In this issue of Blood, Flamm et al combine high-throughput experimental methods and multiscale computer simulations to predict patient-specific thrombus formation potential.1 Their studies reveal a novel thromboxane receptor mutation (TP-V241G) in humans that confers resistance to indomethacin.
Blood platelets integrate both biochemical stimuli and biomechanical cues to regulate their adhesive phenotype. At sites of injury, initial platelet attachment is mediated by von Willebrand factor (VWF), which forms a molecular bridge between platelets and extracellular matrix proteins. Subsequent thrombus formation rate is regulated by platelet response to multiple stimuli including collagen, ADP, thromboxane (TXA2), thrombin, epinephrine, and endothelial cell–secreted inhibitors like nitric oxide and prostacyclin. Fluid flow plays an important role in this process in part by regulating the binding interaction between platelets and VWF, initiating mechanotransduction, and controlling cell and solute transport in the vicinity of the thrombi.2 Platelet-platelet cohesion is strengthened by activated platelet integrin GPIIb-IIIa, fibrin network formation, and signaling through integrins and receptor tyrosine kinases.3 Thus, despite the absence of a nucleus, these seemingly simple cell fragments have developed complex and calculating signal transduction pathways to integrate a variety of environmental cues and signals. The question then arises: Can computer models be designed to mimic in silico the calculations being performed by platelets as they respond to biochemical and biomechanical stimuli? Can these models be personalized to capture the phenotype of individual donors?
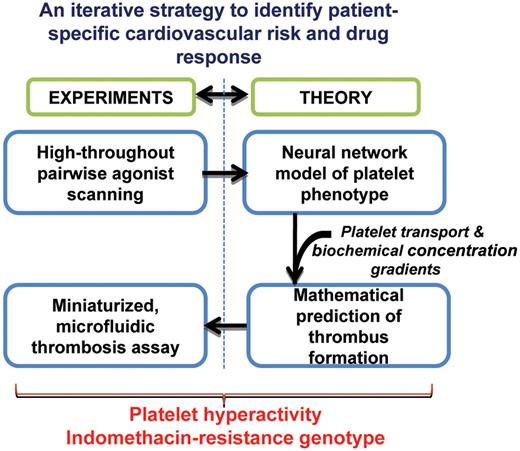
Platelet responsiveness to agonists can be highly variable among healthy human beings.4 Thus, a variety of candidate gene and genome-wide association studies are under way to identify the source of platelet hyperactivity.5 Further, platelets from certain patients exhibit diminished reactivity to drug or therapy.6 These features regulate patient clotting time and bleeding propensity. In this context, the work by Flamm et al suggests that understanding platelet phenotype regulation at the “systems-level,” by combining high-throughput experiments and related computations, can enable the evaluation of patient-specific cardiovascular risk and drug response (see figure).1
High-throughput experiments and mathematical modeling applied to define platelet phenotype. Pair-wise agonist scanning captures the response of platelets to complex, combinatorial stimulus input. The results are stored in an artificial neutral network. The neutral network is embedded in a multiscale model of platelet adhesion and thrombus growth. Mathematical model predictions are validated using cell adhesion assays performed in a microfluidic flow cell.
High-throughput experiments and mathematical modeling applied to define platelet phenotype. Pair-wise agonist scanning captures the response of platelets to complex, combinatorial stimulus input. The results are stored in an artificial neutral network. The neutral network is embedded in a multiscale model of platelet adhesion and thrombus growth. Mathematical model predictions are validated using cell adhesion assays performed in a microfluidic flow cell.
Here, Flamm and colleagues perform high-throughput experiments to monitor platelet response to agonists against the platelet purinergic (ADP), thromboxane (U46619), and GPVI receptors (convulxin). Time-dependent changes in intra-cellular calcium are measured under a variety of conditions, and this is used to quantify platelet response to agonist. By varying the agonists over a wide range (0-10 times EC50) in pairs using a pair-wise agonist scanning (PAS) scheme,7 calcium mobilization is measured in response to 74 different treatments for each donor sample. The large amounts of data generated in this manner are used to construct a donor-specific computer model of the platelet response (ie, a “neural network” model). While this type of modeling is empirical in that it does not provide mechanistic or biochemical insights, it does represent a relatively simple yet powerful approach to capture the input-output response of platelets to both single and multiple complex agonists. Importantly, a separate platelet response model is generated for each blood donor, and thus subsequent experimental and modeling steps capture individual platelet phenotype.
To relate the measured platelet activation response to thrombus formation potential, multiscale computer simulations are performed. Here, mathematical equations of fluid transport, platelet motion, and cell adhesion are included in order to simulate thrombus growth rate under a variety of conditions. While mathematical modeling of platelet adhesion has been performed for some time now,8 a major advance of this work by Flamm et al lies in its ability to link individual donor platelet phenotype to cell-adhesion kinetics. This is done by embedding the neural network platelet response model into the thrombus growth simulations. Validation of this approach is performed by the authors using microfluidic experiments. Together, the modeling and experiments reveal a novel indomethacin-resistance human thromboxane receptor mutation.
The methodology developed by Flamm and colleagues is significant in that it links high-throughput data collection and mathematical models to gain new biologic insight. Although only 3 donors were examined in this study, it is apparent that this approach may be readily extended to more healthy donors. Such an application of pair-wise agonist scanning and related modeling may complement more traditional genetic approaches that aim to identify single nucleotide polymorphisms that regulate platelet function. Further, while the current study uses PPACK anticoagulated blood and low shear rates, extensions to include the effects of hydrodynamic shear, thrombin, and outside-in signaling in the model formulation remain possible. Finally, while the current approach is focused on hematology, the general methodology developed here is also likely to find broader application in other studies related to systems and integrative biology.9,10
Conflict-of-interest disclosure: The author declares no competing financial interests. ■
REFERENCES
National Institutes of Health

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal