Abstract
The earliest stages of natural killer (NK)–cell development are not well characterized. In this study, we investigated in different fetal hematopoietic tissues how NK-cell progenitors and their mature NK-cell progeny emerge and expand during fetal development. Here we demonstrate, for the first time, that the counterpart of adult BM Lin−CD122+NK1.1−DX5− NK-cell progenitor (NKP) emerges in the fetal liver at E13.5. After NKP expansion, immature NK cells emerge at E14.5 in the liver and E15.5 in the spleen. Thymic NK cells arise at E15.5, whereas functionally competent cytotoxic NK cells were present in the liver and spleen at E16.5 and E17.5, respectively. Fetal NKPs failed to produce B and myeloid cells but sustained combined NK- and T-lineage potential at the single-cell level. NKPs were also found in the fetal blood, spleen, and thymus. These findings show the emergence and expansion of bipotent NK/T-cell progenitor during fetal and adult lymphopoiesis, further supporting that NK/T-lineage restriction is taking place prethymically. Uncovering the earliest NK-cell developmental stages will provide important clues, helping to understand the origin of diverse NK-cell subsets, their progenitors, and key regulators.
Introduction
Natural killer (NK) cells are large granular lymphocytes that represent the third lymphoid lineage, distinct from B and T cells. NK cells are a major component of the innate immunity and play an important role in eliminating tumor cells as well as in the defense against viral infections.1 Despite significant progress in understanding NK-cell biology, the early stages of NK-lineage commitment and development remain unclear.
In adult mice and man, the BM is the main site of NK-cell development1,2 ; however, a distinct population of NK cells develops in the thymus.3 These thymic-dependent NK cells play a regulatory function by producing a variety of cytokines and display poor cytotoxic activity compared to conventional BM NK cells.3 The earliest progenitor committed to the NK-cell lineage (NKP) has been identified in adult mouse BM as having the Lin−CD122+NK1.1−DX5− phenotype.4 However, we have recently shown that a sizable fraction of Lin−CD122+NK1.1−DX5− cells in adult mouse BM sustains the ability to also generate T cells, thus representing a bipotent NK/T-cell progenitor.5
The generation of all blood cell lineages depends on a small population of HSCs.6 The liver is the main site of hematopoiesis during mouse fetal development.7 HSCs in the embryo originate within the intraembryonic sites: aorta-gonad-mesonephros region derived from the paraaortic splanchnopleura and placenta, as well as in the extra embryonic site in the yolk sac.7-10 The fetal liver starts to be colonized by hematopoietic progenitors between E10 and E12, and from E12 to E15 the number of HSCs increases exponentially.11,12 The thymic rudiment develops in the mouse fetus by E10, and its colonization starts between E11 and E12, whereas the spleen develops as a hematopoietic organ at E13 and hematopoietic progenitors are first detected around E15.12,13 Hematopoietic progenitor cells appear in the femoral BM at E17, and after birth the BM becomes the main site for hematopoiesis while the hematopoietic activity in the liver and spleen declines.11,12
Multiple fetal hematopoietic tissues, including the liver, thymus, and spleen, contain hematopoietic progenitors shown to have potential to generate NK cells.14-22 However, when and where NK cell–restricted progenitors emerge and expand during ontogeny has yet to be established. Previous studies have shown that NK1.1+ cells were first detected in the fetal thymus and liver at E14 and in the spleen at E16; while the proportion of NK cells expanded in the spleen with age, these cells decreased in the fetal thymus and liver.23 More recent studies identified NK1.1+ NK cells in the fetal thymus at E13 but not in the fetal liver between E13 and E16.24 Importantly, in previous work only NK1.1 expression, which is not NK-cell specific,25 was used to identify NK cells, and the emergence and expansion of distinct NK-cell subsets in different hematopoietic tissues were not investigated nor IL-7Rα+ thymic NK (tNK) cells.
In this study, we investigated in different fetal hematopoietic tissues when NK-cell progenitors and their mature NK-cell progeny emerge and expand during fetal development. We demonstrate here that Lin−CD122+NK1.1−DX5− NKPs with combined NK- and T-cell potential emerge first in the liver at E13.5, followed by the generation and expansion of distinct subsets of mature NK cells in different hematopoietic tissues.
Methods
Mice
C57BL/6 mice were used at 6-12 weeks of age for isolation of hematopoietic progenitors and mature NK cells. Timed pregnancies were determined as described previously26 ; the day the vaginal plug was observed was considered as E0.5. All mice were maintained under specific pathogen-free conditions at Lund University Animal Facility. The Ethical Committee at Lund University approved all performed experiments.
Tissues
Tissues were isolated from fetuses with the use of dissecting microscope as previously described.26 Cell suspensions were prepared from fetal livers, spleens, and thymuses by gently breaking up the tissues in PBS containing 5% FCS (Sigma-Aldrich) and filter through a 70-μm cell strainer (BD Biosciences). Total tissue cellularities were counted by automated hematology analyzer (Sysmex KX21; Sysmex Corporation) or with the use of the Neubauer hematocytometer.
Abs, flow cytometry, and cell sorting
Before staining single-cell suspensions with mAbs against specific surface markers, Fc receptors were blocked by incubation with 2.4G2 (anti-FcRIII). Abs were purchased from BD Biosciences, eBioscience, or BioLegend unless otherwise indicated. mAbs (conjugated with different fluorochromes) used to stain cell surface markers were: B220 (RA3-6B2), CD19 (1D3), TCRβ (H57-597), CD11c (N418), CD3 (17A2), CD122 (TM-β1), CD49b (DX5), NK1.1 (PK136), CD11b/Mac-1 (M1/70), CD127 (A7R34), Ly49D (4E5), CD25 (3C7, 7D4, PC61), Ter119 (LY-76), NKp46 (29A1.4), CD43 (S7), Gr-1 (RB6-8C5), CD4 (H129.19), Thy1.2 (53-2.1), CD8α (53-6.7), and CD107a (1D4B). Fluorescence minus one controls were used to determine the positive signal. Biotinylated Abs were visualized by streptavidin-PE or streptavidin-PECy7 and purified Abs by polyclonal goat anti–rat-Tricolor (both from Caltag). 7-aminoactinomycin D (Sigma-Aldrich), TO-PRO-1 iodide (1mM; Invitrogen), or propidium iodide (1.0 mg/mL; Invitrogen) were used to exclude dead cells from the analysis. For the analysis of NK-cell progenitors and NK-cell compartments, the mixture of the following Abs was used to define the lineage negative (Lin−) population in the fetal liver: anti-CD19, Gr-1, CD3, CD8, and Ter119; and in the fetal thymus: anti-CD19, Gr-1, CD3, CD4, CD8, and Ter119. Samples were analyzed on LSR II (BD Biosciences), between 50 000 and 2 000 000 events were collected, and analysis was performed with FlowJo software (Version 8.8.6; TreeStar).
The Lin−CD122+NK1.1−DX5− NKPs were sorted from 14.5-day-old fetal liver as previously described.5 Cells were first enriched for CD122+ cells by incubation with anti-CD122 biotinylated Ab, anti-biotin Micro beads (Miltenyi Biotec), and then positive selection was done on 25 LS MACS column (Miltenyi Biotec). The enriched CD122+ cells were next stained with directly conjugated Abs against the following lineage markers: anti-CD19, Gr-1, CD3, CD8, and Ter119 as well as anti-NK1.1 and DX5.
For sorting Lin−CD122+NK1.1−DX5− NKPs, liver cells from 18.5-day-old fetuses were first depleted of Lin+ cells by incubating with a cocktail of purified antilineage Abs (anti-CD3, CD8, CD19, Gr-1, Ter119) and subsequent incubation with sheep anti-rat IgG (Fc) conjugated immunomagnetic beads (Dynal). The Lin− cells were isolated on a magnetic particle concentrator (MPC-6; Dynal). To visualize the remaining Lin+ cells, Lin− cells were stained with goat F(ab)2 anti–rat Ig(H+L) Tricolor conjugate (Caltag), as well as directly conjugated Abs against the T and NK-cell lineage markers CD3, NK1.1 and DX5. Finally, cells were stained with directly labeled anti-CD122-PE or CD122-biotinylated Abs that were visualized by streptavidin-PE. The Lin−CD122+NK1.1−DX5− cells were purified using double-sorting strategy: first, CD122+ cells were sorted, followed by sorting of the CD122+DX5−NK1.1− cell population.
The Lin− and Lin−CD122− fetal liver cell populations containing HSCs and hematopoietic progenitor cells were used as a positive control. All sorts for indicated cell phenotypes were performed on a BD FACSAria (BD Biosciences) with average purity of 93%.
OP9 and OP9-DL1 cultures
OP9 and OP9-DL1 stroma cells were kindly provided by Dr J. C. Zúñiga-Pflücker (Department of Immunology, University of Toronto, Sunnybrook Research Institute, Toronto), and cultures were performed as previously described.5,27 NKPs purified from liver from 14.5- and 18.5-day-old fetuses were plated on previously established ∼ 80% confluent stroma cell monolayers in 48- or 96-well plates in Opti MEMplus GlutaMax medium (Invitrogen) containing 10% FCS (Sigma-Aldrich), 1% penicillin/streptomycin (Sigma-Aldrich), 1% 10−2M 2-Mercaptoethanol (Sigma-Aldrich) supplemented with cytokines with the following final concentrations: Flt3 ligand (FL; 25 ng/mL), IL-7 (20 ng/mL), KIT ligand (KL; 25 ng/mL), IL-15 (25 ng/mL), and IL-2 (50 ng/mL); (all cytokines purchased from PeproTech). Cells were cultured at 37°C for 21 days with half medium changes weekly. Cells were harvested and analyzed by flow cytometry for B (CD19+), T (NK1.1−CD25+Thy1.2+TCRβ+ or NK1.1−CD25+Thy1.2+), conventional NK (cNK; TCRβ−NK1.1+DX5+, or Lin−CD122+CD25−NK1.1+IL-7Rα−, or TCRβ−NK1.1+DX5+NKp46+), NK.1.1+ T (TCRβ+NK1.1+CD25+Thy1.2+), and tNK (CD122+CD25−NK1.1+Ly49D−IL-7Rα+) cell committed progeny.
Myeloid lineage potential assays
Purified single Lin−CD122+NK1.1−DX5− NKPs and Lin−CD122− cells (used as positive controls) from liver from 18.5-day-old fetuses were sorted into serum-free medium (X-vivo 15; Lonza Walkersville) with 0.5% BSA (StemCell Technologies), 10% FCS (Sigma-Aldrich), 1% penicillin/streptomycin (Sigma-Aldrich), 1% l-glutamine (Sigma-Aldrich), 1% 10−2M 2-mercaptoethanol (Sigma-Aldrich), supplemented with cytokines with final concentrations as follows: FL (50 ng/mL), KL (50 ng/mL), TPO (50 ng/mL), IL-3 (10 ng/mL), G-CSF (50 ng/mL) and GM-CSF (20 ng/mL) and cultured at 1 cell per well in Terasaki plates for 12 days at 37°C. After 7 and 12 days, cultures were evaluated under the inverted microscope and the size of proliferating clones was scored.
Gene expression analysis by RT-PCR
Clones generated from single Lin−CD122+NK1.1−DX5− fetal liver NKPs in OP9-DL1 stroma co-cultures that were positive for T-cell lineage potential by flow cytometry were also analyzed for T-lineage gene expression, Ptcra. Thymocytes from adult mice were used as a positive control, and OP9-DL1 stroma cells cultured without hematopoietic cells were used as negative control. RT-PCR was performed as previously described.5 The primers used for nested muliplex RT-PCR were as follows: Hprt-1, 5′-GGGGGCTATAAGTTCTTTGC-3′; Hprt-2, 5′-GTTCTTTGCTGACCTGCTGG-3′; Hprt-3, 5′-TGGGGCTGTACTGCTTAACC-3′; Hprt-4, 5′-TCCAACACTTCGAGAGGTCC-3′; Ptcra-1, 5′-CTCTACCATCAGGCATCG-3′; Ptcra-2, 5′-GGCAGTGCCCTAGACGCC-3′; Ptcra-3, 5′-CTCCTGGCTGTCGAAGATTCC-3′; and Ptcra-4, 5′-GAAGCAGTTTGAAGAGGAGC-3′. Primers 1 plus 4 and 2 plus 3 are outer and inner primer pairs, respectively.
Functional NK-cell analysis
Cells from livers and spleens isolated from fetuses at indicated times were plated at a cell concentration of 2 × 106 cells/mL in round-bottomed 96-well plates in RPMI medium (PAA) containing 10% FCS (Sigma-Aldrich), 1% penicillin/streptomycin (Sigma-Aldrich), 1% l-glutamine (Sigma-Aldrich), and 1% 10−2M 2-mercaptoethanol (Sigma-Aldrich). NK cells were specifically activated by 2-hour incubation with a purified anti-NK1.1 Ab at 37°C. Cells incubated in medium alone were used as a negative, whereas cells activated with phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich) and ionomycin (Sigma-Aldrich) were used as a positive control. Afterwards, cells were washed and stained with anti-CD3, CD19, Gr-1, CD8, Ter119, CD122, NKp46, DX5, and CD107a Abs. The activation of Lin−CD3−CD122+DX5+NKp46+ NK cells was evaluated based on the surface expression of the cytotoxic marker CD107a (lysosomal associated membrane protein 1; LAMP-1) by flow cytometry (LSRII; BD Biosciences).
Statistics
Results were expressed as means ± SD. The statistical significances between groups were determined with the Student t test.
Results
Emergence of fetal Lin−CD122+NK1.1−DX5− NKPs
Studies in adult mouse BM have shown that up-regulation of the IL-2Rβ (CD122) marks the commitment to the NK lineage, and NKPs have been shown to have the Lin−CD122+NK1.1−DX5− phenotype.4 So far, the identity of NK-cell lineage-committed progenitor during fetal development has not been established. In search for a candidate NKP in the mouse embryo, we performed time course studies and found that the fetal equivalent of adult BM Lin−CD122+NK1.1−DX5− NKPs emerged in the fetal liver at E13.5 (Figure 1A). The total number of NKPs expanded between E13.5 and E17.5 (Figure 1A-C). In agreement with the finding that hematopoietic progenitors start to colonize fetal BM around E17,11,12 NKPs were clearly present in the peripheral blood (Figure 1D) and in the BM at E18.5 (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The fact that the total number of NKPs slightly decreased at E18.5 in the fetal liver coincided with NKPs being found in the fetal BM as well as in circulation most probably reflects the process of NKPs seeding into the BM. In agreement with the finding in adult mice,1,3,4,28 NKPs were also present at E18.5 in the fetal thymus and fetal spleen (supplemental Figure 1).
Emergence of Lin−CD3−CD122+NK1.1−DX5− NK-cell progenitors in different embryonic sites. (A) Typical FACS profiles of Lin−CD3−CD122+NK1.1−DX5− NK-cell progenitors analyzed in the liver from 12.5- to 18.5-day-old fetuses. (B) Total number of fetal liver cells and (C) total number of Lin−CD3−CD122+NK1.1−DX5− NK-cell progenitors. Data represent mean ± SD values from 5 to 14 livers from fetuses from 2 to 3 litters. Text above the plots and arrows indicate the gating strategies. Numbers in FACS profiles show mean frequencies of populations within the indicated gates relative to total fetal liver cells. Profile from the adult BM is included as a control. The 0 indicates values below the detection level. (D) Representative profiles of Lin−CD3−CD122+NK1.1−DX5− NK-cell progenitors in peripheral blood from 12.5- to 18.5-day-old fetuses. Numbers show mean frequencies from 5 fetuses from 2 litters analyzed per time point in 2-3 replicates. *P < .05, **P < .01, and ***P < .0001.
Emergence of Lin−CD3−CD122+NK1.1−DX5− NK-cell progenitors in different embryonic sites. (A) Typical FACS profiles of Lin−CD3−CD122+NK1.1−DX5− NK-cell progenitors analyzed in the liver from 12.5- to 18.5-day-old fetuses. (B) Total number of fetal liver cells and (C) total number of Lin−CD3−CD122+NK1.1−DX5− NK-cell progenitors. Data represent mean ± SD values from 5 to 14 livers from fetuses from 2 to 3 litters. Text above the plots and arrows indicate the gating strategies. Numbers in FACS profiles show mean frequencies of populations within the indicated gates relative to total fetal liver cells. Profile from the adult BM is included as a control. The 0 indicates values below the detection level. (D) Representative profiles of Lin−CD3−CD122+NK1.1−DX5− NK-cell progenitors in peripheral blood from 12.5- to 18.5-day-old fetuses. Numbers show mean frequencies from 5 fetuses from 2 litters analyzed per time point in 2-3 replicates. *P < .05, **P < .01, and ***P < .0001.
The earliest time when the Lin−CD122+NK1.1−DX5− NKPs were found in the peripheral blood was E14.5 (Figure 1D); that is a day later than in fetal liver (Figure 1A), indicating that NKPs arise in the fetal liver from HSCs or multipotent progenitors rather than already committed NKPs migrating through the circulation from the other hematopoietic sites in the early embryo.
Collectively, these data suggest that fetal NKPs emerge in the liver, expand, and migrate to the other embryonic sites through the circulation.
Fetal NKPs have robust NK potential and sustain ability to produce T cells
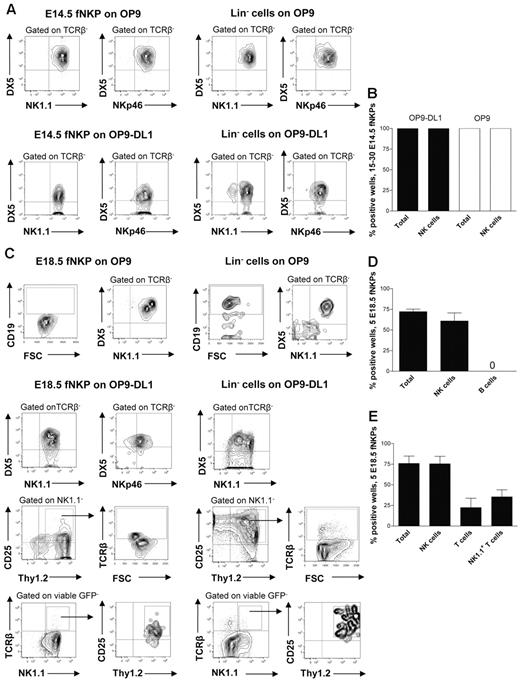
At E13.5 Lin−CD122+NK1.1−DX5− NKPs were too few to allow purification for functional evaluation (E.S. and P.C., unpublished observation, July 2011); however, phenotypically defined NKPs could be purified from the fetal liver at E14.5 and co-cultured on OP9 and OP9DL-1 stroma to determine to what degree they can produce NK cells (supplemental Figure 2B).5,30 It has been previously established that OP9 and OP9-DL1 cell lines efficiently support generation of NK cells from fetal and adult mouse hematopoietic progenitors.5,30 When 15-30 E14.5 NKPs were cultured on OP9 and OP9-DL1 cells, all cultures contained large numbers of TCRβ−NK1.1+DX5+ NK cells (Figure 2A-B). The fact that the same proportion of fetal NKPs plated either on OP9 or OP9-DL1 stroma expressing Notch ligand produced NK cells suggests that the Notch signaling does not restrict the NK-cell generation. To further confirm the NK-cell phenotype of generated progeny, the expression of the most specific NK-cell marker NKp46 was investigated.31 Clones derived from E14.5 NKPs contained TCRβ−DX5+NKp46+ NK cells, further supporting their NK-cell identity and final maturation in the in vitro culture system (Figure 2A-B). These results suggest that the Lin−CD122+NK1.1−DX5− NKPs that emerge in the fetal liver already at E13.5 have considerable NK-lineage potential.
Fetal liver Lin−CD3−CD122+NK1.1−DX5− NK-cell progenitors efficiently generate NK and T cells in vitro. Fifteen to thirty and 5 Lin−CD3−CD122+NK1.1−DX5− NKPs as well as 100 Lin− cells (control) sorted from the livers of 14.5- and 18.5-day-old fetuses were cultured on OP9 and OP9-DL1 stroma cell lines with the following cytokines: IL-7 (first week only), KL, FL, IL-2, and IL-15. After 21 days, cells were harvested and evaluated for the presence of NK (TCRβ-NK1.1+DX5+ or TCRβ−NKp46+), B (CD19+), T (NK1.1−CD25+Thy1.2+, or NK1.1−CD25+Thy1.2+TCRβ+) and NK1.1+ T cells (TCRβ+NK1.1+) by FACS. TO-PRO-1 was used to exclude dead cells, and GFP was used to exclude stroma cells from the analysis. (A) Representative FACS profiles of cells generated from fetal liver E14.5 NKPs and Lin− cells cultured on OP9 and OP9-DL1 stroma. The specific gates are indicated in the text above the plot and by arrows. (B) Mean ± SD proportion of total positive wells and wells containing NK cells generated from 15 to 30 fetal liver E14.5 NKPs cultured on OP9 (white bars) and OP9-DL1 (black bars) stroma. Data represent mean ± SD values from 3 independent experiments (2 with OP9-DL1 and 1 with OP9 stroma) with 17-24 wells analyzed in each experiment (total 58 wells). (C) Representative FACS profiles of cells generated from fetal liver E18.5 NKPs and Lin− cells cultured on OP9 and OP9-DL1 stroma. The specific gates are indicated in the text above the plot and by arrows. Mean ± SD proportion of total positive wells and wells containing NK, B, T, and NK1.1+ T cells generated from 5 fetal liver NKPs cultured on (D) OP9 and (E) OP9-DL1 stroma. Data represent mean ± SD values from 5 independent experiments with 24-72 wells analyzed in each experiment (total 220 wells).
Fetal liver Lin−CD3−CD122+NK1.1−DX5− NK-cell progenitors efficiently generate NK and T cells in vitro. Fifteen to thirty and 5 Lin−CD3−CD122+NK1.1−DX5− NKPs as well as 100 Lin− cells (control) sorted from the livers of 14.5- and 18.5-day-old fetuses were cultured on OP9 and OP9-DL1 stroma cell lines with the following cytokines: IL-7 (first week only), KL, FL, IL-2, and IL-15. After 21 days, cells were harvested and evaluated for the presence of NK (TCRβ-NK1.1+DX5+ or TCRβ−NKp46+), B (CD19+), T (NK1.1−CD25+Thy1.2+, or NK1.1−CD25+Thy1.2+TCRβ+) and NK1.1+ T cells (TCRβ+NK1.1+) by FACS. TO-PRO-1 was used to exclude dead cells, and GFP was used to exclude stroma cells from the analysis. (A) Representative FACS profiles of cells generated from fetal liver E14.5 NKPs and Lin− cells cultured on OP9 and OP9-DL1 stroma. The specific gates are indicated in the text above the plot and by arrows. (B) Mean ± SD proportion of total positive wells and wells containing NK cells generated from 15 to 30 fetal liver E14.5 NKPs cultured on OP9 (white bars) and OP9-DL1 (black bars) stroma. Data represent mean ± SD values from 3 independent experiments (2 with OP9-DL1 and 1 with OP9 stroma) with 17-24 wells analyzed in each experiment (total 58 wells). (C) Representative FACS profiles of cells generated from fetal liver E18.5 NKPs and Lin− cells cultured on OP9 and OP9-DL1 stroma. The specific gates are indicated in the text above the plot and by arrows. Mean ± SD proportion of total positive wells and wells containing NK, B, T, and NK1.1+ T cells generated from 5 fetal liver NKPs cultured on (D) OP9 and (E) OP9-DL1 stroma. Data represent mean ± SD values from 5 independent experiments with 24-72 wells analyzed in each experiment (total 220 wells).
To further investigate the lineage potentials of the fetal NKP, Lin−CD122+NK1.1−DX5− cells were prospectively purified from the liver of 18.5-day-old fetuses (supplemental Figure 2B) and co-cultured on OP9 and OP9-DL1 stroma.5,30 We selected E18.5 based on the high total NKPs number as well as due to the fact that at this time NKPs were also present in the BM and mature NK cells were already generated.
When 5 E18.5 NKPs were co-cultured on OP9 and OP9-DL1 cells, essentially all positive wells contained large numbers of TCRβ−NK1.1+DX5+ as well as TCRβ−DX5+NKp46+ NK cells (Figure 2C-E).
The NKPs from E18.5 fetal liver cultured on OP9 stroma did not produce any CD19+ B cells, in contrast to the Lin− hematopoietic progenitors used as a positive control (Figure 2C-D).
Notably, ∼ 20% of the wells seeded with 5 Lin−CD122+NK1.1−DX5− cells from E18.5 fetal liver on OP9-DL1 stroma contained NK1.1−CD25+Thy1.2+ T cells (43% of these cells also expressed TCRβ) and 30% of the wells contained TCRβ+ T cells that were positive for NK1.1 receptor (NK1.1+TCRβ+ T cells; Figure 2C,E). Previous studies identified a subset of T cells that co-expressed the TCR with the NK1.1 and DX5 molecules25 ; however, NK T cells represent a heterogeneous population, and the exact characteristics of the NK1.1+ TCR+ T cells produced from fetal liver NKPs remain to be established.
To test the myeloid potential, a myeloid culture assay was performed in which the Lin−CD122− hematopoietic progenitors were used as a positive control. No myeloid cells were generated from E18.5 NKPs regardless of the different time points evaluated, confirming their loss of myeloid potential (Table 1).
Lin−CD122+NK1.1−DX5− fetal liver NKPs lack myeloid potential
| Experiment . | Cell population . | No. of cells plated per well . | No. of wells with small size clones (3-50 cells) . | No. of wells with medium size clones (50 cells; 10%) . | No. of wells with big clones > 10% . | Total no. of wells with clones . |
|---|---|---|---|---|---|---|
| 1 | NKP | 5 | 0 | 0 | 0 | 0/120 |
| 1 | Lin−CD122− | 10 | 11 | 6 | 35 | 52/60 |
| 2 | NKP | 5-10 | 0 | 0 | 0 | 0/240 |
| 2 | Lin−CD122− | 10 | 7 | 5 | 39 | 51/60 |
| Experiment . | Cell population . | No. of cells plated per well . | No. of wells with small size clones (3-50 cells) . | No. of wells with medium size clones (50 cells; 10%) . | No. of wells with big clones > 10% . | Total no. of wells with clones . |
|---|---|---|---|---|---|---|
| 1 | NKP | 5 | 0 | 0 | 0 | 0/120 |
| 1 | Lin−CD122− | 10 | 11 | 6 | 35 | 52/60 |
| 2 | NKP | 5-10 | 0 | 0 | 0 | 0/240 |
| 2 | Lin−CD122− | 10 | 7 | 5 | 39 | 51/60 |
Indicated cell populations were sorted from fetal liver at E18.5 and plated at the specified cell numbers into wells containing X-vivo medium containing 10% FCS and cytokines: KL+GM-CSF+G-CSF+IL-3+FL+TPO. The plates were scored at day 7 (not shown) and day 12 (data in the table) of the culture with similar results.
Taken together these data show that the fetal counterpart of adult BM Lin−CD122+NK1.1−DX5− NK-cell progenitors prospectively purified from E18.5 liver have no B and myeloid-lineage potentials, but, in addition to having an extensive NK-cell potential, NKPs sustain the ability to generate conventional T and NK1.1+TCR+ T cells.
To determine whether the combined NK- and T-cell potentials of the fetal liver NKPs reflects the existence of separate NK and T cell–restricted progenitors within the Lin−CD122+NK1.1−DX5− population or the existence of bipotent NK/T-cell progenitors in the fetal liver, single cell experiments were performed.
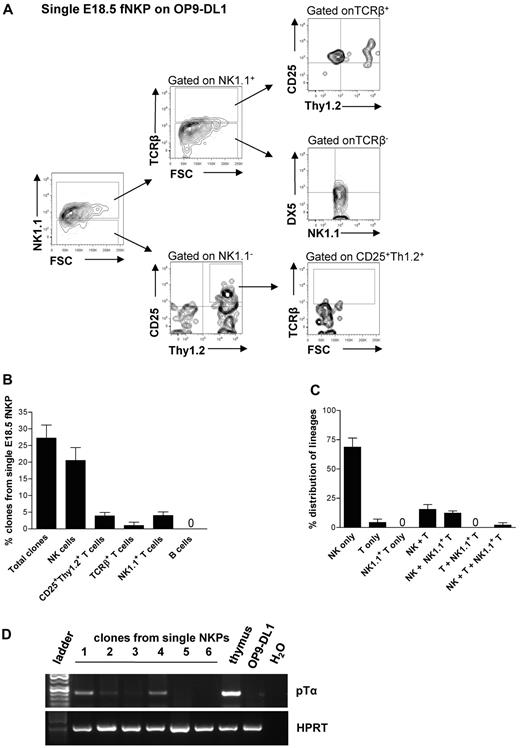
Approximately 29% of single NKPs cultured on OP9-DL1 stroma cells in the presence of FL, KL, IL-15, and IL-2 generated sizable clones that could be analyzed by flow cytometry (Figure 3A-C; supplemental Figure 3A-B). As much as 25.8% of the clones derived from single fetal NKPs contained TCRβ−NK1.1+DX5+ NK cells, 4.2% of clones had detectable NK1.1−CD25+Thy1.2+ T cells, and 4.3% of clones contained NK1.1+TCR+ T cells (Figure 3B-C). Because the levels of TCRβ expression detected by flow cytometry were low, the T-cell identity of the clones containing NK1.1−CD25+Thy1.2+ T cells was also verified based on the expression of the pre-TCRα gene (Ptcra; present in early T-cell progenitors) by RT-PCR (Figure 3D). Importantly, almost all single NKPs generating T cells also produced NK cells (Figure 3C), showing the existence of Lin−CD122+NK1.1−DX5− bipotent NK/T-cell progenitors in the fetal liver.
Single fetal liver Lin−CD122+NK1.1−DX5− NK-cell progenitors have combined NK and T-cell potential in vitro. Single Lin−CD122+NK1.1−DX5− NKPs from the liver from 18.5-day-old fetuses were directly sorted into OP9-DL1 stroma layers and cultured with IL-7 (first week), KL, FL, IL-2, and IL-15. After 21 days clones were harvested and analyzed by FACS. TO-P-RO1 was used to eliminate dead cells and GFP to exclude stroma cells from the analysis. (A) FACS profiles of representative clone generated from single fetal liver Lin−CD122+NK1.1−DX5− NKP. Text above profiles indicates gating. (B) Values are mean ± SD proportion of total proliferating clones and clones containing NK (TCRβ−NK1.1+DX5+), T (NK1.1−CD25+Thy1.2+ or NK1.1−CD25+Thy1+TCRβ+), and NK1.1+ T (NK1.1+TCRβ+) cells generated from single fetal liver NKPs. Data expressed in relation to total plated cells from 4 independent experiments, with 375 plated cells and 102 clones analyzed. (C) Values are mean ± SD proportion of clones generated from single fetal liver NKPs containing NK (TCRβ−NK1.1+DX5+), T (NK1.1−CD25+Thy1.2+ or NK1.1−CD25+Thy1+TCRβ+), and NK1.1+ T (NK1.1+TCRβ+) cells, or a combination of these. Data are expressed in relation to the total clones, from 4 independent experiments, with 375 plated cells and 102 clones analyzed. (D) Clones generated from single fetal liver NKPs (shown in panels A-C), shown to have combined NK and T-cell (CD25+Thy1.2+) potential detected by FACS were analyzed by RT-PCR for Ptcra and Hprt gene expression. Thymocytes from adult mouse were used as a positive control, and OP9-DL1 cells cultured without hematopoietic cells were used as negative control. Gel Red-stained agarose gels with resulting PCR products from RT-PCR analysis of Ptcra and Hprt. The photo has been taken using Gel logic 100 (Kodak). Representative data from 1 of 5 experiments with similar results (29 clones derived from single fetal NKPs were tested).
Single fetal liver Lin−CD122+NK1.1−DX5− NK-cell progenitors have combined NK and T-cell potential in vitro. Single Lin−CD122+NK1.1−DX5− NKPs from the liver from 18.5-day-old fetuses were directly sorted into OP9-DL1 stroma layers and cultured with IL-7 (first week), KL, FL, IL-2, and IL-15. After 21 days clones were harvested and analyzed by FACS. TO-P-RO1 was used to eliminate dead cells and GFP to exclude stroma cells from the analysis. (A) FACS profiles of representative clone generated from single fetal liver Lin−CD122+NK1.1−DX5− NKP. Text above profiles indicates gating. (B) Values are mean ± SD proportion of total proliferating clones and clones containing NK (TCRβ−NK1.1+DX5+), T (NK1.1−CD25+Thy1.2+ or NK1.1−CD25+Thy1+TCRβ+), and NK1.1+ T (NK1.1+TCRβ+) cells generated from single fetal liver NKPs. Data expressed in relation to total plated cells from 4 independent experiments, with 375 plated cells and 102 clones analyzed. (C) Values are mean ± SD proportion of clones generated from single fetal liver NKPs containing NK (TCRβ−NK1.1+DX5+), T (NK1.1−CD25+Thy1.2+ or NK1.1−CD25+Thy1+TCRβ+), and NK1.1+ T (NK1.1+TCRβ+) cells, or a combination of these. Data are expressed in relation to the total clones, from 4 independent experiments, with 375 plated cells and 102 clones analyzed. (D) Clones generated from single fetal liver NKPs (shown in panels A-C), shown to have combined NK and T-cell (CD25+Thy1.2+) potential detected by FACS were analyzed by RT-PCR for Ptcra and Hprt gene expression. Thymocytes from adult mouse were used as a positive control, and OP9-DL1 cells cultured without hematopoietic cells were used as negative control. Gel Red-stained agarose gels with resulting PCR products from RT-PCR analysis of Ptcra and Hprt. The photo has been taken using Gel logic 100 (Kodak). Representative data from 1 of 5 experiments with similar results (29 clones derived from single fetal NKPs were tested).
Emergence of conventional NK-cell subsets in different embryonic sites
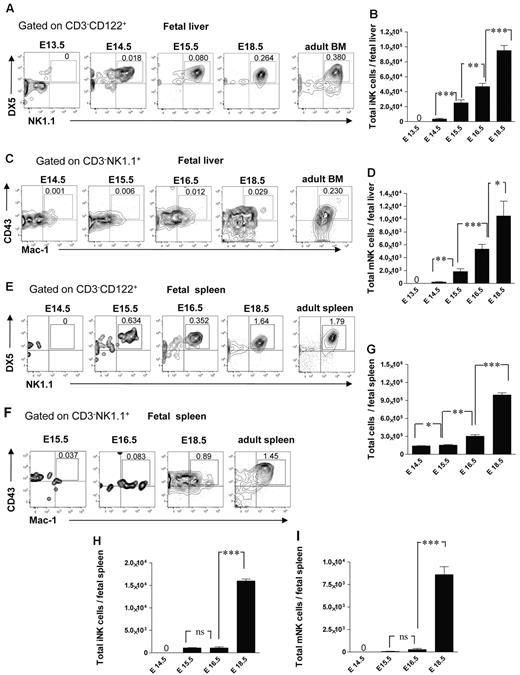
It has not been established when and where different NK-cell subsets arise during fetal development. In agreement with the finding that NKPs emerge in the fetal liver at E13.5, the CD3−CD122+NK1.1+DX5+ immature NK (iNK) cells were present in the fetal liver at E14.5 (∼ 3500 cells per liver), and their number expanded with time (Figure 4A-B). The CD3−CD122+NK1.1+DX5+CD43+Mac-1+ mature NK (mNK) cells32 emerged in the fetal liver later, ∼ 240 and 1900 mNK cells were present at E14.5 and E15.5 in the liver, respectively, and they expanded further by E16.5 (Figure 4C-D). The emergence of mNK cells in the fetal spleen occurred later compared with the fetal liver; iNK cells were first found at E15.5 (Figure 4E-F,H), only a few mNK cells (∼ 280 cells per spleen) were detectable at E16.5 but their number expanded by E18.5 (Figure 4F-G,I).
NK-cell subsets in the fetal liver and spleen. Representative FACS profiles (A) and total numbers (B) of CD3−CD122+NK1.1+DX5+ iNK and (C-D) CD3−NK1.1+CD43+Mac-1+ mNK cells at the different time points in the fetal liver. Data represent mean ± SD values from 5 to 14 livers analyzed in 12.5- to 18.5-day-old fetuses from 3 to 4 litters. Numbers in FACS profiles are mean frequencies of cell populations within the indicated gates, relative to total liver cells. Profile from the adult BM is included as a control. Representative FACS profiles of (E) CD3−NK1.1+DX5+ iNK cells and (F) CD3−NK1.1+CD43+Mac-1+ mNK cells in the fetal spleen. (G) Total number of fetal spleen cells, and (H) total number of iNK and (I) total number of mNK cells in the fetal spleen. Data represent mean ± SD values from 5 to 6 spleens analyzed in 14.5- to 18.5-day-old fetuses from 2 litters. Two spleens were pooled for the analysis. Profile from the adult spleen is included as a control; 0 indicates values below the detection level. *P < .05, **P < .01, and ***P < .0001. ns indicates not significant.
NK-cell subsets in the fetal liver and spleen. Representative FACS profiles (A) and total numbers (B) of CD3−CD122+NK1.1+DX5+ iNK and (C-D) CD3−NK1.1+CD43+Mac-1+ mNK cells at the different time points in the fetal liver. Data represent mean ± SD values from 5 to 14 livers analyzed in 12.5- to 18.5-day-old fetuses from 3 to 4 litters. Numbers in FACS profiles are mean frequencies of cell populations within the indicated gates, relative to total liver cells. Profile from the adult BM is included as a control. Representative FACS profiles of (E) CD3−NK1.1+DX5+ iNK cells and (F) CD3−NK1.1+CD43+Mac-1+ mNK cells in the fetal spleen. (G) Total number of fetal spleen cells, and (H) total number of iNK and (I) total number of mNK cells in the fetal spleen. Data represent mean ± SD values from 5 to 6 spleens analyzed in 14.5- to 18.5-day-old fetuses from 2 litters. Two spleens were pooled for the analysis. Profile from the adult spleen is included as a control; 0 indicates values below the detection level. *P < .05, **P < .01, and ***P < .0001. ns indicates not significant.
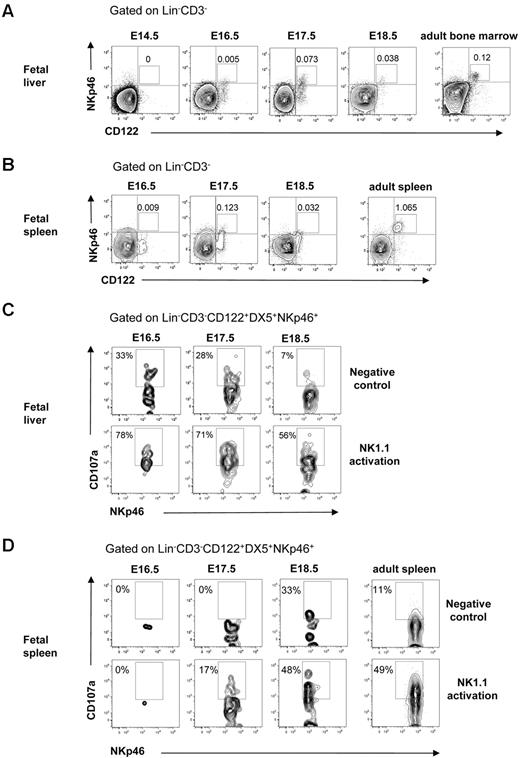
NK cells mediate killing after interaction with virus-infected or transformed tumor target cells that trigger their lytic machinery and lead to the release of granzyme and perforin from their cytotoxic granules. To determine whether NK cells in the fetal liver and spleen were functionally competent, we analyzed the expression of natural cytotoxicity triggering receptor 1 (NKp46) involved in target cell killing31 and performed the CD107a degranulation assay.33 In agreement with the finding that NK cells with mature Mac-1+CD43+ phenotype32 emerged at E16.5 in the fetal liver (Figure 4C-D), NKp46+ NK cells were present at E16.5 in the fetal liver and at E17.5 in the fetal spleen (Figure 5A-B). Furthermore, the Lin−CD122+CD3−DX5+NKp46+ NK cells expressing LAMP-1 (CD107a) on their surface after activation through the NK1.1 receptor were clearly detectable at E16.5 in the liver and at E17.5 in the spleen (Figure 5C-D). Taken together, these results show that functionally competent NK cells are present in the fetal liver at E16.5 and in the spleen at E17.5.
Cytotoxic activity of NK cells in the fetal liver and spleen. FACS profiles of CD122 and NKp46 co-expression in (A) liver and (B) spleen in 14.5- to 18.5-day-old fetuses. BM and spleen cells from adult mice were used as a positive control. Numbers in FACS profiles indicate the mean frequencies of populations within the indicated gates. Text above plots indicates gating. Data from representative analysis, 3-5 fetuses were analyzed from 2 to 3 different litters; 2 spleens were pooled for the analysis. FACS profiles of CD107a expression by Lin−CD3−CD122+DX5+NKp46+ NK cells after activation with purified anti-NK1.1 Ab in (C) liver and (D) spleen cells in 14.5- to 18.5-day-old fetuses. Nonactivated cells were used as negative control, and cells from the adult spleen were used as positive control. Numbers in FACS profiles show frequencies of populations within the indicated gates of total cells. Data from one representative experiment with the use of cells from 2 litters, 3 replicates were performed, all giving similar results.
Cytotoxic activity of NK cells in the fetal liver and spleen. FACS profiles of CD122 and NKp46 co-expression in (A) liver and (B) spleen in 14.5- to 18.5-day-old fetuses. BM and spleen cells from adult mice were used as a positive control. Numbers in FACS profiles indicate the mean frequencies of populations within the indicated gates. Text above plots indicates gating. Data from representative analysis, 3-5 fetuses were analyzed from 2 to 3 different litters; 2 spleens were pooled for the analysis. FACS profiles of CD107a expression by Lin−CD3−CD122+DX5+NKp46+ NK cells after activation with purified anti-NK1.1 Ab in (C) liver and (D) spleen cells in 14.5- to 18.5-day-old fetuses. Nonactivated cells were used as negative control, and cells from the adult spleen were used as positive control. Numbers in FACS profiles show frequencies of populations within the indicated gates of total cells. Data from one representative experiment with the use of cells from 2 litters, 3 replicates were performed, all giving similar results.
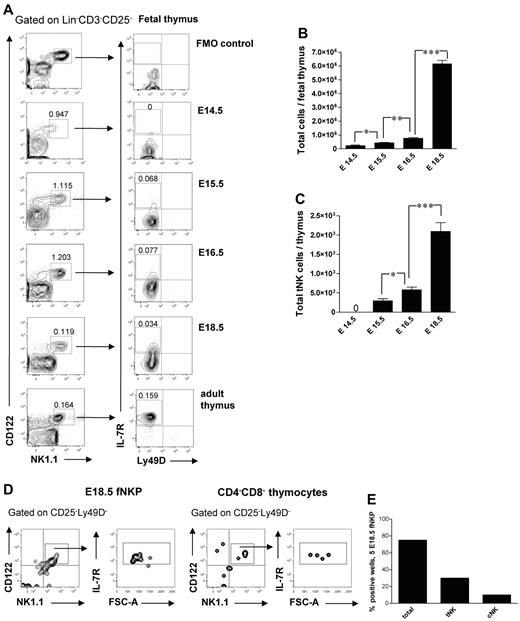
Emergence of thymic-dependent NK cells
It has been previously shown that a subset of NK cells develops in the fetal24 and adult34 thymus. In agreement with previous studies,24 Lin−CD3−CD25−CD122+NK1.1+ cells were present in the thymus already at E14.5 (Figure 6A). Interestingly, their frequency and total number decreased after E16.5 (Figure 6A), most likely due to increased number of generated double-positive and single-positive thymocytes not present at E14.5, suggesting that the NK-cell development preceded the T lymphopoiesis in the thymus.24 The thymic-dependent NK-cell developmental pathway has been characterized in adult mice; however, it has not been studied during fetal stages.3 The Lin−CD3−CD25−CD122+NK1.1+Ly49D−IL-7Rα+ tNK cells emerged in the fetal thymus at E15.5 and expanded by E18.5 (Figure 6A,C).
Emergence and generation of tNK cells in the mouse embryo. (A) Representative FACS profiles of Lin−CD3−CD25−CD122+NK1.1+Ly49D−IL-7Rα+ tNK cells in the fetal thymus. (B) Total number of fetal thymic cells and (C) total number of tNK cells. Data represent mean ± SD values from 5 to 13 thymi analyzed in 14.5- to 18.5-day-old fetuses from 2 to 3 litters. Text above the plots and arrows indicate the gating. Numbers in FACS profiles show mean frequencies of populations within the indicated gates of total fetal thymocytes. Profile from the adult thymus is included as a control; 0 indicates the values below the detection level. Five Lin−CD122+NK1.1−DX5− NKPs were sorted from the liver from 18.5-day-old fetuses and cultured on OP9-DL1 stroma for 21 days with KL, IL-7, FL, and IL15. Double-negative fetal thymocytes (CD4/CD8-depleted) were used as a positive control. Generated clones were evaluated by FACS for the presence of tNK cells (TCRβ−CD25−Ly49D−NK1.1+CD122+IL-7Rα+) and BM-dependent cNK (TCRβ−NK1.1+CD122+IL-7Rα−) cells. (D) Representative FACS profiles of cells generated from fetal liver NKPs and CD4−CD8− thymocytes cultured on OP9-DL1 stroma. The specific gates are indicated in the text above the plot and by arrows. (E) Proportion of total positive wells and wells containing thymic NK (tNK) and conventional NK (cNK) cells generated from E18.5 fetal liver NKPs. Data from one experiment, 20 wells were analyzed. *P < .05, **P < .01, and ***P < .0001.
Emergence and generation of tNK cells in the mouse embryo. (A) Representative FACS profiles of Lin−CD3−CD25−CD122+NK1.1+Ly49D−IL-7Rα+ tNK cells in the fetal thymus. (B) Total number of fetal thymic cells and (C) total number of tNK cells. Data represent mean ± SD values from 5 to 13 thymi analyzed in 14.5- to 18.5-day-old fetuses from 2 to 3 litters. Text above the plots and arrows indicate the gating. Numbers in FACS profiles show mean frequencies of populations within the indicated gates of total fetal thymocytes. Profile from the adult thymus is included as a control; 0 indicates the values below the detection level. Five Lin−CD122+NK1.1−DX5− NKPs were sorted from the liver from 18.5-day-old fetuses and cultured on OP9-DL1 stroma for 21 days with KL, IL-7, FL, and IL15. Double-negative fetal thymocytes (CD4/CD8-depleted) were used as a positive control. Generated clones were evaluated by FACS for the presence of tNK cells (TCRβ−CD25−Ly49D−NK1.1+CD122+IL-7Rα+) and BM-dependent cNK (TCRβ−NK1.1+CD122+IL-7Rα−) cells. (D) Representative FACS profiles of cells generated from fetal liver NKPs and CD4−CD8− thymocytes cultured on OP9-DL1 stroma. The specific gates are indicated in the text above the plot and by arrows. (E) Proportion of total positive wells and wells containing thymic NK (tNK) and conventional NK (cNK) cells generated from E18.5 fetal liver NKPs. Data from one experiment, 20 wells were analyzed. *P < .05, **P < .01, and ***P < .0001.
The origin of tNK cells is not clear, and it has been debated whether they are generated from NKPs or from thymic progenitors.3,24,34 Fetal liver E18.5 NKPs cultured on OP9-DL1 stroma with KL, FL, IL-7, and IL-15 for 21 days generated progeny that contained both Lin−CD122+NK1.1+CD25−IL-7Rα+Ly49D− tNK cells and Lin−CD25−CD122+NK1.1+IL-7Rα− cNK cells (Figure 6D-E). This is in agreement with NKPs being present in E18.5 fetal thymus (supplemental Figure 1).
Discussion
Despite extensive progress in delineation of mature blood cell development, the early stages of NK-cell lineage commitment are less understood compared to B and T lymphopoiesis.
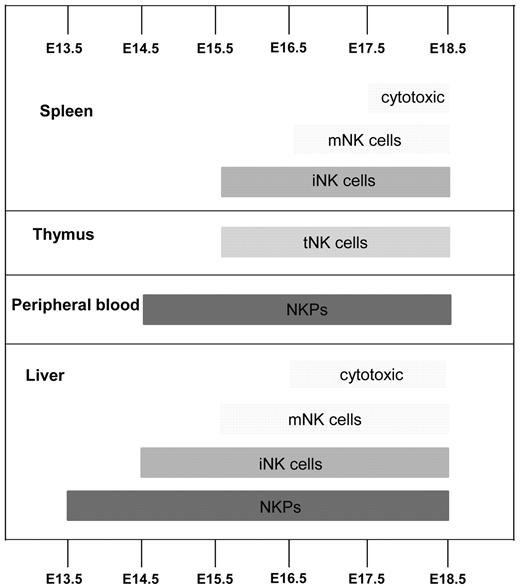
The present studies investigated and established several important and novel aspects of NK-cell commitment and development in mouse embryo (Figure 7). Although previous studies identified multiple fetal hematopoietic progenitors with NK-lineage potential, the identity of NK lineage–restricted progenitors, the time, and the site of their emergence, as well as expansion and contribution to different mature NK-cell progeny, have not been well characterized. Studies in adult mice have shown that up-regulation of the IL-2Rβ chain (CD122) marks the commitment to the NK-cell lineage, and NKPs have been identified as having a Lin−CD122+NK1.1−DX5− phenotype.4 However, the fetal counterpart of adult NKPs has not been studied. Thus, we investigated in different hematopoietic tissues how NKPs emerge, expand, and generate different mature NK-cell subsets during embryonic development.
Emergence of NK-cell progenitors and NK cells in the mouse embryo. Schematic representation of where and when NK-cell progenitors and different NK-cell populations arise during fetal development. iNK indicates immature NK cells (CD3−NK1.1+DX5+), mNK, mature NK cells (CD3−NK1.1+DX5+CD43+Mac-1+), and tNK, thymic NK cells (CD3−CD25−CD122+NK1.1+Ly49D−IL-7R+). Cytotoxic NK cells were identified as CD107a+ using the degranulation assay.
Emergence of NK-cell progenitors and NK cells in the mouse embryo. Schematic representation of where and when NK-cell progenitors and different NK-cell populations arise during fetal development. iNK indicates immature NK cells (CD3−NK1.1+DX5+), mNK, mature NK cells (CD3−NK1.1+DX5+CD43+Mac-1+), and tNK, thymic NK cells (CD3−CD25−CD122+NK1.1+Ly49D−IL-7R+). Cytotoxic NK cells were identified as CD107a+ using the degranulation assay.
We show here for the first time that the fetal equivalent of adult BM Lin−CD122+NK1.1−DX5− NKPs emerged in the liver at E13.5 and was also found in the fetal blood, BM, spleen, and thymus, suggesting that NKPs migrate through the circulation to the other tissues that support NK-cell development. Functional evaluation showed that fetal liver NKPs, already at E14.5, have robust NK-lineage potential, generating NK cells that express NKp46 (the most specific NK-cell marker31 ). Importantly, the absence of NK cells and their progenitors in the circulation at E13.5 suggests that they are not generated in other embryonic sites prior to their generation in the fetal liver, further supporting that fetal NKPs are committed to the NK lineage. It is currently unclear whether the fetal NKPs first expand in the liver and then migrate to the other tissues or whether they arise independently at different embryonic sites; however, their presence in the peripheral blood supports the first possibility. The fact that NKPs were detected for the first time in the circulation at E14.5 that is 1 day later than the time when NKPs emerged in the fetal liver suggests that NKPs are not colonizing fetal liver through the peripheral blood, but, most likely, are generated from hematopoietic stem cells or multipotent progenitors already present in the fetal liver at that time.11,12
In agreement with our recent work showing that a fraction of adult BM NKPs sustain the ability to generate T cells,5 we here established using a highly specific single-cell assay that at least a fraction of the Lin−CD122+NK1.1−DX5− cells in the fetal liver, although having lost B and myeloid potential, sustain combined NK and T-cell potential. These findings show that, as their adult BM counterpart, the Lin−CD122+NK1.1−DX5− population in the fetal liver represents a bipotent NK/T-cell progenitor. Bipotent NK/T-cell progenitors have been previously identified in fetal liver as having the B220lowKit+CD19− phenotype.21 Since a significant fraction of Lin−CD122+NK1.1−DX5− cells expresses KIT receptor and B220 as well as lack CD19 expression, it is very likely that these 2 phenotypes define overlapping cell populations. Importantly, in contrast to the previous work that focused on E15 fetal liver,21 we investigated the emergence of NKPs in multiple hematopoietic tissues at different time points.
By performing time course phenotypic analysis at different embryonic sites we established here that the first cells with the NK-cell phenotype (Lin−CD122+ cells) emerged in the liver at E13.5, whereas the CD3−NK1.1+DX5+ immature NK cells were first found in the liver at E14.5 and spleen at E15.5. Although one previous report did not detect NK1.1+ cells in the fetal liver at E15,24 our findings are in agreement with work by Koo et al,23 detecting NK1.1+ cells at E14 in the fetal thymus and liver and at E15 in the spleen. Importantly, in contrast to the previous study where only anti-NK1.1 serum was used to identify NK cells, we applied multiple NK lineage–specific markers as well as performed functional analysis to conclusively establish the development of NK cells in the embryo. The NKp46 receptor has been identified as the most specific NK-cell marker, and its expression pattern has been established in different NK-cell compartments in adult mice but has not been investigated during fetal development.31 Here, we show, for the first time, that NKp46+ NK cells emerged in the fetal liver at E16.5 and spleen at E17.5, coinciding with the presence of cytotoxic NK cells in the fetal liver at E16.5 and spleen at E17.5.
Previous studies provided the evidence that NK cells develop within the fetal thymus17,18,20,24 and that at E15 the fetal thymus contains progenitors that give rise to NK and T cells as well as NK1.1+ cells with cytotoxic activity.24 However, the emergence of a distinct population of Lin−CD25−CD122+NK1.1+Ly49D−IL-7Rα+ tNK cells that, thought to be thymic dependent, has not yet been studied in the fetal thymus.3 Here, we established that cells with the tNK-cell phenotype arose in the fetal thymus at E15.5
Although there is strong evidence that the lineage commitment of HSCs occurs in a stepwise manner, the cellular pathways and restriction sites to generate the bipotent and unipotent progenitors of lymphoid lineages are the subject of debate and investigation.35,36 Furthermore, the hematopoietic hierarchy and lineage restriction might be different during fetal and adult blood cell development.37 For example, it has been shown that the fetal equivalent of the BM common lymphoid progenitor, in contrast to the adult counterpart, maintains significant macrophage potential.38 Of relevance to our study it has remained an unresolved question whether multipotent hematopoietic progenitors become NK/T-cell lineage restricted before or after entering the thymus.39 In agreement with previous studies,21 our work supports that NK/T lineage restriction, at least in part, takes place prethymically.
It has been a subject of debate whether the tNK cells develop from BM NKPs or early T-cell progenitors in the thymus. The existence of a bipotent NK/T progenitor strongly supports a close developmental relationship between the NK and T-cell lineages in the thymus.
Here, we show that fetal liver NKPs contribute to both BM and thymic-dependent NK-cell pathways in vitro, similar to their counterparts in adult BM.5 The fact that NKPs were present both in the fetal liver and in the thymus further supports that tNK cells are derived from NKPs.
Previous studies have shown that IL-15 and IL-2 have differential effects on thymic bipotent NK/T progenitors, supporting NK- and T-cell generation, respectively.40 It has been suggested that a decisive division between T- and NK-cell fate occurs at the double-negative 2 stage during the early T-cell development in the thymus and that the transcription factors Nfil3, Id2, and Bcl11b mutually inhibit the T and NK pathways.41-47
The Notch signaling pathway is important for development of fetal hematopoiesis in the aorta-gonad-mesonephros region but is dispensable for the later stages.48,49 Notch signaling is critical for T-cell lymphopoiesis at multiple stages; but its role in NK-cell development is less understood.50 Here, we show that the generation of NK cells from fetal NKPs in vitro was not restricted by Notch signaling, as we have shown for the adult BM NKPs.5
Uncovering NK-cell developmental stages and sites where NK cells and NK-cell progenitors arise during fetal development will provide important insights toward a better understanding of the origin of diverse NK-cell subsets, their progenitors, and key regulators.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Tadepally Lakshmikanth and Petter Brodin for helpful discussions and Lillian Wittmann and Zhi Ma for expert advice and technical assistance.
This work was supported by the Swedish Medical Research Council, the Swedish Pediatric Cancer Foundation, the Swedish Cancer Foundation, and the Hemato-Linne grant. E.S. has an Associate Professor Position supported by the Swedish Pediatric Cancer Foundation.
Authorship
Contribution: Y.T., C.P., H.N.C., M.C., and E.S., designed and conceptualized the research, performed experiments, and analyzed the data; P.C. performed experiments and analyzed the data; Y.T., C.P., and E.S. wrote the manuscript; and S.E.W.J. participated in study design, analysis of data, and writing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ewa Sitnicka, Hematopoietic Stem Cell Laboratory, Lund Research Center for Stem Cell Biology and Cell Therapy, Lund University, BMC B10, Klinikgatan 26, 221-84 Lund, Sweden, e-mail: ewa.sitnicka@med.lu.se.
References
Author notes
Y.T., C.P., and H.N.C. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal