Abstract
The nucleoporin gene NUP98 is fused to several genes including HOXD13 in patients with myelodysplastic syndromes (MDS), acute myeloid leukemia, and chronic myeloid leukemia, blast crisis. Genetically engineered mice that express a NUP98-HOXD13 (NHD13) transgene (Tg) display the phenotypic features of MDS, including cytopenias, bone marrow dysplasia, and transformation to acute leukemia. Here we show that short-term treatment with the p53 inhibitor Pifithrin-α partially and transiently rescued the myeloid and lymphoid abnormalities found in NHD13+ Tg mice, with no improvement in the anemia, while the genetic deletion of 2 alleles of p53 rescued both the myeloid progenitor cell and long-term hematopoietic stem cell compartments. Nonetheless, loss of one or both alleles of p53 did not rescue the MDS phenotype, but instead exacerbated the MDS phenotype and accelerated the development of acute myeloid leukemia. Our studies suggest that while targeting p53 may transiently improve hematopoiesis in MDS, over the long-term, it has detrimental effects, raising caution about abrogating its function to treat the cytopenias that accompany this disease.
Introduction
The myelodysplastic syndromes (MDS) are clonal stem cell disorders characterized by ineffective hematopoiesis leading to cytopenias and a high rate of progression to acute myelogenous leukemia (AML).1 MDS is generally incurable unless treated with allogeneic bone marrow transplantation, an option not generally available for persons over age 60, who are most often affected by this disease. MDS is a heterogeneous disease and includes both low-risk and high-risk variants, each with a range of median survivals and a variable risk of progression toward AML.2 MDS represents an excellent model to study the progression to acute leukemia, yet the underlying molecular mechanisms that lead to the evolution to AML are still not identified.
A variety of mouse models have been generated that mimic aspects of MDS with the most accurate being perhaps the NUP98-HOXD13 transgenic (NHD13 Tg) mouse, which uses Vav1 regulatory elements to direct expression of the NHD13 oncogene in hematopoietic tissues. NHD13+ Tg mice develop anemia, neutropenia, and lymphopenia at 4-7 months despite a hypercellular or normocellular bone marrow, indicating ineffective hematopoiesis.3 Similar to the progression pattern observed in patients with MDS, approximately half of the NHD13+ Tg mice with MDS develop acute leukemia, typically at 10-14 months of age and most commonly AML.
Loss of the p53 signaling pathway, either by mutation or deletion, or loss of upstream or downstream signaling components, occurs in the majority of human cancers. Overall, p53 mutations occur in 8%-14% of MDS cases, with a higher frequency in cases with complex karyotypes,4 or in the refractory anemia with excess of blasts (RAEB-I or RAEB-II) subtype.5-7 p53 mutations have been found in approximately 15% of the low-risk, del (5q) MDS patients at presentation and they are associated with an increased risk of leukemia progression.8
Several studies have demonstrated an important pathophysiologic role for p53 in the 5q− syndrome, a subtype of MDS where haploinsufficiency of the RPS14 ribosomal protein appears to drive the anemia that accompanies this disease.9-11 An elevated level of p53 activity appears to trigger the excessive apoptosis and dysplastic morphology seen in the erythroid cells of patients with this disorder, as loss of p53 in “5q− mice,” that lack one copy of the chromosomal region syntenic to the human 5q region that contains the RPS14 gene, show improved blood counts and disappearance of the dysplasia.10 This suggests that the cytopenias in some MDS patients may result from defective ribosomal biosynthesis, leading to activation of p53 and excessive apoptosis.
We observed increased intracellular p53 levels in the Lin−Sca-1+c-Kit+ (LSK) cells isolated from NHD13+ Tg mice and investigated the role of p53 in NHD13-driven MDS and AML. By treating these mice with the p53 inhibitor Pifithrin-α (PFT-α), we partially and transiently rescued the myeloid and lymphoid lineage differentiation defects, with no improvement in the hemoglobin level, and by generating NHD13+p53+/− and NHD13+p53−/− mice, we also found that p53 deficiency did not rescue the anemia in this MDS mouse model. Rather, p53 deficiency accelerated not only the MDS phase of the disease but also the development of acute leukemia. Our results suggest distinct roles for p53 in different types of MDS, and raise caution about the use of p53 inhibitors to treat the cytopenias that characterize this disease.
Methods
Mouse strains, transplantation studies, and assessment of hematopoietic tissues
NUP98-HOXD13 transgenic mice (NHD13+Tg) on a C57BL/6 background were kindly provided by P.D.A.3 p53+/− mice (C57BL/6, CD45.2+), wild-type C57BL/6 (CD45.2+), and B6.SJL (CD45.1+) mice were purchased from The Jackson Laboratory. NHD13+p53+/− mice were generated by breeding NHD13+ Tg mice with p53+/− mice and NHD13+p53−/− mice were generated by breeding p53+/− mice with NHD13+p53+/− mice. All mice were maintained in the Memorial Sloan-Kettering Cancer Center (MSKCC) Animal Facility according to institutional animal care and use committee–approved protocols, and kept in Thorensten units with filtered germ-free air. For the in vivo Pifithrin-α (PFT-α; Sigma-Aldrich) treatment experiments, 5 mice in each group were injected intraperitoneally on a daily basis with PFT-α (2 mg/kg body weight) or with the DMSO vehicle as a control. Mice were euthanized if they exhibited signs of ill health. Complete blood counts (CBCs) were performed using tail vein blood and a HEMAVET Multispecies Hematology Analyzer (CDC Technologies). Transplantation of cells from leukemic animals was performed by intravenous or intrafemoral injection into sublethally irradiated (5 Gy) CD45.1+ congenic recipient mice. The immunophenotype of single-cell suspensions—prepared from thymus, spleen, lymph nodes, and/or bone marrow—was analyzed by flow cytometry.
Flow cytometric analysis and cell sorting
For the lineage staining of BM cells, we used a cocktail of biotinylated antibodies including anti-CD11b (M1/70), anti-B220 (RA3-6B2), anti-CD5 (53-7.3), anti–Gr-1 (RB6-8C5), anti-Ter119 (TER-119), anti-CD3 (500A2), anti-CD19 (ID3), anti-CD4 (GK1.5), and anti-CD8a (53-6.7). For detection and sorting, we used streptavidin-allophycocyanin (APC) Cy7. Directly conjugated antibodies included anti–Sca-1–phycoerythrin (PE)–Cy7 or PerCP-Cy5.5 (clone D7; eBioscience), anti–c-Kit–APC (clone 2B8) or PE/Cy7 (clone 2B8; BioLegend), anti–CD34-FITC (clone RAM34; eBioscience), anti-CD135-PE (clone A2F10.1), anti–CD48-fluorescein isothiocyanate (clone HM48−; eBioscience), anti–CD150-PE (clone TC15-12F12.2; BioLegend) or APC (clone 9D1; eBioscience), anti–CD16/32 eFouor 450 (clone 93; eBioscience), anti-CD127 APC (clone A7R34; eBioscience), anti-CD45R APC-Cy7 (clone RA3-6B2), anti-IgM (clone 11/41; eBioscience), anti-CD3 (clone 17A2; eBioscience), anti-CD4 PerCPCy5.5 (clone RM-4-5), anti-CD8a Pacific Blue, anti-CD25 PE, anti-CD44 (clone IM7; eBioscience), anti-CD11b PE, anti-Gr1 APC, anti-CD71 (C2), and anti-Ter119 APC Ab. For congenic strain discrimination, anti–CD45.1-PE and anti–CD45.2-FITC antibodies were used. To measure intracellular p53 protein levels, cells were stained for cell-surface markers first, then fixed with 1.6% paraformaldehyde for 10 minutes at room temperature, permeabilized with ice-cold methanol for 30 minutes at 4°C, and incubated for 30 minutes with PE-conjugated mouse anti-p53 Ab (G59-12). Unless specified, all antibodies were from BD Biosciences/BD Pharmingen. Fluorescence-activated cell sorting (FACS) was performed on a FACSAria (BD Biosciences) or MoFlo (Beckman Coulter) cell sorter; analysis was performed with a LSRII (BD Biosciences).
Histologic analysis
Hematoxylin-eosin (H&E)–stained sections from tissues such as spleen, liver, and tibia were generated using conventional staining techniques. Bone marrow cells were harvested from both femurs by flushing with Iscove media and assessed by both thin smears and cytospins. Photomicrographs were taken by a Zeiss Axio2 Imaging wide-field microscope (Carl Zeiss MicroImaging) with a Zeiss AxioCam MRm Camera, Axiovision acquisition software, and a 20 × 0.8 NA DRY DIC objective lens at room temperature.
Statistical analysis
The log-rank (Mantel-Cox) test was used to determine P values for all Kaplan-Meier survival curve analyses. Differences between groups were analyzed using an unpaired 2-sided t test. Data are expressed as the mean ± SEM. A probability (P) value < .05 was considered significant.
Results
The most primitive HSC pool is increased in NHD13+ Tg mice
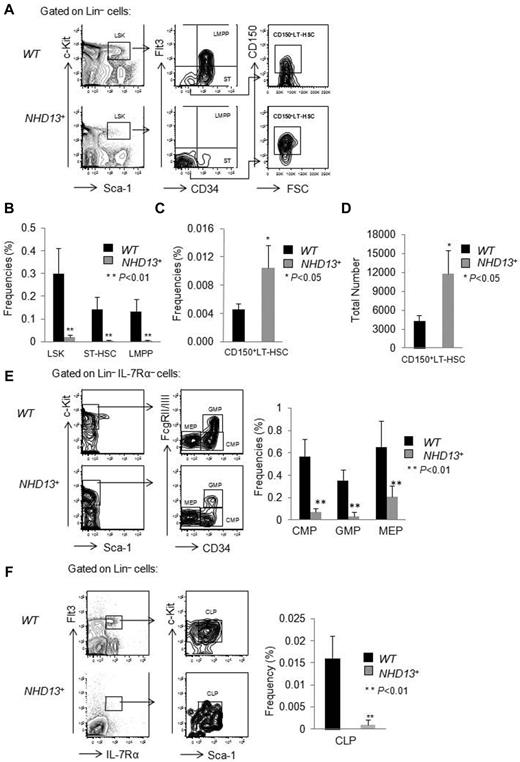
The NUP98-HOXD13 (NHD13) fusion gene was initially identified in a patient with therapy-related MDS that progressed to AML.12 Although the NUP98-HOXD13 fusion is rare in human MDS, the NHD13 fusion induces the expression of HOXA cluster genes13 which are commonly overexpressed in human MDS.14,15 To study its function, the Aplan laboratory generated NHD13+ Tg mice that express the NHD13 transgene under control of the Vav1 promoter.3 We first confirmed their findings that the transgenic C57Bl6 NHD13+ mice develop cytopenias after approximately 4 months. To define the pathogenesis of the MDS that develops, we analyzed the hematopoietic stem cell (HSC) and progenitor (HSPC) compartments of these mice, comparing the frequency of HSPCs in the bone marrow (BM) of wild-type (WT) and NHD13+ Tg mice at 4 months of age, when they first develop MDS. We observed a significant decrease (∼ 10-fold) in the LSK cell compartment of the NHD13+ mice, a compartment highly enriched for HSCs, compared with WT mice (0.02% ± 0.01% in NHD13+ mice vs 0.30% ± 0.11% in WT controls, P < .01; Figure 1A-B). However, the LSK compartment can be separated into long-term HSCs (LT-HSCs), short-term HSCs (ST-HSCs), and lymphoid-primed multipotent progenitors (LMPPs) on the basis of CD150, CD34, and Flt3 cell-surface expression, with the CD150+CD34−Flt3− cells comprising the most primitive CD150+LT-HSC compartment,16 the CD34+Flt3− population representing the ST-HSCs and the CD34+Flt3+ compartment containing LMPPs. While both the frequency (0.010% ± 0.003% in NHD13+ mice vs 0.004% ± 0.001% in WT controls, P < .05; Figure 1A,C) and the total number of CD150+ LT-HSCs (CD150+CD34−Flt3−LSK cells; 11 700 ± 3800 in NHD13+ mice compared with 4300 ± 960 in WT controls, P < .05; Figure 1D) are increased, the frequency of ST-HSCs and LMPPs is significantly decreased in NHD13+ mice (Figure 1A-B). Thus, the CD150+ LT-HSC compartment is spared from the process that depletes the other stem and pluripotent progenitor cell populations.
HSPC frequencies in NHD13+ Tg and control mice. Bone marrow (BM) cells from wild-type and NHD13+ Tg mice were harvested at 4 months of age and prepared for flow cytometric analysis. (A) Representative FACS plots show LSK, CD34, Flt3, and CD150 expression within WT and NHD13+ Tg mice BM cells. (B) The frequencies of LSK cells, ST-HSCs (LSKCD34+Flt3−) and LMPPs (LSKCD34+Flt3+) in WT and NHD13+ Tg mouse BM (mean ± SD, n = 7, *P < .05, **P < .01). (C) The frequency and (D) absolute number of CD150+ LT-HSCs (CD150+LSKCD34−Flt3−) in WT and NHD13+ Tg mouse BM are shown (mean ± SD, n = 7, *P < .05). (E left panel) The FACS plots and gating strategies used to identify CMPs (Lin−Sca-1− cKit+ IL-7Rα−CD34+FcgRlow), GMPs (Lin−Sca-1−cKit+IL-7Rα−CD34+FcgRhigh), and MEPs (Lin−Sca-1−cKit+IL-7Rα−CD34−FcgRlow) within the Lin−IL-7Rα−cKit+ population. Right panel, The frequencies of CMPs, GMPs, and MEPs in WT and NHD13+ Tg mice BM (mean ± SD, n = 8, **P < .01). (F left panel) Representative FACS plots of the gating strategy to identify the CLPs (Flt3+Lin−IL-7Rα+ckitintSca-1int) population. (Right panel) The frequency of CLPs in WT and NHD13+ Tg mice BM (mean ± SD, n = 8, **P < .01).
HSPC frequencies in NHD13+ Tg and control mice. Bone marrow (BM) cells from wild-type and NHD13+ Tg mice were harvested at 4 months of age and prepared for flow cytometric analysis. (A) Representative FACS plots show LSK, CD34, Flt3, and CD150 expression within WT and NHD13+ Tg mice BM cells. (B) The frequencies of LSK cells, ST-HSCs (LSKCD34+Flt3−) and LMPPs (LSKCD34+Flt3+) in WT and NHD13+ Tg mouse BM (mean ± SD, n = 7, *P < .05, **P < .01). (C) The frequency and (D) absolute number of CD150+ LT-HSCs (CD150+LSKCD34−Flt3−) in WT and NHD13+ Tg mouse BM are shown (mean ± SD, n = 7, *P < .05). (E left panel) The FACS plots and gating strategies used to identify CMPs (Lin−Sca-1− cKit+ IL-7Rα−CD34+FcgRlow), GMPs (Lin−Sca-1−cKit+IL-7Rα−CD34+FcgRhigh), and MEPs (Lin−Sca-1−cKit+IL-7Rα−CD34−FcgRlow) within the Lin−IL-7Rα−cKit+ population. Right panel, The frequencies of CMPs, GMPs, and MEPs in WT and NHD13+ Tg mice BM (mean ± SD, n = 8, **P < .01). (F left panel) Representative FACS plots of the gating strategy to identify the CLPs (Flt3+Lin−IL-7Rα+ckitintSca-1int) population. (Right panel) The frequency of CLPs in WT and NHD13+ Tg mice BM (mean ± SD, n = 8, **P < .01).
We also quantified the percentages of myeloerythroid and lymphoid progenitors in the BM of NHD13+ Tg mice and observed a dramatic decrease in common myeloid progenitors (CMPs; Lin−IL-7Rα−cKit+Sca-1−CD34+FcgRlow), granulocyte-macrophage progenitors (GMPs; Lin−IL-7Rα−cKit+Sca-1−CD34+FcgRhigh), and megakaryocyte-erythroid progenitors (MEPs; Lin−IL-7Rα−cKit+Sca-1−CD34−FcgRlow), as well as a significant decrease in the frequency of common lymphoid progenitors (CLPs; Flt3+Lin−IL-7Rα+ckitintSca-1int) compared with WT control mice (Figure 1E-F). Because the cellularity of WT and NHD13+ BM mice is similar (data not shown), it appears that the NHD13+ Tg mice have more CD150+ LT-HSCs, but fewer ST-HSCs, LMPPs, CMPs, GMPs, MEPs, and CLPs at the MDS stage.
Increased p53 levels in NHD13+ LSK cells and CD71+Ter119− cells
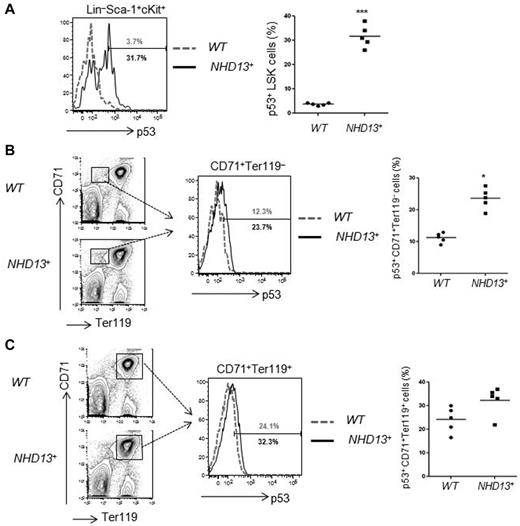
Haploinsufficiency of ribosomal protein genes, such as RPS19 and RPS14, appear to underlie the inherited, Diamond-Blackfan anemia (DBA) marrow failure syndrome, and acquired 5q− MDS, respectively.17 RPS protein deficiency leads to elevated levels of RPL11, which impair the ability of HDM2 to down-regulate p53 protein levels,18,19 leading to p53 activation. Given this finding, we examined LSK, CD71+Ter119−, and CD71+Ter119+ cells isolated from WT and NHD13+ Tg mice BM at the MDS stage (at 4 months), and found a significantly increased percentage of p53+ cells in the NHD13+ LSK cell population (31.7% ± 4.6% in NHD13+ LSKs vs 3.7% ± 0.5% in WT controls, P < .001; Figure 2A). The immature CD71+Ter119− erythroid cell compartment had increased p53 levels (23.7% ± 4.4% in NHD13+ CD71+Ter119− cells vs 12.3% ± 2.4% in WT controls, P < .05; Figure 2B), while p53 expression in the mature CD71+Ter119+ cell compartment was mildly increased compared with WT controls, and of borderline significance (32.3% ± 6.0% in NHD13+ CD71+Ter119+ cells vs 24.1% ± 5.4% in WT controls, P = .052; Figure 2C). Thus, p53 expression is increased in the early stem and progenitor cells found in NHD13-driven MDS.
Increased p53 level in NHD13+ bone marrow cells. The intracellular p53 level was measured by flow cytometry. (A) Representative FACS profiles of p53 protein levels in WT (gray) and NHD13+ Tg (black) mouse bone marrow (BM) LSK (Lin−Sca-1+cKit+) cells. The graph on the right indicates the mean percentage of p53+ cells present (n = 5, ***P < .001). (B) Representative FACS profiles of p53 protein levels in WT (gray) and NHD13+ Tg (black) mice BM CD71+Ter119− cells. The bar graph on the right indicates the mean percentage of p53+ cells present (n = 5, *P < .05). (C) Representative FACS profiles of p53 protein levels in WT (gray) and NHD13+ Tg (black) mice BM CD71+Ter119+ cells. Data shown on the right are the mean percentage of p53+ cells present (n = 5, P = .052).
Increased p53 level in NHD13+ bone marrow cells. The intracellular p53 level was measured by flow cytometry. (A) Representative FACS profiles of p53 protein levels in WT (gray) and NHD13+ Tg (black) mouse bone marrow (BM) LSK (Lin−Sca-1+cKit+) cells. The graph on the right indicates the mean percentage of p53+ cells present (n = 5, ***P < .001). (B) Representative FACS profiles of p53 protein levels in WT (gray) and NHD13+ Tg (black) mice BM CD71+Ter119− cells. The bar graph on the right indicates the mean percentage of p53+ cells present (n = 5, *P < .05). (C) Representative FACS profiles of p53 protein levels in WT (gray) and NHD13+ Tg (black) mice BM CD71+Ter119+ cells. Data shown on the right are the mean percentage of p53+ cells present (n = 5, P = .052).
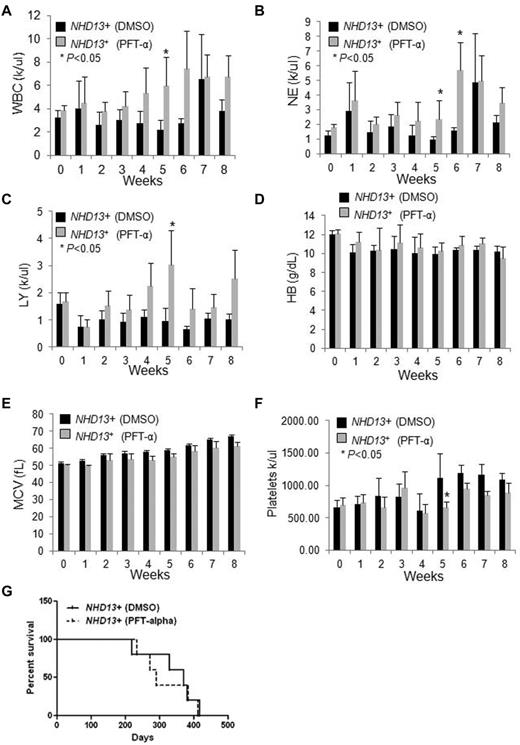
To determine whether the increase in p53 contributes to the anemia or other hematologic manifestations seen in NHD13+ mice, we examined whether PFT-α, an inhibitor of p53 function20 could rescue its hematopoietic defects. NHD13+ Tg mice with MDS (at 5 months of age) and age-matched WT controls were intraperitoneally injected daily with PFT-α (2 mg/kg body weight) as in Li et al,21 or with DMSO as a control for 8 weeks. Peripheral blood (PB) counts were checked weekly and the NHD13+ mice treated with PFT-α showed a trend toward increased white blood cell (WBC), lymphocyte, and neutrophil counts compared with NHD13+ mice treated with DMSO vehicle (Figure 3A-C). Significant differences were found during week 5 (for WBC, lymphocyte, neutrophil, and platelet counts) and week 6 (for neutrophil counts) between the control and the PFT-α–treated NHD13+ mice (Figure 3A-C,F). However, neither the hemoglobin level nor the mean corpuscular volume (MCV) remained elevated (Figure 3D-E). Lastly, we found no significant difference in the overall survival between the PFT-α–treated and the vehicle-treated NHD13+ mice (Figure 3G, Table 1): 3 mice died from MDS, and 2 from AML in the PFT-α–treated group, while 4 mice died from MDS, and 1 mouse died from AML in the control group (Figure 3G, Table 1). Among the CBC parameters, there was no statistically significant difference in WBC, lymphocyte, neutrophil counts, hemoglobin level, and platelet counts between the day 0 PFT time point and the week 5 time point (Figure 3A-F). These data show that PFT-α can partially and transiently rescue the myeloid and lymphoid defects seen in NHD13+ mice at the MDS stage, but not the anemia. Probably because of the transient and partial nature of the rescue, PFT-α treatment did not prolong the survival of NHD13+ mice.
The effects of PFT-α treatment on NHD13+ Tg mice.NHD13+ Tg mice at the MDS stage (∼ 5 months of age) were intraperitoneally injected daily with DMSO vehicle or PFT-α (2 mg/kg body weight) for 8 weeks. Peripheral blood was collected weekly. (A) The white blood cell (WBC) counts, (B) neutrophil (NE) counts, (C) lymphocyte (LY) counts, (D) hemoglobin (HB) value, (E) mean corpuscular volume (MCV), and (F) platelet counts are shown (mean ± SD, n = 5 mice/group, *P < .05). (G) Kaplan-Meier survival curves of NHD13+ mice treated with DMSO vehicle or PFT-α (2 mg/kg body weight) for 8 weeks (n = 5 mice/group).
The effects of PFT-α treatment on NHD13+ Tg mice.NHD13+ Tg mice at the MDS stage (∼ 5 months of age) were intraperitoneally injected daily with DMSO vehicle or PFT-α (2 mg/kg body weight) for 8 weeks. Peripheral blood was collected weekly. (A) The white blood cell (WBC) counts, (B) neutrophil (NE) counts, (C) lymphocyte (LY) counts, (D) hemoglobin (HB) value, (E) mean corpuscular volume (MCV), and (F) platelet counts are shown (mean ± SD, n = 5 mice/group, *P < .05). (G) Kaplan-Meier survival curves of NHD13+ mice treated with DMSO vehicle or PFT-α (2 mg/kg body weight) for 8 weeks (n = 5 mice/group).
Survival of NHD13+ mice treated with PFT-α or with the DMSO vehicle
| NHD13+ . | Died from MDS, day . | Died from AML, day . |
|---|---|---|
| DMSO | 219, 329, 370, 416 | 380 |
| PFT-alpha | 272, 290, 383 | 234, 412 |
| NHD13+ . | Died from MDS, day . | Died from AML, day . |
|---|---|---|
| DMSO | 219, 329, 370, 416 | 380 |
| PFT-alpha | 272, 290, 383 | 234, 412 |
PFT-α indicates Pifithrin-α; MDS, myelodysplastic syndrome; and AML, acute myeloid leukemia.
We also examined whether PFT-α affects HSPC frequencies in the BM of NHD13+ mice at 5 months of age when the mice have MDS. We found no effect of 5 weeks of treatment with PFT-α on BM cellularity (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). While the frequencies of LSK, CD150+ LT-HSCs, ST-HSCs, and LMPPs did not differ significantly between PFT-α–treated mice and control mice (supplemental Figure 1B), the frequency of LK cells (Lin−Sca-1−c-Kit+, a population enriched for myeloid progenitors) was increased in the PFT-α–treated mice (supplemental Figure 1C; P < .05). These mice also displayed a trend toward an increased CLP frequency compared with those control mice (supplemental Figure 1D; P = .06), and it is likely that the increased myeloid progenitor and CLP frequency contributed to the increased WBC, lymphocyte, and neutrophil counts observed in the PFT-α–treated NHD13+ mice.
Rescue of HSPC subsets by lack of p53
As a reversible inhibitor of p53, the effects of PFT-α may be different from genetic loss of p53; thus, to further investigate whether the presence or absence of p53 affects the MDS or AML phases of NHD13-driven disease, we generated NHD13+p53+/− and NHD13+p53−/− mice. We first analyzed the effect of loss of p53 on the frequencies of HSPCs within the BM cells isolated from various genotypes at an early, pre-MDS time point (6 weeks of age), and consistent with previous studies,22-25 we observed increased LSK cell frequency in the p53+/− and p53−/− mouse BM compared with WT controls (supplemental Figure 2A-B, P < .05). At this early stage, the NHD13+ mice showed a 50% reduction in the frequency of LSK cells (P < .05), similar frequencies of CD150+ LT-HSCs and ST-HSCs, and a dramatically decreased LMPP frequency (P < .01) compared with WT controls (supplemental Figure 2A-B). The complete, but not partial, absence of p53 partially rescued the LSK cell frequency (P < .05) with minimal effects on the CD150+LT-HSC, ST-HSC, and LMPP frequencies (supplemental Figure 2A-B). The deletion of one or both alleles of p53 did not affect myeloid differentiation (supplemental Figure 3A), B-cell differentiation (supplemental Figure 3B), or T-cell differentiation (supplemental Figure 3C) in NHD13+ mice.
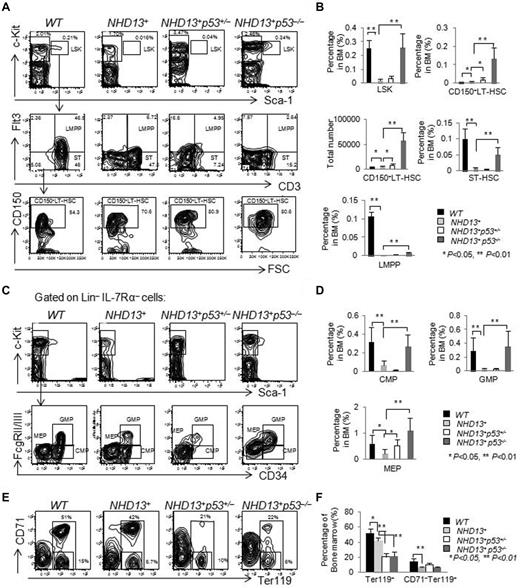
The frequency of LSK cells in the NHD13+ mice declines with age, and reaches its nadir at ∼ 4 to 5 months of age when mice show signs of early MDS. We further analyzed the frequencies of HSPCs in the BM of the WT, NHD13+, NHD13+p53+/−, and NHD13+p53−/− mice at this time period and found elevated frequencies of the LSK cells (P < .01), CD150+LT-HSCs (P < .01), ST-HSCs (P < .01), LMPPs (P < .01), CMPs (P < .01), GMPs (P < .01), and MEPs (P < .01) in the NHD13+p53−/− mice compared with the NHD13+ mice (Figure 4A-D). Deletion of one p53 allele had less of an effect on the NHD13+ mice, although the frequency of CD150+LT-HSCs (P < .05) and MEPs (P < .05) increased compared with NHD13+ mice (Figure 4A-D). The percentage of Ter119+ cells were ∼ 50% reduced in the NHD13+p53+/− and NHD13+p53−/− MDS mice compared with the NHD13+ MDS mice (Figure 4E-F). These data suggest that the deletion of p53 rescues the defects of HSPCs in NHD13+ Tg mice at MDS stage, but aggravates the erythroid differentiation.
The loss of 1 or 2 p53 alleles does not rescue the MDS phenotype induced by NHD13 fusion protein. (A) Representative staining profiles for LSK cells, CD34, Flt3, and CD150 expression within BM cells of NHD13+, NHD13+p53+/−, and NHD13+p53−/− mice with MDS, compared with WT controls. (B) The frequencies of LSK cells, ST-HSCs, LMPPs, CD150+ LT-HSCs, and the total number of CD150+ LT-HSCs in NHD13+, NHD13+p53+/−, and NHD13+p53−/− mice with MDS are compared with WT control mice (mean ± SD, *P < .05, **P < .01). (C) Representative FACS profiles for CMPs (Lin−Sca-1− cKit+ IL-7Rα−CD34+FcgRlow), GMPs (Lin−Sca-1−cKit+IL-7Rα−CD34+FcgRhigh), and MEPs (Lin−Sca-1−cKit+IL-7Rα−CD34−FcgRlow) within the Lin−IL-7Rα−cKit+ population for the NHD13+, NHD13+p53+/−, and NHD13+p53−/− mice with MDS, compared with the WT control mice. (D) The frequencies of CMPs, GMPs, and MEPs in the NHD13+, NHD13+p53+/−, and NHD13+p53−/− mice with MDS, compared with WT control mice (mean ± SD, *P < .05, **P < .01). (E) Representative FACS profiles showing CD71 and Ter119 staining of BM cells isolated from NHD13+, NHD13+p53+/−, and NHD13+p53−/− mice with MDS, compared with WT control. (F) The frequencies of Ter119+ and CD71−Ter119+ cells in the NHD13+, NHD13+p53+/−, and NHD13+p53−/− mice with MDS, compared with WT controls (mean ± SD, *P < .05, **P < .01). The mice used for this figure were between 4 and 5 months old; n = 8 for each genotype.
The loss of 1 or 2 p53 alleles does not rescue the MDS phenotype induced by NHD13 fusion protein. (A) Representative staining profiles for LSK cells, CD34, Flt3, and CD150 expression within BM cells of NHD13+, NHD13+p53+/−, and NHD13+p53−/− mice with MDS, compared with WT controls. (B) The frequencies of LSK cells, ST-HSCs, LMPPs, CD150+ LT-HSCs, and the total number of CD150+ LT-HSCs in NHD13+, NHD13+p53+/−, and NHD13+p53−/− mice with MDS are compared with WT control mice (mean ± SD, *P < .05, **P < .01). (C) Representative FACS profiles for CMPs (Lin−Sca-1− cKit+ IL-7Rα−CD34+FcgRlow), GMPs (Lin−Sca-1−cKit+IL-7Rα−CD34+FcgRhigh), and MEPs (Lin−Sca-1−cKit+IL-7Rα−CD34−FcgRlow) within the Lin−IL-7Rα−cKit+ population for the NHD13+, NHD13+p53+/−, and NHD13+p53−/− mice with MDS, compared with the WT control mice. (D) The frequencies of CMPs, GMPs, and MEPs in the NHD13+, NHD13+p53+/−, and NHD13+p53−/− mice with MDS, compared with WT control mice (mean ± SD, *P < .05, **P < .01). (E) Representative FACS profiles showing CD71 and Ter119 staining of BM cells isolated from NHD13+, NHD13+p53+/−, and NHD13+p53−/− mice with MDS, compared with WT control. (F) The frequencies of Ter119+ and CD71−Ter119+ cells in the NHD13+, NHD13+p53+/−, and NHD13+p53−/− mice with MDS, compared with WT controls (mean ± SD, *P < .05, **P < .01). The mice used for this figure were between 4 and 5 months old; n = 8 for each genotype.
The loss of p53 does not rescue the MDS phenotype induced by NUP98-HOXD13 fusion gene
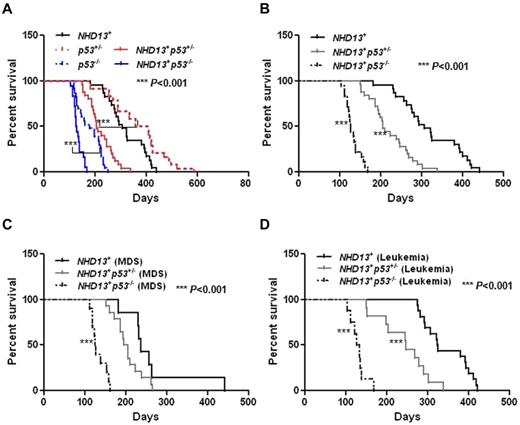
The loss of p53 actually impaired the survival of the NHD13+ mice, as the mice died more rapidly from MDS and from AML (Table 2, Figure 5A-B). The ratio of mice dying from MDS versus AML was reversed by the lack of p53 (NHD13+: 30% of the mice died from MDS and 70% from AML; NHD13+p53−/−: 56% of the mice died from MDS and 44% from AML; Table 2).
Survival of NHD13+, NHD13+p53+/−, and NHD13+p53−/− mice with MDS or AML
| Mice . | NHD13+ . | NHD13+p53+/− . | NHD13+p53−/− . | p53+/− . | p53−/− . |
|---|---|---|---|---|---|
| Median survival, d | |||||
| MDS | 236 | 200 | 126 | ||
| AML | 324 | 246 | 130 | ||
| Overall | 307 | 205 | 127 | 382 | 178 |
| Total number (%) | |||||
| Died from MDS | 7 (30) | 14 (56) | 10 (56) | ||
| Died from AML | 16 (70) | 11 (44) | 8 (44) | ||
| Overall | 23 (100) | 25 (100) | 18 (100) | 24 | 23 |
| Mice . | NHD13+ . | NHD13+p53+/− . | NHD13+p53−/− . | p53+/− . | p53−/− . |
|---|---|---|---|---|---|
| Median survival, d | |||||
| MDS | 236 | 200 | 126 | ||
| AML | 324 | 246 | 130 | ||
| Overall | 307 | 205 | 127 | 382 | 178 |
| Total number (%) | |||||
| Died from MDS | 7 (30) | 14 (56) | 10 (56) | ||
| Died from AML | 16 (70) | 11 (44) | 8 (44) | ||
| Overall | 23 (100) | 25 (100) | 18 (100) | 24 | 23 |
MDS indicates myelodysplastic syndrome; and AML, acute myeloid leukemia.
The inactivation of p53 decreases the survival of NHD13+ Tg mice with MDS or AML. (A) Kaplan-Meier survival curves for the NHD13+ (n = 23), NHD13+p53+/− (n = 25), NHD13+p53−/− (n = 18) mice with MDS or AML, and the p53+/− (n = 24), p53−/− (n = 23) control mice are shown. The P value represents the comparison of survival between the NHD13+p53−/− and p53−/− mice, or a comparison of survival between NHD13+p53+/− and p53+/− mice (***P < .001). (B) Survival curves for the NHD13+ (n = 23), NHD13+p53+/− (n = 25), and NHD13+p53−/− (n = 18) mice with MDS or AML. The P value represents a comparison of survival between the NHD13+p53−/− and NHD13+ mice, or between the NHD13+p53+/− and the NHD13+ mice (***P < .001). (C) Survival curves for the NHD13+ (n = 7), NHD13+p53+/− (n = 14), and NHD13+p53−/− (n = 10) mice with MDS. The P value represents the comparison of survival between NHD13+p53−/− and NHD13+ MDS mice (***P < .001). (D) Survival curves for the NHD13+ (n = 16), NHD13+p53+/− (n = 11), and NHD13+p53−/− (n = 8) mice with AML. The P value represents a comparison of survival between NHD13+p53−/− and NHD13+ mice, or between NHD13+p53+/− and NHD13+ mice (***P < .001).
The inactivation of p53 decreases the survival of NHD13+ Tg mice with MDS or AML. (A) Kaplan-Meier survival curves for the NHD13+ (n = 23), NHD13+p53+/− (n = 25), NHD13+p53−/− (n = 18) mice with MDS or AML, and the p53+/− (n = 24), p53−/− (n = 23) control mice are shown. The P value represents the comparison of survival between the NHD13+p53−/− and p53−/− mice, or a comparison of survival between NHD13+p53+/− and p53+/− mice (***P < .001). (B) Survival curves for the NHD13+ (n = 23), NHD13+p53+/− (n = 25), and NHD13+p53−/− (n = 18) mice with MDS or AML. The P value represents a comparison of survival between the NHD13+p53−/− and NHD13+ mice, or between the NHD13+p53+/− and the NHD13+ mice (***P < .001). (C) Survival curves for the NHD13+ (n = 7), NHD13+p53+/− (n = 14), and NHD13+p53−/− (n = 10) mice with MDS. The P value represents the comparison of survival between NHD13+p53−/− and NHD13+ MDS mice (***P < .001). (D) Survival curves for the NHD13+ (n = 16), NHD13+p53+/− (n = 11), and NHD13+p53−/− (n = 8) mice with AML. The P value represents a comparison of survival between NHD13+p53−/− and NHD13+ mice, or between NHD13+p53+/− and NHD13+ mice (***P < .001).
Based on the diagnostic criteria for MDS,26 we first examined the effect of p53 loss on NHD13-generated MDS: 56% of the NHD13+p53−/− mice (10 of 18 mice) and 56% of the NHD13+p53+/− mice (14 of 25 mice) developed MDS with a median survival of 126 days and 200 days, respectively, compared with 30% of the NHD13+ mice (7 of 23 mice) developing MDS, with a median survival of 236 days (Table 2, Figure 5C, supplemental Table 1). The NHD13+p53+/− and NHD13+p53−/− MDS mice had dysplastic, normocellular to hypercellular BMs and died with no signs of acute leukemia. The blood counts of the NHD13+p53−/− mice (aged 4 to 5 months), the NHD13+p53+/− mice (aged from 4 to 7 months), and the NHD13+ mice (aged 4 to 7 months) all showed leukopenia and anemia (supplemental Table 2). Thus, the partial or complete loss of p53 does not rescue NHD13-driven MDS, and the NHD13+p53+/− and the NHD13+p53−/− mice develop a more rapid and aggressive MDS than the NHD13+ Tg mice.
Haploinsufficiency or loss of p53 accelerates the transformation of NHD13+ mice to AML
In a previous report, approximately 60% of the NHD13+ mice developed acute leukemia, primarily AML with differentiation or AML with some degree of differentiation.3 Thus, while 70% of the NHD13+ mice in this study developed AML, only ∼ 44% of the NHD13+p53+/− mice (11 of 25) and 44% of the NHD13+p53−/− mice (8 of 18) developed AML, with a median survival of 246 days and 130 days, compared with 70% of the NHD13+ mice, whose median survival was 324 days (shown in Table 2, Figure 5D, supplemental Table 1).
Consistent with their shorter survival, NHD13+p53+/− AML mice had higher WBC counts (P = .0005) and more severe anemia (∼ 9.7 g/dL hemoglobin [HB] in NHD13+p53+/− mice vs ∼10.8 g/dL HB in NHD13+ mice, P < .05; supplemental Figure 4A-B) than the NHD13+ mice at 8 months of age, and most of the NHD13+p53−/− mice had an elevated WBC count, consistent with AML by 4 months of age (supplemental Table 3), which was uniformly fatal by 170 days (Table 2, Figure 5D, supplemental Table 1).
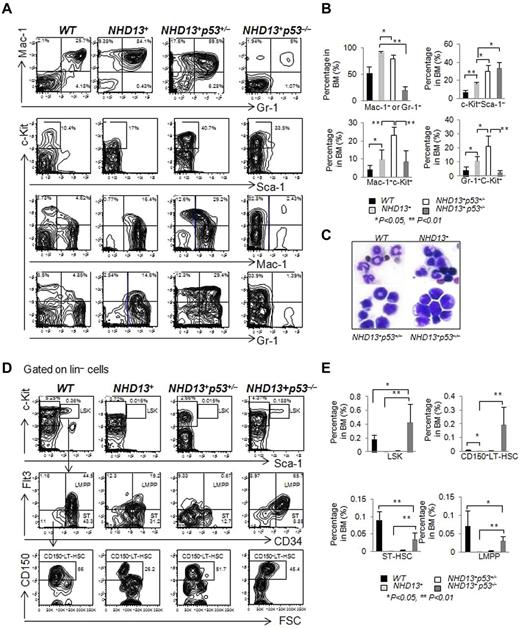
Phenotypically, the AML that developed in the NHD13+p53+/− and NHD13+p53−/− mice was c-Kit positive but Sca-1 negative (Figure 6A), and all mice had an increase in the percentage of c-Kit+ cells in the BM compared with the NHD13+ AML mice (Figure 6B). The NHD13+p53−/− AML showed the least amount of myeloid differentiation, while the NHD13+p53+/− AML showed an intermediate amount of differentiation compared with the NHD13+p53+/+AML, based on the frequency of Mac-1+ + Gr-1+ cells (91.4% ± 2.3% in NHD13+ AML mice BM, 79.5% ± 7.1% in NHD13+p53+/− AML mice BM, 20% ± 6.6% in NHD13+p53−/− AML mice BM and 52.7% ± 12.6% in the WT control mice, Figure 6A-B). The percentage of myeloid blasts (Mac-1+c-Kit+ or Gr-1+c-Kit+ cells) in the NHD13+p53+/− AML was higher than the NHD13+p53+/+ or the NHD13+p53−/− AML (Figure 6A-B). Consistent with the FACS data, morphologically, the leukemic NHD13+p53−/− cells were much less differentiated than the leukemic NHD13+p53+/− or NHD13+ cells (Figure 6C). The spleens and livers of the NHD13+p53+/− leukemic mice were massively enlarged, with extensive extramedullary hematopoiesis and infiltration by myeloblasts (data not shown); splenomegaly and hepatomegaly in the NHD13+p53−/− mice was not massive.
The loss of one or two p53 alleles accelerates the development of AML. (A) Representative FACS profiles show Mac-1 and Gr-1, c-Kit and Sca-1, c-Kit and Mac-1, c-Kit and Gr-1staining of BM cells isolated from NHD13+, NHD13+p53+/−, and NHD13+p53−/− mice with AML, compared with WT control mice. (B) The frequencies of Mac-1+ or Gr-1+, c-Kit+Sca-1−, Mac-1+c-Kit+, and Gr-1+c-Kit+ cells in NHD13+, NHD13+p53+/−, and NHD13+p53−/− mouse BM with AML, compared with WT control mice (mean ± SD, *P < .05, **P < .01). (C) Representative H&E-stained BM cytospin samples from NHD13+, NHD13+p53+/−, and NHD13+p53−/− mice with AML compared with WT control. (D) Representative staining profiles for LSK cells, showing also CD34, Flt3, and CD150 expression within the BM of NHD13+, NHD13+p53+/−, and NHD13+p53−/− mice with AML, compared with WT controls. (E) The frequencies of LSK cells, CD150+ LT-HSCs, ST-HSCs, and LMPPs in NHD13+, NHD13+p53+/−, and NHD13+p53−/− mice with AML, compared with WT control mice (mean ± SD, *P < .05, **P < .01). The mice used for this figure: NHD13+ mice with AML (n = 10, age between 9 months to 12 months), NHD13+p53+/− mice with AML (n = 10, age between 5 months to 10 months), NHD13+p53−/− mice with AML (n = 8, age between 4 and 6 months), and WT control (n = 6, age between 4 and 7 months).
The loss of one or two p53 alleles accelerates the development of AML. (A) Representative FACS profiles show Mac-1 and Gr-1, c-Kit and Sca-1, c-Kit and Mac-1, c-Kit and Gr-1staining of BM cells isolated from NHD13+, NHD13+p53+/−, and NHD13+p53−/− mice with AML, compared with WT control mice. (B) The frequencies of Mac-1+ or Gr-1+, c-Kit+Sca-1−, Mac-1+c-Kit+, and Gr-1+c-Kit+ cells in NHD13+, NHD13+p53+/−, and NHD13+p53−/− mouse BM with AML, compared with WT control mice (mean ± SD, *P < .05, **P < .01). (C) Representative H&E-stained BM cytospin samples from NHD13+, NHD13+p53+/−, and NHD13+p53−/− mice with AML compared with WT control. (D) Representative staining profiles for LSK cells, showing also CD34, Flt3, and CD150 expression within the BM of NHD13+, NHD13+p53+/−, and NHD13+p53−/− mice with AML, compared with WT controls. (E) The frequencies of LSK cells, CD150+ LT-HSCs, ST-HSCs, and LMPPs in NHD13+, NHD13+p53+/−, and NHD13+p53−/− mice with AML, compared with WT control mice (mean ± SD, *P < .05, **P < .01). The mice used for this figure: NHD13+ mice with AML (n = 10, age between 9 months to 12 months), NHD13+p53+/− mice with AML (n = 10, age between 5 months to 10 months), NHD13+p53−/− mice with AML (n = 8, age between 4 and 6 months), and WT control (n = 6, age between 4 and 7 months).
We analyzed the leukemia-HSPC compartments in the AML mice. The absence of both p53 alleles, but not one p53 allele increased the frequency of LSK cells, CD150+ LT-HSCs, ST-HSCs, and LMPPs compared with the NHD13+ leukemic controls (P < .01; Figure 6D-E), suggesting that loss of p53 increased the leukemia-HSC pool and blocked myeloid differentiation in NHD13+ AML mice. Thus, haploinsufficiency of p53 hastens the development of NHD13-driven AML with differentiation, while the absence of p53 accelerates AML development and leads to a less differentiated AML.
The transplantability of NHD13-induced AML has not been previously shown, so we isolated CD45.2+ bone marrow mononuclear cells (BMMNCs) from NHD13+ Tg mice, and injected 2 × 106 cells into sublethally irradiated congenic (CD45.1+) WT recipients by intrafemoral (IF) or intravenous (IV) injection. All recipient mice developed anemia and leukocytosis as early as week 4 (supplemental Table 4) and the mice rapidly develop AML, with a median latency of 60 days (range, 59-60) for the IV group and 67 days (range, 55-72) for the IF group (data not shown). Donor chimerism was ∼ 90% in the PB, BM, and spleen of both groups of mice 2 months posttransplantation, based on CD45.2 expression (supplemental Figure 5B), and the AML that is seen in the recipient mice recapitulates the donor NHD13+ AML (supplemental Figure 5A-B, and data not shown). Of note, the NHD13+p53+/− and NHD13+p53−/− AMLs were also readily transplantable into sublethally irradiated recipient mice (data not shown).
Discussion
MDS represents a heterogeneous group of clonal hematopoietic stem cell diseases, and while the subpopulations of HSPCs present in most patients with MDS is not known, we have found striking reductions in the frequencies and numbers of ST-HSC, LMPPs, CMPs, GMPs, MEPs, and CLPs in NHD13+ Tg mice. In contrast, the frequency and number of CD150+ LT-HSCs is increased, which is associated with myeloid predominant hematopoiesis, and is consistent with previous reports that CD150+ LT-HSCs predominantly give rise to myeloid cells after transplantation.27-30
While p53 mutations are infrequently found in patients with MDS or AML,31,32 their presence is strongly correlated with the transformation of MPNs (myeloproliferative neoplasms) to AML.33 p53 plays an essential role in regulating the cellular response to stress, and evidence is emerging that p53 can promote the cytopenias and dysplasia that characterize MDS associated with ribosomal biogenesis defects10 ; in this setting, loss of p53 reverses the dysplasia and erythroid defects seen in 5q− mice. Although we found increased p53 levels in the NHD13+ LSK cells, inhibition or loss of p53 did not rescue the anemia, nor the megaloblastic appearance or profound nuclear-cytoplasmic asynchrony seen in the erythroid compartment of the NHD13+ MDS mice.
Loss of p53 did rescue the deficient BM HSPC compartment of the NHD13+ mice (including LSK cells, CD150+ LT-HSCs, ST-HSCs, LMPPs, CMPs, GMPs, and MEPs). Loss of p53 enhances the self-renewal of early hematopoietic progenitors,34 and increases mammary stem cell (SC) symmetric divisions in vitro35 ; this combination of increased self-renewal and symmetric divisions might explain the increased LT-HSC pool seen in NHD13+p53−/− MDS mice.
The less differentiated AML observed in NHD13+p53−/− mice may reflect an enhanced self-renewal of the leukemic stem/progenitor cells, or an earlier differentiation block, somehow triggered by the lack of p53, or its downstream target genes. Both short hairpin RNA (shRNA)–mediated suppression and genetic deletion of p53 were recently shown to accelerate the leukemia that develops in a KRAS-driven AML mouse model.36 This cooperativity with oncogenic Ras may similarly reflect an effect of p53 on differentiation. Taken in the aggregate, the available data suggest a distinct role for p53 in nonmalignant MDS, such as in the erythroid compartment of RPS14-deficient mice, versus its role in oncogene-driven MDS, where there is a greater propensity to evolve AML clones. Our data also suggest that defects in the p53 pathway could play a role in the deterioration of hematopoiesis that occurs in patients with MDS that does not progress to AML.
In summary, our data indicate that transient suppression of p53 function may temporarily improve hematopoiesis, but chronic deficiency in p53 function accelerates the progressive ineffective hematopoiesis that characterizes MDS, and its progression to AML. Rather than blocking p53 function, our data suggest that promoting p53 function, or triggering its downstream effects, may help eliminate MDS clones.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the MSKCC Antitumor Assessment Core for help with animal experiments, Dr Michael Kharas for critically reviewing the manuscript, and members of the Nimer laboratory for helpful comments and suggestions.
This work was supported by a Leukemia & Lymphoma Society SCOR grant (S.D.N.), National Institutes of Health RO1 grant (DK52208; S.D.N.), a Translational and Integrative Medicine Research Fund (S.D.N.), and the Intramural Research Program of the National Institutes of Health, National Cancer Institute (P.D.A.).
National Institutes of Health
Authorship
Contribution: H.X. and S.D.N. designed the research and wrote the manuscript; H.X., S.M., B.S., N.B., G.G., and T.R.D. performed the research; H.X. analyzed data; and P.D.A. contributed vital reagents.
Conflict-of-interest disclosure: P.D.A. receives royalties from the National Institutes of Health Technology Transfer Office for the invention of NHD13 Tg mice. The remaining authors declare no competing financial interests.
Correspondence: Stephen D. Nimer, MD, University of Miami Sylvester Comprehensive Cancer Center, University of Miami School of Medicine, 1550 NW 10th Ave, Miami, FL 33136; e-mail: snimer@med.miami.edu.