell cycle (supplemental Figure 3C). Together, these findings demonstrate that the CSF1 pathway is certainly involved in the expansion and the survival of the colonies. However, blockage of CSF1 pathway does not alter the initiation of the macrophage differentiation program, as the percentage of F4/80+ cells was unaltered on the exposure to GW2580 albeit the colony size was much smaller (data not shown).
Macrophages in colonies are polarized toward the M2 subtype and express high levels of VEGFA
Th1 (eg, LPS and IFN-γ) and Th2 (eg, IL-4, IL-13, and IL-10) cytokines can differentially polarize macrophages to M1 (or classically activated) or M2 (or alternatively activated) phenotype, respectively. M1-polarized macrophages produce high levels of IL-1β, IL-12, IL-23, TNFα, and CXCL10, and are markedly proinflammatory and angiostatic, whereas M2 macrophages secrete IL-10, CCL17, and CCL22, which are known to promote tissue repair and angiogenesis.16-18
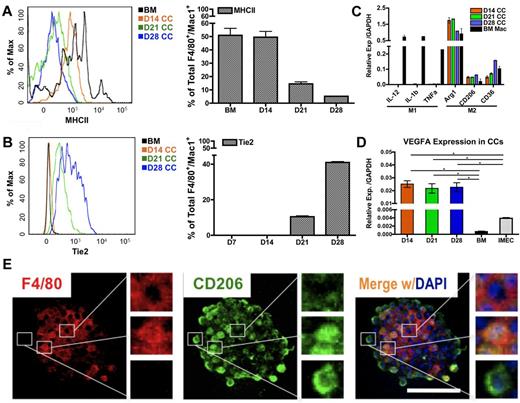
To determine whether the polarization of macrophages occurs in the colonies, we evaluated levels of MHCII, a phenotypic marker for inflammatory/M1–polarized macrophages. Flow cytometric overlay graphs at progressive time points of cocultures showed a continuous reduction of MHCII expression on colony macrophages (Figure 4A) indicating that the colony macrophages were not M1-polarized.
Endothelial cells impart M2 polarity on colony macrophages. (A) MHCII expression in colony cells is reduced on coculture, as determined by FACS overlay graph (left) and quantification (right; n = 3). (B) Tie2 expression in colony cells is gradually increased over time as shown by FACS overlay graph (left) and quantification (right; n = 3). (C) Colony cells exhibit M2 macrophage markers but not M1 markers, as detected by real-time PCR analysis. Ctrl indicates bone marrow–derived macrophages stimulated with 50 ng/mL LPS (for M1 control) or with 5 ng/mL IL-4 (for M2 control) for 24 hours (n = 6). (D) Colony cells express high level of Vegfa transcripts. (E) Immunolocalization of F4/80 (pan macrophage marker, red) and CD206/Mrc1 (M2 macrophage marker, green) in a macrophage colony. Note that cells in the center of the colony are F4/80+ and CD206−; cells on the periphery are F4/80− and CD206+; cells in between are positive for both markers. Colony shown is from day 14 coculture. DAPI indicates staining for nucleus (blue). Scale bar, 50 μm (*P < .05; unpaired Student t test).
Endothelial cells impart M2 polarity on colony macrophages. (A) MHCII expression in colony cells is reduced on coculture, as determined by FACS overlay graph (left) and quantification (right; n = 3). (B) Tie2 expression in colony cells is gradually increased over time as shown by FACS overlay graph (left) and quantification (right; n = 3). (C) Colony cells exhibit M2 macrophage markers but not M1 markers, as detected by real-time PCR analysis. Ctrl indicates bone marrow–derived macrophages stimulated with 50 ng/mL LPS (for M1 control) or with 5 ng/mL IL-4 (for M2 control) for 24 hours (n = 6). (D) Colony cells express high level of Vegfa transcripts. (E) Immunolocalization of F4/80 (pan macrophage marker, red) and CD206/Mrc1 (M2 macrophage marker, green) in a macrophage colony. Note that cells in the center of the colony are F4/80+ and CD206−; cells on the periphery are F4/80− and CD206+; cells in between are positive for both markers. Colony shown is from day 14 coculture. DAPI indicates staining for nucleus (blue). Scale bar, 50 μm (*P < .05; unpaired Student t test).
To evaluate M2-associated markers, we first determined Tie2 levels. Tie2, a tyrosine kinase receptor for angiopoietin, is also considered to be a marker associated with M2-like, proangiogenic macrophages in tumors.12,19 Interestingly, the expression of Tie2 rose significantly in colony macrophages over time (Figure 4B), suggesting that the endothelial-induced macrophages were polarized toward the M2 fate.
Additional transcripts associated with M1 and M2 polarization were measured. We consistently found that colony macrophages expressed high levels of M2 markers, Arg1, CD206/Mrc1, and CD36. In contrast, levels for IL-12, IL-1β, and Tnfα (M1 markers) were not detectable in these macrophages (Figure 4C). Additional phenotypic markers for M1 and M2-associated markers were examined by flow cytometry (supplemental Figure 4A-C). Combined, the data indicate that the colony macrophages were M2 polarized, which is also confirmed by microarray data (GSE39660; supplemental Figure 5A-D). We were also able to detect M2–like macrophages in the supernatant of the cocultures (supplemental Figure 4D-E). We further found that the macrophages expressed high levels of Vegfa (Figure 4D), which may explain why the endothelial monolayer is able to be retained under the coculture conditions for longer periods of time (up to 2 months), compared with in the absence of the coculture (1-2 weeks).
We further confirmed the M2-like polarization of macrophages by immunofluorescence and noticed a hierarchical organization within the colonies. Expression of the pan-macrophage marker F4/80 was concentrated in the center of the colonies; whereas levels of CD206/Mrc1 were gradually increased toward the periphery of the colony (Figure 4E). This indicates that maturation toward an M2-skewed phenotype occurred as the cells migrated toward the edge of the colony.
Endothelial cells induce M2-like macrophage differentiation regardless of the developmental stage of the initiating myeloid progenitor
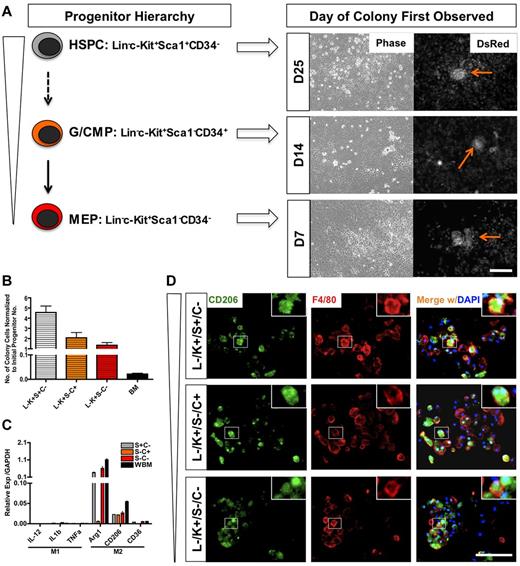
To identify the progenitor cell that could induce the generation of the M2-like macrophages on contact with the endothelium, we sorted HSPCs, G/CMPs (granulocyte-macrophage and common myeloid progenitors), and MEPs (megakaryocyte-erythroid progenitors) from whole bone marrow and proceeded to test their ability to generate colonies on coculture with endothelial cells. These 3 populations were sorted based on lineage markers, c-Kit, Sca1, and CD34 expression (supplemental Figure 6A). We found that all of the progenitor populations were able to generate colonies on coculture, but at different times. Colonies from MEP culture emerged first at day 7, followed by G/CMP at day 14. Cocultures with HSPCs were the last to show emergence of colonies, at day 25 (Figure 5A).
IMECs support macrophage differentiation from hematopoietic progenitors. (A) Schematic representation of hematopoietic progenitors evaluated and the time of emergence of macrophage colonies (right). HSPC indicates hematopoietic stem/progenitor cell; G/CMP, granulocyte-macrophage and common myeloid progenitor; and MEP, megakaryocyte-erythroid progenitor. Arrows indicate colonies. Scale bar, 200 μm. (B) Colony cell number at day 28 of cocultures was normalized to the initial number of progenitor cells plated (n = 6). (C) Colony cells from all progenitors moderately display M2 macrophage markers but lack M1 markers. Ctrl indicates colony cells from the coculture of whole bone marrow (WBM) cells and IMECs (n = 6). (D) Colonies from all progenitors expressed CD206/Mrc1 (M2 macrophage marker, green) and F4/80 (pan macrophage marker, red). Colonies shown are from d14 coculture. DAPI indicates staining for nucleus (blue). Scale bar, 100 μm.
IMECs support macrophage differentiation from hematopoietic progenitors. (A) Schematic representation of hematopoietic progenitors evaluated and the time of emergence of macrophage colonies (right). HSPC indicates hematopoietic stem/progenitor cell; G/CMP, granulocyte-macrophage and common myeloid progenitor; and MEP, megakaryocyte-erythroid progenitor. Arrows indicate colonies. Scale bar, 200 μm. (B) Colony cell number at day 28 of cocultures was normalized to the initial number of progenitor cells plated (n = 6). (C) Colony cells from all progenitors moderately display M2 macrophage markers but lack M1 markers. Ctrl indicates colony cells from the coculture of whole bone marrow (WBM) cells and IMECs (n = 6). (D) Colonies from all progenitors expressed CD206/Mrc1 (M2 macrophage marker, green) and F4/80 (pan macrophage marker, red). Colonies shown are from d14 coculture. DAPI indicates staining for nucleus (blue). Scale bar, 100 μm.
Although unsorted bone marrow gave rise to the highest number of colonies (supplemental Figure 6D), normalization to the original number of seeded cells revealed that HSPCs had the greatest fold of expansion (Figure 5B). The findings imply that on contact with endothelium, HSPCs were able to survive, self-renew, and further differentiate into the M2-like subtype more effectively than other progenitors. These results also showed that the adult endothelium is able to directly program the differentiation of macrophages from any hematopoietic progenitors. Similar to the effect of Csf1r inhibition on whole bone marrow, GW2580 also impaired the expansion of colony cells from all progenitors (data not shown).
Evaluation of transcripts and immunofluorescence for M2-associated markers confirmed that the colony macrophages emerging from the coculture with each progenitor population were also alternatively activated (Figure 5C-D). This suggested that IMECs were able to support M2-like macrophage colony differentiation from all hematopoietic progenitors.
We also demonstrated that the colonies could be promptly induced from CD45+ cells rather than CD45− cells. Surprisingly, Gr-1+/Mac-1+ cells were not able to induce colony formation on coculture, yet the Gr-1− population was able to generate macrophages on exposure to endothelial cells (supplemental Figure 6B-C).
Macrophages from colonies promote tumor growth and angiogenesis
Tumor associated macrophages (TAMs) have been generally considered as M2-polarized.19 These cells have been shown to promote tumor growth and angiogenesis.12,19 To test whether the colony macrophages could function as M2 macrophages in the tumor microenvironment, we coinjected these cells together with RM1 cells (murine prostate cancer cells) into wild type C57BL/6 mice. Resulting subcutaneous tumors were palpable by 6 days after injection and reached terminal point around day 12.
Tumors injected with colony macrophages exhibited faster growth kinetics compared with the control group. Although tumors from both groups were similar by day 7, the tumors coinjected with macrophages showed higher tumor volume at subsequent time points (Figure 6A). The proangiogenic function of colony macrophages in the tumors was also evaluated. CD31 staining revealed greater vascular density in the tumors that were injected with colony macrophages (Figure 6B-C). Quantification of the vascular area and vessel numbers also supported the proangiogenic role of these macrophages in vivo (Figure 6D-E).
Macrophages derived from coculture promote tumor growth and angiogenesis. (A) Colony cells promote tumor growth in wild-type C57BL/6 mice. Mice were injected with RM1 tumor cells (1 × 105) alone or with colony cells (1 × 104; n = 3∼5). (B) Representative pictures of tumor sections stained for CD31 (red). Tumors that were co-injected with colony cells showed a higher vascular density. Arrowhead indicates representative vessel. DAPI indicates staining for nuclei (blue). Scale bar, 100 μm. (C) Quantification (percentage) of vascular area in tumor sections shown in (B; n = 3). (D) Quantification of vessel area in tumor sections from indicated days (n = 3). (E) Quantification of vessel number in tumor sections from indicated days (n = 3). (F) Immunolabeling (i,ii) and 3D surface rendering (iii,iv) of colony cells (DsRed) and vessels (MECA, green) after colony cells and IMECs were injected with matrigel for 7 days. Colony cell (arrowhead) is bridging 2 endothelial cells. Scale bar, 50 μm. (G) Model of IMEC-induced M2 macrophage differentiation (*P < .05; **P < .01; unpaired Student t test).
Macrophages derived from coculture promote tumor growth and angiogenesis. (A) Colony cells promote tumor growth in wild-type C57BL/6 mice. Mice were injected with RM1 tumor cells (1 × 105) alone or with colony cells (1 × 104; n = 3∼5). (B) Representative pictures of tumor sections stained for CD31 (red). Tumors that were co-injected with colony cells showed a higher vascular density. Arrowhead indicates representative vessel. DAPI indicates staining for nuclei (blue). Scale bar, 100 μm. (C) Quantification (percentage) of vascular area in tumor sections shown in (B; n = 3). (D) Quantification of vessel area in tumor sections from indicated days (n = 3). (E) Quantification of vessel number in tumor sections from indicated days (n = 3). (F) Immunolabeling (i,ii) and 3D surface rendering (iii,iv) of colony cells (DsRed) and vessels (MECA, green) after colony cells and IMECs were injected with matrigel for 7 days. Colony cell (arrowhead) is bridging 2 endothelial cells. Scale bar, 50 μm. (G) Model of IMEC-induced M2 macrophage differentiation (*P < .05; **P < .01; unpaired Student t test).
The association of colony macrophages with capillaries was clearly noted in matrigel assays. Macrophages isolated from the cocultures were coinjected with IMECs in matrigel plugs. After 7 days, vascular networks were found highly decorated with macrophages (Figure 6F) simulating the intimate interaction between macrophages and blood vessels described in vivo.20
Discussion
In the last decade, interactions between endothelial cells and macrophages have been characterized in detail and their biologic consequences have started to be understood. Yet, as we gain further appreciation of the physical and functional relationships between macrophages and endothelial cells, additional questions arise. In particular: How do these interactions take place and what are the relevant molecular players? How and when does macrophage polarization occur? And does proliferation/expansion of macrophages take place in situ or does it require individual cell recruitment and differentiation? The work presented here demonstrates that endothelial cells can provide a selective niche for the differentiation and functional polarization of M2-like macrophages. We found that several types of adult endothelial cells shared this capacity and that endothelial-mediated macrophage differentiation could be triggered in hematopoietic progenitors at several stages of developmental progression. The growth-mediated properties imparted by endothelial cells were greatly dependent on their ability to activate CSF1R through production of CSF1. In turn, colony macrophages increased the stability of endothelial cell monolayers and provided a local source of VEGFA. We also found that macrophage colonies were M2-polarized and that the colonies exhibited a well-organized structure with poorly differentiated cells in the center and M2-polarized cells in the periphery (Figure 6G). More importantly, these endothelial-induced macrophages were able to associate with blood vessels and promote angiogenesis and tumor growth similarly to M2-like macrophages in vivo. These findings imply that the differentiation of macrophages, particularly M2-polarized cells such as TEMs, might be mediated by cell-cell interactions with the endothelium as these cells extravasate and gain residence in the vessel wall. Such instructive role of the vascular niche in promoting macrophage differentiation toward an M2-like, proangiogenic phenotype may well explain the reported association of Tie2+CD206/Mrc1+ TEMs with tumors blood vessels.12,19
The recent realization that the diversity of macrophage phenotypes also impacts their interaction and functional relationship with the endothelium has resolved some ongoing controversies.21 In particular, there is increasing evidence that M1-like macrophages have angiostatic properties and do not necessarily associate with vessels. In contrast, M2-like macrophages tightly associate with endothelial cells and promote angiogenesis.20 In this context, our findings that endothelial cells could effectively induce macrophage differentiation and M2-like polarization in vitro is consistent with a productive and symbiotic relationship between these 2 cell types. The present studies now raise the question as to whether endothelial contact is largely (or solely) responsible for the induction of the M2 phenotype, whereas the M1 phenotype might be induced by different cells or through distinct mechanisms. Furthermore, if vessel-associated M2 macrophages continuously promote angiogenesis, what is the counteracting factor that inhibits the abnormal angiogenic response? In addition, it is unknown, at present, whether these macrophages are constitutively associated with the vasculature or only recruited by the endothelium under pathologic settings. If constitutively associated, are they inactive in their proangiogenic capacity during normal conditions?
Our understanding of macrophage heterogeneity is only rudimentary. The topic has been on the spotlight because of the widely diverse and important effects that each different subset of macrophages has on autoimmune disorders, microbial infections, and cancer.22-24 The initial inflammatory response is typically conducted by classically activated (M1-like) macrophages.23 In contrast, the resolution phase is driven by alternatively activated (M2-like) macrophages. The latter are known to be hyporesponsive to inflammatory stimuli, to drive tissue remodeling, promote wound healing, and accelerate angiogenesis.20 Previous studies have shown that M2 polarization can be induced by specific cytokines and exposure of adherent macrophages to IL4.25-27 Our findings indicate that direct contact with the endothelium was necessary and sufficient for the induction of macrophage colonies and M2-like polarization; therefore it is unlikely that in this setting secreted factors alone would be sufficient. Instead, our findings demonstrate that endothelial cell contact is essential to initiate, support, and maintain M2-like macrophage colonies. Nonetheless, the identity of the cell adhesion molecules that drive the process is still unclear and subject to current investigations.
Cell-surface interactions between endothelial and HCs play essential roles in immune function and homeostasis.5 In fact, heterotypic molecular activation plays a pivotal role in hematopoietic homing and in the inflammatory response. More recently endothelial cells have been proposed to constitute the bone marrow niche and support stem cell maintenance; as well as to regulate their differentiation.6-8,28,29 Although soluble factors have been shown to promote HSCs, a rapid attrition is shown to occur in cell-free cultures.30-35 On the other hand, the direct interaction of HSCs with stroma, and in particularly with endothelial cells has shown to be necessary to support long-term self-renewal and maintenance of these hematopoietic progenitors,36-39 and more recently of HSCs.7 Consistent with the latter, our data provided solid evidence to support that endothelial cells were able to capture, retain, and regulate the differentiation of macrophages through direct cell contact.
Our findings also indicate that in addition to cell contact, CSF1 is required for the expansion of the macrophage colonies. CSF1 is a key cytokine to support the differentiation of expansion of monocyte lineages. Deletion of Csf1 in mice (op/op mutant) greatly reduces the number of monocytes40 ; interestingly, macrophages are still present in various organs of op/op mice, such as liver, lung, and brain.41 This has provoked the speculation as to the presence of alternative factors that could partially compensate for the crucial role of Csf1 in macrophage formation. In our study, the administration of GW2580, a small molecule that blocks CSF1R function, significantly reduced the expansion of macrophage colonies but did not inhibit their formation. Together the findings indicate that CSF1 is necessary for robust proliferation of macrophages but it does not appear to affect the process of differentiation.
The question as to whether macrophages proliferate while at the tissue of residence or are terminally differentiated without proliferative capacity has been subject to discussion. Several studies have provided support to the concept that tissue-resident macrophages proliferate locally.42-44 In particular, a recent paper demonstrated that in response to IL-4, macrophages can replicate in situ in the absence of recruitment from the hematopoietic pool.44 Interestingly, this is the same cytokine known to promote polarization toward the M2-like phenotype. Along these lines, the coculture system described here shows that endothelial-induced M2-like macrophages had proliferative capacity.
Finally, the actual source of macrophages is also an interesting puzzle. Although macrophages are believed to differentiate directly from monocytes during (or shortly after) extravasation from the blood stream, macrophages do exist in the embryo before the production of monocytes in the fetal liver.45 This opens the argument: What is the origin of these macrophages? Can other progenitors give rise to macrophages? Is this only developmental or can it occur in adult settings? And can macrophage differentiation be “programmed” by the surrounding cells, particularly endothelial cells, on angiogenic sites? In this study, we have shown that in fact, monocytes (Gr-1+/Mac-1+) are not largely responsible for the formation of M2-polarized macrophage colonies; but rather, other progenitors at prior stages of differentiation gave rise to macrophage colonies when in contact with endothelium.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Howard Lo and Ryan Freshman for their technical assistance, Calvin Pan for his bioinformatic assistance, the Tissue Procurement Core Laboratory Shared Resource, the Electron Microscopy Core Facility, the FACSCalibur Flow Cytometric Facility at the University of California, Los Angeles.
This work was supported by the California Institute for Regenerative Medicine Award (RB1-01328), CTSI (NIH/NCATS Grant UL1TR000124) and the grant from the Jonsson Cancer Center Foundation.
National Institutes of Health
Authorship
Contribution: H.H., J.X., L.W., and M.L.I.A. designed the research; H.H., C.M.W., D.D., J.X., and X.L. performed the experiments; H.H., X.L., L.W., and M.L.I.A. analyzed the data; and H.H. and M.L.I.A. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: M. Luisa Iruela-Arispe, Department of Molecular, Cell, and Developmental Biology, Biomedical Sciences, Rm 447, UCLA, Los Angeles, CA; e-mail: arispe@mcdb.ucla.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal