Abstract
Increased expression of Kruppel-like factor 7 (KLF7) is an independent predictor of poor outcome in pediatric acute lymphoblastic leukemia. The contribution of KLF7 to hematopoiesis has not been previously described. Herein, we characterized the effect on murine hematopoiesis of the loss of KLF7 and enforced expression of KLF7. Long-term multilineage engraftment of Klf7−/− cells was comparable with control cells, and self-renewal, as assessed by serial transplantation, was not affected. Enforced expression of KLF7 results in a marked suppression of myeloid progenitor cell growth and a loss of short- and long-term repopulating activity. Interestingly, enforced expression of KLF7, although resulting in multilineage growth suppression that extended to hematopoietic stem cells and common lymphoid progenitors, spared T cells and enhanced the survival of early thymocytes. RNA expression profiling of KLF7-overexpressing hematopoietic progenitors identified several potential target genes mediating these effects. Notably, the known KLF7 target Cdkn1a (p21Cip1/Waf1) was not induced by KLF7, and loss of CDKN1A does not rescue the repopulating defect. These results suggest that KLF7 is not required for normal hematopoietic stem and progenitor function, but increased expression, as seen in a subset of lymphoid leukemia, inhibits myeloid cell proliferation and promotes early thymocyte survival.
Introduction
Although current therapeutic protocols have achieved a cure rate of > 85% in pediatric patients with acute lymphoblastic leukemia (ALL), outcomes for patients with persistent disease after induction chemotherapy or who have relapsed disease are much worse. Currently, more than half of children with relapsed ALL die from the disease, and a study from the Children's Oncology Group demonstrated that the 5-year survival rate for children with an early relapse (< 18 months from initial diagnosis) is only 21.0% ± 1.8%.1 The identification of patients with high-risk features and the need for intensification of therapy is crucial to the management of such persons.
In an analysis of bone marrow cells of 187 children with ALL, Flotho et al showed that the expression of Kruppel-like factor 7 (KLF7) was significantly elevated in patients with minimal residual disease at day 19 of therapy and was an independent predictor of leukemic relapse in a separate cohort of 99 patients.2 Minimal residual disease at day 19 of induction therapy is highly predictive of high-risk disease and is a reliable marker of chemotherapy-refractory leukemia.3 In addition, a 5.5-fold increase in KLF7 expression was found in the bone marrow cells of patients with imatinib-resistant CML compared with sensitive controls.4
The KLF family of transcription factors are involved in regulating cellular growth and differentiation in multiple tissue types. Furthermore, members of this family are involved in numerous aspects of hematopoiesis, including the regulation of erythropoiesis, lymphopoiesis, and myelopoiesis.5-7 KLF7 is important for neurogenesis, and mice lacking KLF7 die perinatally with severe neurologic defects.8 Although no specific role for KLF7 in hematopoiesis has been previously reported, the loss of the closely related family member KLF6 is associated with defective blood cell production.9 Targets of KLF7 include known regulators of hematopoietic stem and progenitor cell (HSPC) function, including TRKA, CEBP/α, and CDKN1A (p21Cip1/Waf1).8,10,11 Given the association of KLF7 with resistant leukemia and the fact that other members of the KLF family are involved in various aspects of hematopoiesis, we hypothesized that KLF7 may play a role in regulating normal HSPC function and may contribute to leukemogenesis or resistance to therapy. To test this hypothesis, we have established Klf7−/− fetal liver chimeras to address the effects of loss of KLF7 on HSPC function. To model the overexpression of KLF7 seen in resistant leukemia, we generated retroviral and lentiviral vectors to overexpress KLF7 in HSPCs.
Methods
Mice
C57BL/6 mice (B6.SJL-Ptprc* Pep3b BoyJ) carrying Ly5.1 were obtained from the The Jackson Laboratory. Cdkn1a-deficient mice (Ly5.2) were generated as previously described12 and backcrossed > 10 generations onto a C57BL/6 background. Klf7+/− mice (Ly 5.2) were a kind gift of Dr Susan Dorsey at the University of Maryland. They were generated as previously described.8 Sex- and age-matched mice 6-9 weeks of age were used in transplant experiments, unless otherwise indicated, in accordance with the guidelines of the Washington University Animal Studies Committee. Mice were housed in a specific pathogen-free environment.
Generation of fetal liver chimeras
Wild-type (WT; Ly5.1) or Klf7−/− (Ly5.2) fetal liver cells were obtained by performing timed matings and harvesting fetal livers at 13.5-14.5 days after conception. Fetal liver cells were then passed through an 18-gauge needle to obtain a single-cell suspension and frozen at −80°C in freezing media (90% FBS, 10% DMSO). Genotyping was performed on fetal tissue using PCR to distinguish the mutant and WT alleles using Taq DNA polymerase (Invitrogen) and the following cycling conditions: 95°C for 5 minutes, then 40 cycles of 95°C for 1 minute, 63°C for 1 minute and 72°C for 1 minute, then 72°C for 10 minutes. Primers for the mutant and WT products, respectively, were as follows: Neo/F: 5′-GGAGAGGCTATTCGGCTATGACTG-3′; Neo/R: 5′-CTCTTCGTCCAGATCATCCTGATC-3′; EX1/F: 5′-TTTCCTGGCAGTCATCTGCAC-3′; EX1/R: 5′- GGGTCTGTTTGTTTGTCAGTCTGT-3′). Thawed cells were then washed in MEM-α (Invitrogen) with 15% FBS and counted on a hemacytometer with trypan blue to exclude dead cells. A total of 2 million live cells (from 2 or 3 pooled donors) were mixed in a 1:1 ratio with competitor (Ly5.2) cells and injected retro-orbitally into lethally irradiated (1000 cGy) recipient (Ly5.1/Ly5.2) mice. Prophylactic antibiotics (trimethoprim-sulfamethoxazole; Alpharma) were given for 2 weeks after transplantation. For serial transplants, bone marrow was harvested from the femurs of primary recipients 3-6 months after transplantation. Bone marrow from similar genotypes was pooled, analyzed by flow cytometry for Ly5.1 and Ly5.2 to determine input chimerism, and transplanted (3-4 million cells/recipient) into lethally irradiated secondary or tertiary recipients.
KLF7 virus production
KLF7 overexpression was achieved using 2 different viral vectors: the MSCV-IRES-GFP retroviral vector and the HIV-MND-GFP lentiviral vector.13 The HIV-MND-GFP vector was modified to contain an internal ribosomal entry site (IRES) upstream of GFP. The murine Klf7 coding sequence (nucleotides 389-1294; NM_033563.2) was inserted into each vector. For retrovirus production, 293T cells were transfected with the Ecopak plasmid and the MSCV vector using the Calcium Phosphate Transfection Kit (Invitrogen), following the manufacturer's recommendations. A similar procedure was used to generate lentivirus, except that the packaging plasmids pMD.G, pRSV-Rev and PMDLg were cotransfected with the HIV-MND vectors.14 Viral supernatant was collected 48 and 72 hours after transfection and, for lentivirus, concentrated by centrifugation at 76 000g for 1.5 hours at 4°C (SW32 rotor in Optima LE-80K ultracentrifuge; Beckman Coulter).
Retroviral transduction and transplantation of primary murine hematopoietic progenitors
For experiments using MSCV virus, donor mice were treated with 5-fluorouracil (Sigma-Aldrich; 100 mg/kg intraperitoneally), and bone marrow was harvested by centrifugation of femurs and tibias (3300g for 1 minute) 24 hours later. Cells were then plated in 6-well plates in transfection media (MEM-α, Invitrogen; with 15% FCS and a cytokine cocktail [TPO 10 ng/mL, IL-3 10 ng/mL, SCF 100 ng/mL, and Flt3-L 50 ng/mL]). Virus was added at a multiplicity of infection of ∼ 10:1, and cells were spun at (966g) for 90 minutes at 30°C in a Sorvall RT 7 centrifuge. They were then placed in a 37°C incubator overnight, washed, and spun again with fresh media and virus. After the second spinfection, cells were allowed to rest at 37°C for at least 6 hours, then washed 1 time in media, filtered, and resuspended in PBS with 0.1% BSA for transplantation. For transplants, 2-3 million cells were injected retro-orbitally into lethally irradiated (1000 cGy) recipient mice.
For the HIV-MND-GFP lentivirus, bone marrow cells were harvested from donor mice by crushing leg bones, pelvis, and sternum in PBS + 10% FCS using a mortar and pestle. Cells were then filtered, counted, and incubated with anti-CD117 beads (c-Kit, Miltenyi) following the manufacturer's instructions. Kit+ cells were isolated on an AutoMacs cell separator (Miltenyi). The c-Kit+ cells were cultured in transfection media and transduced by spinfection over 2 days, using an multiplicity of infection of ∼ 10:1. Approximately 500 000 cells were transplanted per recipient.
CFU-C assay
Transduced bone marrow cells or fetal liver cells (25 000) were plated in MethoCult 3434 (StemCell Technologies) and cultured for 7 days at 37°C in a humidified chamber with 5% CO2. For colony formation (CFU), CFU-G, CFU-GM, CFU-M, and BFU-E, cells were plated in base methylcellulose (M3231, StemCell Technologies) with the addition of appropriate cytokines (PeproTech; for CFU-G; G-CSF 100 ng/mL; for CFU-GM, GM-CSF 25 ng/mL and IL-3 10 ng/mL; for CFU-M, M-CSF 10 ng/mL; and for BFU-E, Epo 2 U/mL, SCF 100 ng/mL and IL-3 10 ng/mL). The total number of colonies and the percentage of GFP+ colonies were assessed using an Olympus IX51 scope.
Flow cytometry
Peripheral blood was obtained by retro-orbital sampling, and blood counts determined using a Hemavet (Drew Scientific) automated cell counter. Spleen and thymus were harvested, and single-cell suspensions generated using a dounce homogenizer. After lysis of red blood cells using Tris-buffered ammonium chloride buffer, pH 7.2, cells were then stained in PBS containing 1mM EDTA and 0.2% (weight/volume) BSA (FACS buffer) with 1 or more of the following antibodies: anti–mouse CD45.1 (clone A20), anti–mouse CD45.2 (clone 104), anti–mouse CD115 (clone AFS98), anti–mouse/human CD45R (B220; clone RA3-6B2), anti–mouse CD3e (clone 145-2C11), anti–mouse Ly-6G (Gr-1; clone RB6-8C5), anti–mouse CD8a (clone 53-6.7, BioLegend), anti–mouse CD4 (clone GK1.5, BioLegend), and anti–mouse CD3e (clone 145-2C11). All antibodies were obtained from eBioscience, unless otherwise noted.
For the analysis of apoptosis, cells were stained for surface markers, then resuspended in binding buffer (20mM HEPES, pH 7.4, 132mM NaCl, 6mM KCl, 2.5mM CaCl2, 1mM MgCl2, 1.2mM Kh2PO4, 5.5mM glucose, and 0.5% BSA) with the addition of annexin V (BD Biosciences). After binding 15 minutes, cells were washed and resuspended in binding buffer with DAPI for flow cytometry.
For the analysis of hematopoietic progenitor populations, bone marrow cells were incubated with the following progenitor antibody panel: anti–mouse Ly6A/E (Sca-1) PerCP-Cy5.5 (clone D7), anti–mouse CD117 (c-Kit) APC–eFluor 780 (clone 2B8), anti–mouse CD34 PE (clone HM34, BioLegend), anti–mouse CD16/32 eFluor 450 (clone 93), anti–mouse CD45.1 PE-Cy7 (clone A20), anti–mouse/human CD45R (B220) biotin (clone RA3-6B2), anti–mouse CD3e biotin (clone 145-2C11), anti–mouse Ly-6G (Gr-1) biotin (clone RB6–8C5), anti–mouse Ter119 biotin (clone TER-119), and streptavidin eFluor 605NC. To measure common lymphoid progenitors (CLP), bone marrow cells were incubated with anti–mouse Ly6D PE (clone 49-H4; BD Biosciences PharMingen), anti–mouse CD27 PerCp-Cy5.5 (clone LG.3A10), anti–mouse/human CD45R (B220) PE-Cy7 (clone RA3-6B2), anti–mouse CD3e PE-Cy7 (clone 145-2C11), anti–mouse Ly-6G (Gr-1) Pe-Cy7 (cone RB6-8C5), anti–mouse Ter119 PE-Cy7 (clone TER-119), anti–mouse CD135 APC (clone A2F10), anti–mouse IL7Rα biotin (clone A7R34; a kind gift of Dr Deepta Bhattacharya, Department of Immunology & Pathology, Washington University, St Louis, MO), anti–mouse CD45.1 APC-eFluor 780 (clone A20), and streptavidin eFluor 450). All staining was done on ice for 30 minutes, and cells were then washed and incubated with secondary antibody, where indicated, for an additional 30 minutes. Cells were analyzed on the Gallios Flow Cytometer (Beckman Coulter) and analyzed using the FloJo Version 9.3.4 software (TreeStar).
RNA expression profiling
For RNA expression profiling of HSCs, bone marrow was pooled from 10 mice for each of the 3 experimental replicates. Cells were stained with the progenitor antibody panel (with exception of the use of anti–mouse CD150 [clone TC15-12F12.2, BioLegend], anti–mouse CD41 [clone MWReg30; BD Biosciences PharMingen], and anti–mouse CD48 [clone HM48-1, eBioscience]), and then c-Kit+ Sca-1+ lineage− CD150+ CD41− CD48 −cells were sorted using an iCyt Reflection cell sorter directly into Trizol LS reagent (Invitrogen). For RNA expression profiling of Klf7−/− HSPCs, bone marrow was harvested from fetal liver chimeras and stained with the progenitor antibody panel. Kit+ Sca+ lineage− (KLS) cells were sorted using a MoFlo cell sorter (Dako North America) directly into lysis buffer, and RNA was prepared using the RNA XS column kit (Macherey-Nagel) according to the manufacturer's directions. For expression profiling of KLF7 overexpressing HSPCs, bone marrow cells exposed to MSCV or HIV-MND virus expressing KLF7 or GFP alone were harvested 72 hours after transduction. KLS GFP+ cells were sorted using a MoFlo cell sorter directly into lysis buffer, and RNA was prepared using the RNA XS column kit. RNA was amplified using the NuGen Ovation system (NuGen), and hybridized to the Affymetrix MoGene Version 1.0 ST array. Data normalization using the Robust Multichip Average algorithm, and detection P value determinations were made using the Affymetrix Expression Console Version 1.1 software. Gene set enrichment was performed using the gene set enrichment analysis Version 2.0 software (Broad Institute). Submission of expression data can be found by following the link, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE40331.
Real-time quantitative RT-PCR
Real-time RT-PCR was performed using the TaqMan One-Step RT-PCR Master Mix Reagents kit (Applied Biosystems) on a GeneAmp 5700 Sequence Detection System (Applied Biosystems). The reaction mix consisted of 0.5-1 μL RNA, 10 μL RT-PCR mix, primer/probe, and 0.5 μL MultiScribe reverse transcription and RNase inhibitor in a total reaction volume of 20 μL. Reactions were repeated in the absence of reverse transcription to confirm that DNA contamination was not present. RNA content was normalized to murine β-actin. PCR conditions were 48°C for 30 minutes and 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute.
Primers and probes were as follows: β-actin: forward: 5′-ACCAACTGGGACGATATGGAGAAGA-3′; reverse: 5′-TACGACCAGAGGCATACAGGGACAA-3′; probe: 5′-AGCCATGTACGTAGCCATCCA-3′ (FAM/TAMRA). KLF7: forward: 5′-CCTGGCAGCAGACATGCCTTGA-3′; reverse: 5′-AGGCGCCGGAAGCTCTCCTC-3′; probe: 5′-CGGCGGATCTCGGAGACCTT-3′ (FAM/TAMRA).
The following TaqMan Gene Expression Assay (Applied Biosystems) primer/probe reagents were used: KLF4: Mm00516104_m1; KLF6: Mm00516184_m1; KLF9: Mm00495172_m1; KLF10: Mm00449812_m1; CDKN1a: Mm00432448_m1; BCL2A1: Mm03646861_mH.
Statistics
Significance was determined using Prism Version 5.0 software (GraphPad). Statistical significance of differences was calculated using 2-tailed Student t tests, or 1- or 2-way ANOVA. P < .05 was considered significant. All data are presented as mean ± SEM except as noted.
Results
Loss of KLF7 does not impair HSPC repopulating activity
To assess KLF family gene expression in murine HSCs, we isolated RNA from sorted KLS CD150+ CD48− CD41− bone marrow cells and performed RNA expression profiling. This cell population is highly enriched for HSCs, with nearly one half of the cells having long-term repopulating activity.15 Of the 17 KLF family members, the majority were expressed at low but detectable levels, including KLF7 (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In agreement with these data, an RNA profiling study of HSCs and their differentiated progeny by Chambers et al demonstrated expression of KLF7 in the side population+ KLS cells.16
We next quantified KLF7 mRNA expression in different HSPC subsets and selected mature hematopoietic lineages (supplemental Figure 1). KLF7 expression was detected in all hematopoietic lineages and HPSCs subsets. Interestingly, an ∼ 5-fold increase in KLF7 expression was observed in the transition from CD4− CD8− (double-negative; DN) thymocytes to CD4+ CD8+ (double-positive, DP) thymocytes.
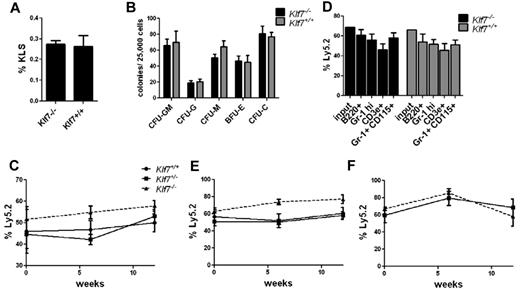
Loss of KLF7 is embryonic lethal because of neurologic defects.8 To assess the role of KLF7 in normal hematopoiesis, we first analyzed HSPCs in Klf7−/− fetal livers. The percentage of KLS cells in Klf7−/− fetal livers and their capacity to generate myeloid colonies in vitro were similar to that observed in the fetal liver of Klfl7+/+ siblings (Figure 1A-B). To assess in vivo HSPC function, we generated Klf7−/− fetal liver chimeras. The contribution of Klf7−/− cells to multilineage engraftment was similar to control cells at up to 12 weeks after transplantation (Figure 1C-D). To assess the effects of KLF7 loss on HSC self-renewal, serial transplantations were performed. Again, Klf7−/− HSCs engrafted secondary and tertiary recipients with equal efficiency to Klf7+/+ HSCs (Figure 1E-F).
Loss of KLF7 does not affect bone marrow repopulating activity. The frequency of KLS (A) and colony-forming cells (B) in the fetal liver from 13.5-14.5-dpc Klf7−/− and Klf7+/+ embryos is shown. Data represent 2 or 3 independent experiments with 2 or 3 livers per genotype. Fetal liver chimeras were generated by transplanting Klf7+/+, Klf7−/+, or Klf7−/− fetal liver (Ly5.2) cells with an equal number of WT competitor fetal liver (Ly5.1) cells into irradiated congenic (Ly5.1/5.2) mice. (C) The percentage of donor leukocytes at the indicated times. (D) The contribution of donor cells to B cells (B220+), T cells (CD3e+), neutrophils (Gr-1 high), and monocytes (Gr-1+ CD115+) assessed 12 weeks after transplantation is shown for 1 representative experiment. Bone marrow was harvested from primary recipients at 12 weeks and serially transplanted. Shown is donor contribution in secondary (E) or tertiary (F) recipients. Data represent 2 or 3 independent experiments for each genotype, with 4 or 5 mice per genotype per experiment.
Loss of KLF7 does not affect bone marrow repopulating activity. The frequency of KLS (A) and colony-forming cells (B) in the fetal liver from 13.5-14.5-dpc Klf7−/− and Klf7+/+ embryos is shown. Data represent 2 or 3 independent experiments with 2 or 3 livers per genotype. Fetal liver chimeras were generated by transplanting Klf7+/+, Klf7−/+, or Klf7−/− fetal liver (Ly5.2) cells with an equal number of WT competitor fetal liver (Ly5.1) cells into irradiated congenic (Ly5.1/5.2) mice. (C) The percentage of donor leukocytes at the indicated times. (D) The contribution of donor cells to B cells (B220+), T cells (CD3e+), neutrophils (Gr-1 high), and monocytes (Gr-1+ CD115+) assessed 12 weeks after transplantation is shown for 1 representative experiment. Bone marrow was harvested from primary recipients at 12 weeks and serially transplanted. Shown is donor contribution in secondary (E) or tertiary (F) recipients. Data represent 2 or 3 independent experiments for each genotype, with 4 or 5 mice per genotype per experiment.
To address the possibility that other KLF family members may compensate for the loss of KLF7, we isolated Klf7−/− and Klf7+/+ KLS cells from primary recipients 12 weeks after transplantation and performed RNA expression profiling. Although modest differences in expression of certain KLF family members were initially identified by expression profiling (supplemental Table 2), none of these changes was validated by real time RT-PCR (supplemental Figure 2). Thus, at least at the RNA level, compensatory expression of other KLF family members was not induced by the loss of KLF7. Collectively, these results suggest that KLF7 does not play an essential role in normal hematopoiesis because HSCs lacking KLF7 are able to engraft irradiated recipients and support multlineage hematopoiesis to a similar extent as WT HSCs.
Overexpression of KLF7 inhibits in vitro myeloid colony formation
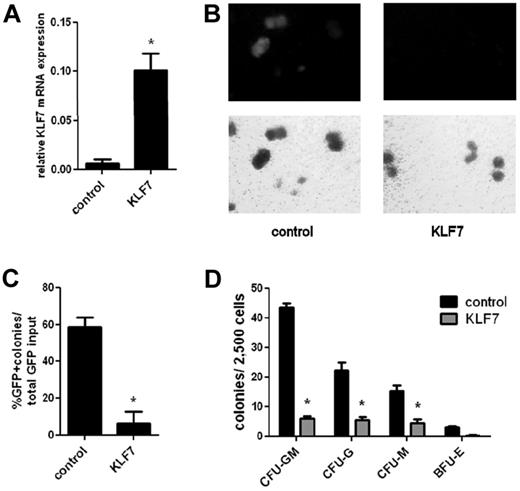
To model the effects of overexpression of KLF7 on HSPC function, we generated retroviral and lentiviral vectors that express KLF7. First, we used a retroviral vector (MSCV-IRES-GFP) expressing the murine KLF7 coding sequence to transduce primary mouse bone marrow cells. Expression of KLF7 in transduced (GFP+) WT bone marrow cells was increased approximately 10-fold compared with control cells (Figure 2A). After viral transduction of whole bone marrow from 5-fluorouracil–treated WT mice, cells were plated, unsorted, onto complete methylcellulose media to assess in vitro myeloid colony-forming ability. Relative to the initial transduction efficiency, the number of myeloid colonies produced from KLF7-transduced cells compared with vector-alone transduced cells was reduced 5.7 ± 1.9–fold, demonstrating that overexpression of KLF7 suppresses myeloid cell growth (Figure 2B-C). To further characterize the effects of KLF7 overexpression on myeloid development, WT c-Kit+ selected cells were transduced with lentivirus expressing KLF7 or GFP alone, and sorted GFP+ cells were plated on complete methylcellulose media (Figure 2D). Again, enforced expression of KLF7 inhibited the formation of all myeloid colony types, including CFU-G, CFU-M, CFU-GM, and BFU-E.
KLF7 overexpression inhibits in vitro myeloid colony formation. WT bone marrow cells from 5-fluorouracil–treated donors were transduced with MSCV-IRES-GFP expressing KLF7 or vector alone. (A) Transduced (GFP+) cells were isolated 72 hours after transduction, and the expression of KLF7 (relative to β-actin) was measured by real-time RT-PCR. Data represent 2 independent experiments. *P = .033. Unsorted bone marrow cells exposed to control or KLF7 retrovirus were plated in methylcellulose and cultured for 7 days. (B) Representative photomicrograph showing the GFP+ colonies (top panel) and total colonies (bottom panel). Original magnification ×10. (C) The percentage of GFP+ colonies relative to the input percentage of GFP+ cells. *P = .026. Data represent the average ± SEM of 2 independent experiments, each plated in duplicate. (D) GFP+ sorted cells from lentivirally transduced c-Kit+ cells were plated in complete methylcellulose, and colonies were scored by morphology after 7 days of growth.
KLF7 overexpression inhibits in vitro myeloid colony formation. WT bone marrow cells from 5-fluorouracil–treated donors were transduced with MSCV-IRES-GFP expressing KLF7 or vector alone. (A) Transduced (GFP+) cells were isolated 72 hours after transduction, and the expression of KLF7 (relative to β-actin) was measured by real-time RT-PCR. Data represent 2 independent experiments. *P = .033. Unsorted bone marrow cells exposed to control or KLF7 retrovirus were plated in methylcellulose and cultured for 7 days. (B) Representative photomicrograph showing the GFP+ colonies (top panel) and total colonies (bottom panel). Original magnification ×10. (C) The percentage of GFP+ colonies relative to the input percentage of GFP+ cells. *P = .026. Data represent the average ± SEM of 2 independent experiments, each plated in duplicate. (D) GFP+ sorted cells from lentivirally transduced c-Kit+ cells were plated in complete methylcellulose, and colonies were scored by morphology after 7 days of growth.
Overexpression of KLF7 inhibits the repopulating ability of HPSCs
We next assessed the repopulating activity of cells transduced with KLF7 MSCV retrovirus by transplanting unsorted cells into irradiated congenic recipients. Transduction efficiency was assessed in vitro by measuring the percentage of KLS cells that expressed GFP at 72 hours after transduction (Figure 3A, time zero). The contribution of KLF7 transduced cells was markedly suppressed at 6 and 12 weeks after transplantation (Figure 3A). Furthermore, whereas control transduced cells produced both myeloid and lymphoid lineage cells, minimal contribution of KLF7-transduced cells was observed in all lineages, except T cells (Figure 3B).
KLF7 overexpression inhibits bone marrow reconstitution. (A) Bone marrow cells transduced with MSCV expressing KLF7 or vector alone were transplanted into lethally irradiated recipients, and peripheral blood was analyzed 6 and 12 weeks after transplantation. Transduction efficiency was determined by measuring the percentage of GFP+ KLS cells just before transplantation (72 hours after viral exposure, time = 0). Data represent 2 individual experiments with 3-5 mice per genotype per experiment. *P < .001. (B) Shown is the percentage of donor B cells (B220+), T cells (CD3e+), and neutrophils (Gr-1 high) that were GFP+ from 1 representative experiment. *P < .01. (C) Bone marrow cells were transduced with HIV-MND–expressing KLF7 or vector alone at a low multiplicity of infection were transplanted into irradiated congenic recipients and the percentage GFP+ leukocytes in the blood measured. Data represent 3 independent experiments. *P < .001. **P < .05. (D) The percentage of donor B cells (B220+), T cells (CD3e+), and neutrophils (Gr-1 high) that were GFP+ at 6 weeks after transplantation. *P < .05. **P < .01. ***P < .0001. (E) GFP+ cells were sorted 72 hours after viral transduction and RNA analyzed by real-time RT-PCR for KLF7 expression. Data are normalized to β-actin and represent 2 independent experiments. *P = .043. (F) To assess homing, GFP+ KLS cells were transplanted into irradiated recipients and the number of GFP+ cells recovered from the bone marrow 20 hours later determined. Shown are the number of GFP+ cells per 106 whole bone marrow cells, corrected for the number of cells injected. Data represent 3 individual experiments, with 3-6 mice per condition.
KLF7 overexpression inhibits bone marrow reconstitution. (A) Bone marrow cells transduced with MSCV expressing KLF7 or vector alone were transplanted into lethally irradiated recipients, and peripheral blood was analyzed 6 and 12 weeks after transplantation. Transduction efficiency was determined by measuring the percentage of GFP+ KLS cells just before transplantation (72 hours after viral exposure, time = 0). Data represent 2 individual experiments with 3-5 mice per genotype per experiment. *P < .001. (B) Shown is the percentage of donor B cells (B220+), T cells (CD3e+), and neutrophils (Gr-1 high) that were GFP+ from 1 representative experiment. *P < .01. (C) Bone marrow cells were transduced with HIV-MND–expressing KLF7 or vector alone at a low multiplicity of infection were transplanted into irradiated congenic recipients and the percentage GFP+ leukocytes in the blood measured. Data represent 3 independent experiments. *P < .001. **P < .05. (D) The percentage of donor B cells (B220+), T cells (CD3e+), and neutrophils (Gr-1 high) that were GFP+ at 6 weeks after transplantation. *P < .05. **P < .01. ***P < .0001. (E) GFP+ cells were sorted 72 hours after viral transduction and RNA analyzed by real-time RT-PCR for KLF7 expression. Data are normalized to β-actin and represent 2 independent experiments. *P = .043. (F) To assess homing, GFP+ KLS cells were transplanted into irradiated recipients and the number of GFP+ cells recovered from the bone marrow 20 hours later determined. Shown are the number of GFP+ cells per 106 whole bone marrow cells, corrected for the number of cells injected. Data represent 3 individual experiments, with 3-6 mice per condition.
To obtain a more durable overexpression of KLF7, we repeated these experiments using lentivirus expressing KLF7 or GFP alone. We again observed an ∼ 10-fold increased expression of KLF7 in transduced cells (Figure 3E). In experiments using a high multiplicity of infection, resulting in high transduction efficiency (≥ 60% transduced cells), overexpression of KLF7 was associated with impaired radioprotection. Whereas all (12 of 12) recipients transplanted with control-transduced cells survived, only 42% (5 of 12) of recipients of KLF7 transduced cells survived more than 2 weeks after transplantation (P < .003). An analysis of moribund recipients revealed marked bone marrow hypocellularity indicative of engraftment failure (data not shown).
When we performed these experiments using a reduced multiplicity of infection to achieve a lower transduction efficiency, all recipient mice survived at least 6 weeks. Again, whereas control-transduced cells were readily detected at near input levels (on average, 40% of nucleated blood cells), minimal contribution of KLF7-transduced cells was observed in all lineages, except T cells (Figure 3C-D; see Figure 6). Importantly, no cases of leukemia were observed during the 3-month observation period. Thus, overexpression of KLF7 is not sufficient to induce leukemia in mice in the short term.
To investigate whether KLF7 overexpression impairs homing to the bone marrow, we sorted GFP+ KLS bone marrow cells from cultures transduced with KLF7 or control lentivirus, and injected the cells into lethally irradiated recipient mice. Twenty hours later, a similar number of GFP+ cells were recovered from the bone marrow of mice transplanted with KLF7 or control lentivirus-transduced cells (Figure 3F). Thus, the lack of engraftment of KLF7 overexpressing cells is not the result of impaired homing of KLF7 overexpressing cells to the bone marrow.
Overexpression of KLF7 suppresses bone marrow stem and progenitor cells
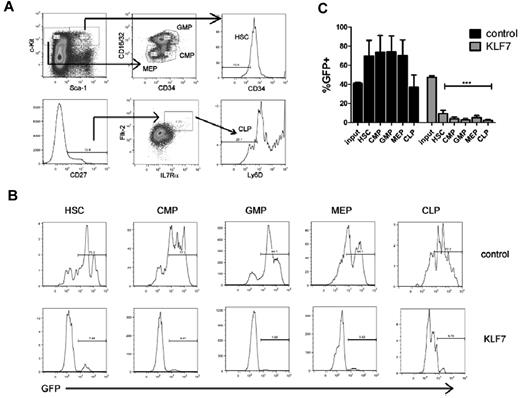
To determine whether KLF7 overexpression suppresses hematopoiesis at the hematopoietic stem cell or more committed progenitor level, we characterized the contribution of KLF7-transduced cells to defined hematopoietic progenitor cell populations in mice 6 weeks after transplantation. KLF7 overexpression was associated with a marked suppression of all hematopoietic progenitor populations, including HSCs (Figure 4). Of note, despite the relatively high contribution of KLF7-transduced cells to the T-cell lineage, KLF7-transduced common lymphoid progenitors (CLPs) were markedly reduced.
KLF7 overexpression suppresses hematopoietic stem and progenitors. Bone marrow stem and progenitor populations were analyzed by flow cytometry 6 weeks after transplantation with control or KLF7 lentivirally transduced cells. (A) Gating strategy used to identify HSCs (KLS CD34−), common myeloid progenitors (CMP, Lin− c-Kit+ Sca-1− CD16/32lo CD34+), granulocyte-monocyte progenitors (GMPs, Lin− C-Kit+ Sca-1− CD16/32hi CD34+), and megakaryocytic-erythroid progenitors (MEP, Lin− c-Kit+ Sca-1− CD16/32lo CD34−; top panel) and common lymphoid progenitors (CLP, Lin− CD27+ IL7R+ Flk-2+ Ly6D−; bottom panel). Cells are first gated on live, donor, and lineage-negative populations. (B) Representative histograms showing GFP expression in the indicated progenitor cell population. (C) A summary of the percentages of donor cells that are GFP+ for the indicated progenitor population is provided. Input refers the percentage of bone marrow cells that were GFP+ at the time of transplantation. Data represent the mean ± SEM of 2 independent experiments. ***P < .001.
KLF7 overexpression suppresses hematopoietic stem and progenitors. Bone marrow stem and progenitor populations were analyzed by flow cytometry 6 weeks after transplantation with control or KLF7 lentivirally transduced cells. (A) Gating strategy used to identify HSCs (KLS CD34−), common myeloid progenitors (CMP, Lin− c-Kit+ Sca-1− CD16/32lo CD34+), granulocyte-monocyte progenitors (GMPs, Lin− C-Kit+ Sca-1− CD16/32hi CD34+), and megakaryocytic-erythroid progenitors (MEP, Lin− c-Kit+ Sca-1− CD16/32lo CD34−; top panel) and common lymphoid progenitors (CLP, Lin− CD27+ IL7R+ Flk-2+ Ly6D−; bottom panel). Cells are first gated on live, donor, and lineage-negative populations. (B) Representative histograms showing GFP expression in the indicated progenitor cell population. (C) A summary of the percentages of donor cells that are GFP+ for the indicated progenitor population is provided. Input refers the percentage of bone marrow cells that were GFP+ at the time of transplantation. Data represent the mean ± SEM of 2 independent experiments. ***P < .001.
KLF7 suppression of HSPCs is not mediated by Cdkn1a
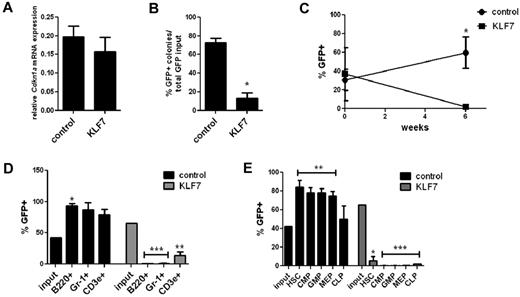
Previous studies have shown that Cdkn1a (p21 cip1/waf1) is a direct transcriptional target of KLF7, and overexpression of KLF7 in neuronal cells results in increased CDKN1A expression.8,17 Given the known role of CDKN1A in regulating HSPC cycling and self-renewal capacity,12,18,19 we next asked whether the growth-suppressing effects of KLF7 on HSPCs are mediated by CDKN1A. Surprisingly, no change in Cdkn1a mRNA expression was observed in KLS cells transduced with KLF7 lentivirus (Figure 5A). We then repeated the in vitro growth assay and bone marrow repopulating assays using Cdkn1a−/− hematopoietic cells. Similar to WT cells, significantly fewer KLF7 transduced Cdkn1a−/− cells formed colonies compared with controls (Figure 5B). Furthermore, hematopoietic reconstitution by KLF7 transduced Cdkn1a−/− cells was markedly impaired. Again, we observed a relative preservation of T cells, which, although still significantly lower that input levels, were less repressed that other lineages (Figure 5C-E). Thus, the effects of KLF7 on the suppression of cell growth and marrow reconstitution are not dependent on CDKN1A.
KLF7 suppression of HSPC function is not mediated by CDKN1A. (A) Cdkn1a mRNA expression relative to β-actin in GFP+ KLS cells 72 hours after transduction with lentivirus expressing KLF7 or vector alone. (B) Bone marrow cells exposed to KLF7 or control lentivirus were plated in methylcellulose and colonies scored on day 7. The percentage of GFP+ colonies is shown, normalized for the transduction efficiency. Data represent 2 independent experiments, plated in duplicate. *P = .0002. Cdkn1a−/− bone marrow was transduced with KLF7 or control lentivirus and transplanted into irradiated congenic recipients. Six weeks after transplantation, the percentage of donor leukocytes (C), cells of the indicated hematopoietic lineage (D), or indicated progenitor cell population (E) that was GFP+ was determined. Time zero in panel C refers to the transduction efficiency. Input indicates the percentage of GFP+ cells at the time of transplantation. Data represent 2 individual experiments, each with 5 or 6 mice per genotype. *P < .01. **P < .05. ***P < .0001.
KLF7 suppression of HSPC function is not mediated by CDKN1A. (A) Cdkn1a mRNA expression relative to β-actin in GFP+ KLS cells 72 hours after transduction with lentivirus expressing KLF7 or vector alone. (B) Bone marrow cells exposed to KLF7 or control lentivirus were plated in methylcellulose and colonies scored on day 7. The percentage of GFP+ colonies is shown, normalized for the transduction efficiency. Data represent 2 independent experiments, plated in duplicate. *P = .0002. Cdkn1a−/− bone marrow was transduced with KLF7 or control lentivirus and transplanted into irradiated congenic recipients. Six weeks after transplantation, the percentage of donor leukocytes (C), cells of the indicated hematopoietic lineage (D), or indicated progenitor cell population (E) that was GFP+ was determined. Time zero in panel C refers to the transduction efficiency. Input indicates the percentage of GFP+ cells at the time of transplantation. Data represent 2 individual experiments, each with 5 or 6 mice per genotype. *P < .01. **P < .05. ***P < .0001.
KLF7 overexpression induces genes associated with lymphocyte development
Because KLF7 is a transcription factor, we performed RNA expression profiling on KLF7 transduced HSPCs to address the mechanisms by which KLF7 inhibits HSPC function. GFP+ KLS cells were isolated by sorting 72 hours after transduction of c-Kit+-selected WT bone marrow cells and RNA expression profiling was performed using the Affymetrix MoGene 1.0ST array. Using a false discovery rate of < 10% and a criteria of a minimum of ≥ 2-fold difference in expression, 75 genes were identified whose expression was increased by KLF7 expression and 14 that were decreased (supplemental Table 3). Notably, the list of up-regulated genes includes the previously described KLF7 targets Tuba1a and L1cam,20,21 both of which are direct transcriptional targets of KLF7 that are important for neurogenesis. However, expression of the most well-characterized growth regulatory factors induced by KLF7, Cdkn1a and Cdkn1b (p27, kip-1),8 were not significantly different between the KLF7-transduced and control samples. Among the enriched gene sets, gene set enrichment analysis identified significant enrichment for NF-κB and T cell receptor signaling pathways (supplemental Figure 3). However, with the exception of the thymic homing receptor CCR7, no significant difference in the expression of genes known to play a significant role in the determination of T-cell fate or thymic homing22,23 was observed between control and KLF7-overexpressing cells (supplemental Table 4).
KLF7 overexpression promotes survival of DN thymocytes
To further elucidate the role of KLF7 in T-cell development, we analyzed T-cell subsets in the blood, spleen, and thymus in mice transplanted with KLF7 or control transduced bone marrow (Figure 6A-B). Although GFP+ T cells in the blood and spleen were significantly lower than input GFP+ values, they were higher than GFP+ CLPs (Figure 4C). In the thymus, none of the T-cell subsets analyzed (DN, DP, CD4 SP, and CD8 SP) was significantly different from input values (Figure 6B). Thus, whereas enforced expression of KLF7 suppresses bone marrow stem and progenitor populations (including CLPs), thymic, and to some extent splenic and blood, T cells are preserved. We next examined the effect of KLF7 overexpression on thymocyte survival (Figure 6C-D). Enforced expression of KLF7 resulted in the significant suppression of apoptosis in DN thymocytes. Conversely, in later stages of thymocyte development (DP, CD4 SP, and CD8 SP), a trend to increased apoptosis is observed.
KLF7 overexpression supports early thymocyte development. Blood, spleen, and thymic T-cell populations were analyzed by flow cytometry 6 weeks after transplantation with control or KLF7 lentivirally transduced cells. (A) Representative flow plot for the thymus, with each of the 4 major thymic populations (CD4+ CD8+ [DP], CD4− CD8− [DN], CD4+ CD8− [CD4 SP], and CD4− CD8+ [CD8 SP]) subsequently gated for GFP. (B) CD4 SP and CD8 SP T-cell subsets in the blood and spleen and DN, DP, CD4 SP, and CD8 SP subsets in the thymus as gated in the representative flow plot (A) were analyzed, and the percentage of GFP+ for each population from each tissue is shown. Input refers to the percentage GFP+ cells at the time of transplantation. *P < .05 compared with input values. (C) GFP+ thymic populations as sorted in panel A were analyzed for annexin V and DAPI staining. The results are summarized in panel D, showing the percentage of the GFP+ fraction of the indicated thymic population that were annexin V+. Data represent 2 independent experiments with 4 mice per condition. *P < .05.
KLF7 overexpression supports early thymocyte development. Blood, spleen, and thymic T-cell populations were analyzed by flow cytometry 6 weeks after transplantation with control or KLF7 lentivirally transduced cells. (A) Representative flow plot for the thymus, with each of the 4 major thymic populations (CD4+ CD8+ [DP], CD4− CD8− [DN], CD4+ CD8− [CD4 SP], and CD4− CD8+ [CD8 SP]) subsequently gated for GFP. (B) CD4 SP and CD8 SP T-cell subsets in the blood and spleen and DN, DP, CD4 SP, and CD8 SP subsets in the thymus as gated in the representative flow plot (A) were analyzed, and the percentage of GFP+ for each population from each tissue is shown. Input refers to the percentage GFP+ cells at the time of transplantation. *P < .05 compared with input values. (C) GFP+ thymic populations as sorted in panel A were analyzed for annexin V and DAPI staining. The results are summarized in panel D, showing the percentage of the GFP+ fraction of the indicated thymic population that were annexin V+. Data represent 2 independent experiments with 4 mice per condition. *P < .05.
To address whether KLF7 is necessary for T-cell development, we analyzed blood, splenic, and thymic T cells in the Klf7−/− fetal liver chimeras. No differences were noted between the frequencies of CD4 SP and CD8 SP populations among the Klf7−/− cells and Klf7+/+ cells in the blood and spleens of the chimeric mice, and similarly there were no significant differences in the frequencies of the T-cell precursors in the thymus (supplemental Figure 4). The possibility remains, however, that more subtle defects in T-cell development may occur in the absence of KLF7, and the possible contribution of KLF7 to T-cell development and/or trafficking warrants further investigation.
Discussion
Although there is considerable evidence that KLF family members regulate specific hematopoietic lineages, there are limited data about their contribution to HSC maintenance. Matsumoto et al reported that loss of KLF6 impaired yolk sac hematopoiesis and, conversely, that overexpression of KLF6 resulted in enhanced hematopoietic potential of embryonic stem cells.9 Moreover, there is evidence that KLF4, KLF2, and KLF5 support self-renewal of embryonic stem cells, and expression of KLF4, in conjunction with Oct4, Sox2, and c-Myc, is widely used to reprogram somatic cells to induced pluripotent stem cells.24,25 Jiang et al demonstrated that KLF4, KLF2, and KLF5 play redundant roles in maintaining embryonic stem cells, sharing many common targets, and loss of all 3 factors simultaneously was required to induce differentiation of embryonic stem cells.25 Herein, we show that enforced expression of KLF7 results in impaired myeloid differentiation and decreased HSC activity in mice. However, KLF7 does not appear to be required for normal HSC function, as the long-term repopulating activity and self-renewal of Klf7−/− cells are comparable to WT cells. Given the redundancy of KLF family members in regulating embryonic stem cells, however, and the fact that we detect expression of the majority of the KLF family members in HSCs, it is quite possible that other KLFs may compensate for the loss of KLF7 in regulating HSC function.
The molecular mechanisms by which KLF7 overexpression suppresses HSC function are not known. KLF7 has been shown to induce the expression of CDKN1A and CDKN1B, and thus we predicted that the growth inhibitory of effects of KLF7 is probably mediated by these factors. Contrary to this idea, we found that neither of these genes were up-regulated on overexpression of KLF7, and we observed a similar suppression of hematopoiesis in the Cdkn1a−/− cells with overexpression of KLF7 compared with WT cells. Therefore, the growth suppressive effects of KLF7 in HSPCs are not mediated by CDKN1A, and other targets of KLF7 in these cells are responsible for inhibiting HSPC proliferation. Interestingly, RNA profiling of KLF7 overexpressing HSPCs did not reveal a dysregulation of other genes known to regulate HSC activity. Given the heterogeneous nature of the KLS HSPC population, the identification of KLF7 targets conferring growth suppression in HSCs may require profiling of a further enriched HSC population after induction of KLF7 expression. Furthermore, as discussed later in the “Discussion,” enforced KLF7 expression in HSPCs is associated with an up-regulation of genes involved in lymphocyte development, and thus perhaps the suppression of HSCs is the result of differentiation of KLF7 overexpressing cells toward the lymphocytic lineage.
Relative to HSCs and CLPs, T-cell production is preserved in mice transplanted with KLF7 overexpressing cells, implicating, for the first time, a role for KLF7 in T-cell development. Interestingly, in a profiling study of thymocyte subpopulations, Tabrizifard et al reported up-regulation of KLF7 during the preDP to smDP transition.26 This transition involves a switch from active cell proliferation to cell cycle arrest, allowing for selection and further differentiation of cells with T-cell receptors of the correct specificity. In agreement with this finding, our own quantitative RT-PCR data show a 5-fold increase in KLF7 mRNA expression from the DN to DP stage (supplemental Figure 1). Further supporting a role for KLF7 in T-cell development, our RNA profiling of KLF7 overexpressing HSPCs identified up-regulation of a number of genes that play a role in lymphocyte development and activation, including Ccl22, Pira2, Pira3, Pira11, Lilrb3, Ccl17, H2-Aa, Ccr1, Cd83, and H2-DMb2, as well as antiapoptotic factors, including Serpinb9, Spp1, Traf1, and Bcl2a1. On the other hand, Klf7−/− cells produce a grossly similar profile of T-cell subsets to Klf7+/+ cells, suggesting that KLF7 is not absolutely necessary for T-cell development. However, the mice used in these analyses were 4-10 months after transplantation; therefore, a role for KLF7 in early thymic development remains a possibility. Moreover, numerous other KLF family members, including KLF2, KLF4, KLF5, KLF6, KLF10, and KLF13, have been implicated in T-cell development.7 Thus, redundancy in KLF function, with compensation for KLF7 loss by another family member, could explain the ability of Klf7−/− cells to produce a normal T-cell profile.
Flotho et al identified KLF7 as 1 of 14 genes that independently predicted therapy resistance and relapse of ALL in a large cohort of pediatric patients.2 Notably, inhibition of the cell cycle and DNA replication was a common function of many of the dysregulated genes in that study, indicating that suppression of leukemic cell growth may contribute to chemotherapy resistance. Of particular note is an up-regulation of B-cell lymphoma 2-related protein A1 (Bcl2a1) on enforced expression of KLF7 (real-time RT-PCR confirmed an ∼ 2-fold increase in expression of Bcl2a1 in KLS cells overexpressing KLF7; data not shown). Bcl2a1 is a pro-survival member of the BCL2 family, and it has been previously been shown to be a pre-T cell receptor–induced regulator of thymocyte survival. Enhanced expression of BCL2A1 has been described in multiple tumor types, including leukemias and lymphomas,27 and overexpression is associated with chemotherapy-resistant and advanced disease.28,29 Furthermore, overexpression of BCL2A1 in cell lines has been shown to confer chemotherapy resistance,30-32 and conversely knockdown of BCL2A1 was found to sensitize malignant B-cell lines to chemotherapy-induced apoptosis.33 Small-molecule inhibitors of BCL2A1 are currently being tested in multiple cancers, including leukemia.27 The up-regulation of BCL2A1 and other antiapoptotic and growth regulatory factors on KLF7 overexpression suggests that KLF7 may contribute to therapy resistant disease by promoting lymphocyte survival. Indeed, our data show that KLF7 overexpression leads to reduced apoptosis of DN thymocytes, further supporting the idea that aberrantly high KLF7 expression in lymphoblastic leukemic cells may contribute to enhanced survival and resistance to chemotherapy. Finally, BCL2A1 is a target of NF-κB, and significant up-regulation of this pathway was identified by gene set enrichment analysis in KLF7-overexpressing HSPCs. Activation of this pathway plays a crucial role in normal lymphopoiesis, and enhanced NF-κB activity is associated with numerous lymphoid and myeloid malignancies.34
Taken together, this study suggests that KLF7, although not necessary for normal HSPC repopulating and self-renewal activities, may play a role in regulating thymocyte development. Overexpression of KLF7, as seen in resistant leukemia, inhibits normal HSPC function and supports early thymocyte survival. Future studies will be aimed at determining the direct targets of KLF7 in hematopoietic cells, identifying the growth suppressing factors mediated by KLF7 in HSPCs and the role of KLF7 in thymocyte development. This information will allow for a better understanding of the role of KLF7 in normal lymphopoiesis and in leukemia. Ultimately, inhibition of KLF7 or its downstream targets could then potentially be used to sensitize chemotherapy-resistant cells, providing a targeted approach to the treatment of refractory or relapsed leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Susan Dorsey (University of Maryland) for the Klf7-deficient mice.
This work was supported by the National Institutes of Health (grants R01 CA136617 and R01 HL60772, D.C.L.). L.G.S. is a Fellow of the Pediatric Scientist Development Program and a Hyundai Hope on Wheels Scholar. The project described was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (award K12HD000850) and the Hyundai Corporation.
National Institutes of Health
Authorship
Contribution: L.G.S. participated in the experimental design, performed the experiments, analyzed the data, and participated in writing the manuscript; P.K.G. participated in the experimental design and assisted with experiments; F.O.G. and M.P.R. assisted with experiments; R.V.O. generated the Cdkn1a−/− mice; and D.C.L. participated in the experimental design and data interpretation and assisted with writing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Daniel C. Link, Washington University School of Medicine, 660 S Euclid Ave, Campus Box 8007, St Louis, MO 63110; e-mail: dlink@dom.wustl.edu.


![Figure 6. KLF7 overexpression supports early thymocyte development. Blood, spleen, and thymic T-cell populations were analyzed by flow cytometry 6 weeks after transplantation with control or KLF7 lentivirally transduced cells. (A) Representative flow plot for the thymus, with each of the 4 major thymic populations (CD4+ CD8+ [DP], CD4− CD8− [DN], CD4+ CD8− [CD4 SP], and CD4− CD8+ [CD8 SP]) subsequently gated for GFP. (B) CD4 SP and CD8 SP T-cell subsets in the blood and spleen and DN, DP, CD4 SP, and CD8 SP subsets in the thymus as gated in the representative flow plot (A) were analyzed, and the percentage of GFP+ for each population from each tissue is shown. Input refers to the percentage GFP+ cells at the time of transplantation. *P < .05 compared with input values. (C) GFP+ thymic populations as sorted in panel A were analyzed for annexin V and DAPI staining. The results are summarized in panel D, showing the percentage of the GFP+ fraction of the indicated thymic population that were annexin V+. Data represent 2 independent experiments with 4 mice per condition. *P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/120/15/10.1182_blood-2012-02-409839/4/m_zh89991297880006.jpeg?Expires=1765906205&Signature=y8MhMUi2L3lwEQWSpbANZWKXr99pttIvC-~wT8KiiNcTjodvyBBKCw3vgWm32IYlczthI15dSmtqjiYQttzCib2DR0eBums8r72IIvtexwHFkCMVEwxxsViQkNaEWAe1hveUvw4s745c2lirXBUlMxfAJsK7BbNLqw2~ufeYocmIdu~DV3ozM64ROnRzgH9c6RrIw2n47e-K2WfXSUoRGvs6UXBOcwvoBZPIxdF3R6QM~b4rlYF7YiKcmgNVA91Edu1A3tyaI4bY~lD6dg8zaYju3g4SVgmSnT2osnBrFscz-3qPVIikDoVYlnaryj~G7Gv3hUFOwzPSIKGEoMKkIg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal