Abstract
Natural killer (NK) cells play a critical role in early host defense to infected and transformed cells. Here, we show that mice deficient in Eri1, a conserved 3′-to-5′ exoribonuclease that represses RNA interference, have a cell-intrinsic defect in NK-cell development and maturation. Eri1−/− NK cells displayed delayed acquisition of Ly49 receptors in the bone marrow (BM) and a selective reduction in Ly49D and Ly49H activating receptors in the periphery. Eri1 was required for immune-mediated control of mouse CMV (MCMV) infection. Ly49H+ NK cells deficient in Eri1 failed to expand efficiently during MCMV infection, and virus-specific responses were also diminished among Eri1−/− T cells. We identified miRNAs as the major endogenous small RNA target of Eri1 in mouse lymphocytes. Both NK and T cells deficient in Eri1 displayed a global, sequence-independent increase in miRNA abundance. Ectopic Eri1 expression rescued defective miRNA expression in mature Eri1−/− T cells. Thus, mouse Eri1 regulates miRNA homeostasis in lymphocytes and is required for normal NK-cell development and antiviral immunity.
Introduction
Natural killer (NK) cells are important lymphocyte effectors that participate in early immune responses against tumors and pathogen-infected cells by secreting cytokines and directly lysing target cells.1 Unlike T and B cells, which become activated through single antigen-specific receptors, NK-cell activation is controlled by the integration of signals from activating and inhibitory receptors.2 The inhibitory mouse Ly49 receptors are a highly polymorphic class of molecules that predominantly recognize MHC class I ligands. In contrast, Ly49 activating receptors can bind MHC class I molecules or ligands expressed on transformed or infected cells. For example, Ly49H recognizes the m157 glycoprotein encoded by mouse CMV (MCMV).3,4 During NK-cell development in the bone marrow (BM), Ly49 receptors are acquired in a stochastic fashion just before cells undergo a major proliferative burst and are released into the periphery. Humans and mice with selective NK-cell deficiencies are susceptible to severe recurrent infections, especially from herpesviruses like CMV.5,6
miRNAs are ∼ 22-nt noncoding RNAs generated from long RNA precursors by serial cleavage steps mediated by the Drosha-DGCR8 complex and Dicer. NK and T cells that lack miRNAs because of the targeted inactivation of Dicer, Drosha, or DGCR8 show dramatic defects in proliferation, survival, and effector function.7,8 In addition, mutations in specific miRNAs, such as miR-181, miR-150, and miR-155, can have dramatic effects on NK-cell development, cytotoxicity, and IFN-γ production.9-11 Individual miRNAs can modestly affect the stability and translation of hundreds of target mRNAs.12,13 Because multiple miRNAs may regulate the same biologic processes, posttranscriptional regulation of miRNAs as a class may profoundly alter gene expression programs.14
Eri1 is a 3′-to-5′ exoribonuclease of the DEDDh family with a deeply conserved role in 5.8S rRNA 3′ end processing.15,16 It has also been repeatedly recruited into species-specific small RNA regulatory pathways over the course of evolution. eri1 mutant Schizosaccharomyces pombe accumulate excess endogenous short-interfering RNAs (endo-siRNAs) that promote heterochromatin formation.17,18 In contrast, Caenorhabditis elegans ERI-1 forms a complex with Dicer that generates worm-specific classes of endo-siRNAs.19,20 eri-1 mutant worms lack these endo-siRNAs, but also display an enhanced RNAi (Eri) phenotype whereby exogenous siRNAs show more robust silencing of mRNA targets.21 Eri1 overexpression suppresses RNAi in mouse and human cell lines,22 but its role in mammalian endogenous small RNA pathways remains undefined. Here, we report that Eri1 negatively regulates global miRNA abundance and is required to promote normal NK-cell homeostasis and immune function.
Methods
Mice and infections
C57BL/6 (JAX; B6), CD45.1+ (Ptprca/a) B6 (NCI), ICR, and Rag2−/−Il2rg−/− mice (Taconic) were purchased. Eri1fl/fl;CD4-cre, Eri1−/−, and Ly49H-deficient (Klr8−/−) B6 mice were described previously.15,23 ICR/B6 mice were generated by crossing ICR to Eri1+/− B6 mice and backcrossing F1 mice to Eri1+/− B6. To create chimeras, embryonic day 14.5 fetal liver cells were harvested and injected intravenously into B6 mice lethally irradiated with a split dose of 1100 rad. For infections, mice were injected intraperitoneally with 5 × 104 PFU MCMV (Smith Strain). All experiments were conducted in accordance with the University of California, San Francisco Institutional Animal Care and Use Committee (IACUC) guidelines with approval from the university IACUC.
NK cytokine production and cytotoxicity
Splenic NK cells were stimulated with Abs against NK1.1, NKp46, Ly49D, Ly49H, or rat IgG2a or were incubated with IL-12 (20 ng/mL) and IL-18 (10 ng/mL; R&D Systems). For LAMP-1 analysis, stimulated NK cells were incubated with anti-CD107a Ab (1D4B; eBioscience). Splenocytes were incubated on Ab-coated plates for 5 hours in the presence of Brefeldin A followed by intracellular cytokine staining for IFN-γ. For cytotoxicity assays, wild-type (WT) and Eri1−/− NK1.1+ TCRβ− NK cells were incubated for 6 hours with Ba/F3 cells or Ba/F3 cells stably expressing m157.3 51Cr release was used as described to measure NK cell–mediated lysis.24
Western blot
Splenic NK1.1+TCRβ−Ly49H+ cells from uninfected or MCMV-infected B6 mice were sorted using a FACSAria (BD Biosciences). Western blots were performed using a mAb against β-actin (AC-74; Sigma Aldrich) and an affinity-purified polyclonal Ab against Eri1 (A28).15
Adoptive transfers
Congenically marked WT and Eri1−/− B6 splenic Ly49H+ NK cells were mixed at a 1:1 ratio, labeled with 10μM CellTrace Violet as per the manufacturer's instructions (Invitrogen), and 6 × 104 Ly49H+ NK equivalents were transferred intravenously into Ly49H-deficient recipients. Twenty-four hours later, mice were injected with MCMV. Alternatively, WT and Eri1−/− B6 splenic NK cells were mixed at a 1:1 ratio, and 0.5 × 106 NK equivalents were transferred intravenously into Rag2−/−Il2rg−/− B6 mice.
MCMV titers
Unmixed chimeras were infected with MCMV and euthanized 3.5 days later. Splenic and hepatic viral titers were determined as described previously.24
T-cell culture and stimulation
CD4+ T cells were purified from spleen and lymph nodes by magnetic bead selection (Dynal; Invitrogen) and activated with anti-CD3 and anti-CD28 Abs. Cells were taken off stimulus on day 3 and expanded in IL-2–containing media for sorting and analysis on day 6. Splenic T cells from MCMV-infected mice (day 8 postinfection [p.i.]) were isolated and stimulated with MCMV peptides (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) or 10 nM phorbol 12-myristate 13-acetate (PMA) and 1μM ionomycin and stained for intracellular IFN-γ as described previously.25
MicroRNA microarrays
Purified CD4+ T cells from one Eri1−/− and 2 Eri1fl/fl;CD4-cre mice and WT littermate controls (Eri1+/+, Eri1fl/fl, and Eri1+/+;CD4-cre) were activated in vitro for 40 hours under Th2 (1000 U/mL IL4 and 5 μg/mL anti–IFN-γ) conditions. Total RNA was isolated (miRNeasy kit; QIAGEN) and used for miRNA analysis using custom one-color Agilent microarrays (8 × 15K Agilent UCSF Custom miRNA V3.1) containing sequences from Sanger miRBase Version 11.26 For each set of 5 replicated probes across arrays, log2-scale average intensities were determined, corrected for background, and quantile-normalized. The false discovery rate was calculated as described.27
Deep sequencing
Small (18-30 bp) RNA libraries were constructed from activated ICR/B6 WT and Eri1−/− CD4+ T cells and sequenced as described.28 Adaptor sequences were trimmed from reads as described,29 and all reads 15-30 nt were mapped to the mouse genome (UCSC mm8 assembly). Only sequences mapping to the genome with up to 2 mismatches were analyzed. Mouse small noncoding RNA annotations were compiled as described.30 For genome-wide analysis, sequences were grouped into independent genomic loci as described,29 and relative reads from each library were compared at every locus. Using an empirical Bayes method,31 we determined that loci with a posterior probability > 0.9 of ≥ 5-fold change were determined to have significantly differential expression.
Accession numbers
Results
Peripheral deficiency and impaired BM expansion of Eri1−/− NK cells
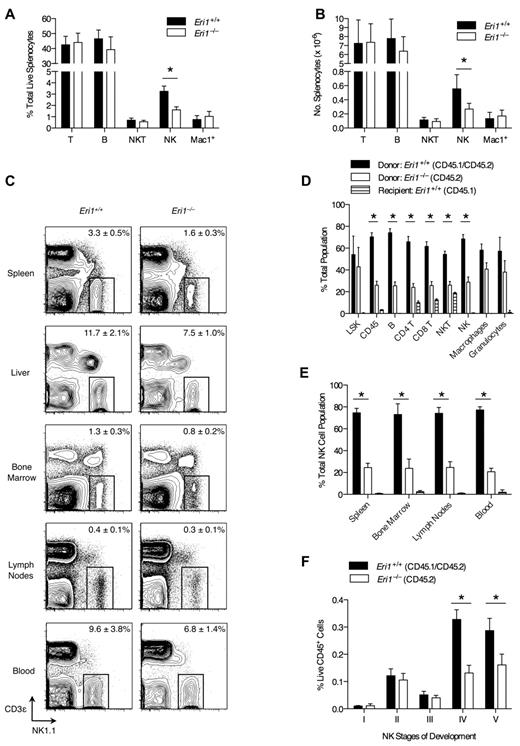
Compared with other tissues, Eri1 is highly expressed in mouse spleen and thymus, suggesting a role in the immune system.15 To determine whether the absence of Eri1 alters mature immune cell homeostasis, we evaluated splenocyte population frequencies in WT and Eri1-deficient (Eri1−/−) animals. The frequency and number of NK cells in the spleens of Eri1−/− animals was reduced by 50% compared with their WT siblings (Figure 1A-B). Other lymphocyte populations were present at normal numbers. NK-cell frequency was also significantly reduced in the liver (P < .01) and BM (P < .05; Figure 1C). Similar results were obtained in hematopoietic chimeras reconstituted with WT or Eri1−/− E14.5 fetal liver cells (supplemental Figure 1). Thus, Eri1−/− NK-cell reduction is intrinsic to the hematopoietic compartment.
NK-cell deficiency in Eri1−/− mice. (A) Frequency and (B) absolute number of spleen T (CD3ϵ+NK1.1−), B (CD19+), NKT (NK1.1+CD3ϵ+), NK (NK1.1+CD3ϵ−), and Mac1+ (CD11b+NK1.1−) cells enumerated by flow cytometry (N = 5, ICR/B6 mice). (C) NK1.1+CD3ϵ− NK cells in the indicated tissues. Numbers are NK-cell frequency ± SD among total lymphocytes (N = 5, ICR/B6 mice). (D-F) CD45.1+ lethally irradiated hosts were reconstituted with a 1:1 mixture of fetal liver cells from WT (CD45.1+ CD45.2+) and Eri1−/− (CD45.2+) donors and analyzed 8-15 weeks later (N = 3). (D) Frequency of donor- and host-derived cells among BM LSK (Lin−c-Kit+ Sca-1+) cells and spleen CD45+, B, CD4+ and CD8+ T, NKT, NK, macrophage (CD11b+NK1.1−Gr1−), and granulocyte (CD11b+ Gr1+) cells. (E) Frequency of donor- and host-derived cells among NK cells in the indicated tissues. (F) Frequency of developing NK-cell subsets (see supplemental Figure 2A for gating strategy) among BM WT (CD45.1+CD45.2+) and Eri1−/− (CD45.1−CD45.2+) cells. Bar graphs show mean ± SD; *P ≤ .05, (A-B) unpaired or (D-F) paired Student t test. All data are representative of at least 2 independent experiments.
NK-cell deficiency in Eri1−/− mice. (A) Frequency and (B) absolute number of spleen T (CD3ϵ+NK1.1−), B (CD19+), NKT (NK1.1+CD3ϵ+), NK (NK1.1+CD3ϵ−), and Mac1+ (CD11b+NK1.1−) cells enumerated by flow cytometry (N = 5, ICR/B6 mice). (C) NK1.1+CD3ϵ− NK cells in the indicated tissues. Numbers are NK-cell frequency ± SD among total lymphocytes (N = 5, ICR/B6 mice). (D-F) CD45.1+ lethally irradiated hosts were reconstituted with a 1:1 mixture of fetal liver cells from WT (CD45.1+ CD45.2+) and Eri1−/− (CD45.2+) donors and analyzed 8-15 weeks later (N = 3). (D) Frequency of donor- and host-derived cells among BM LSK (Lin−c-Kit+ Sca-1+) cells and spleen CD45+, B, CD4+ and CD8+ T, NKT, NK, macrophage (CD11b+NK1.1−Gr1−), and granulocyte (CD11b+ Gr1+) cells. (E) Frequency of donor- and host-derived cells among NK cells in the indicated tissues. (F) Frequency of developing NK-cell subsets (see supplemental Figure 2A for gating strategy) among BM WT (CD45.1+CD45.2+) and Eri1−/− (CD45.1−CD45.2+) cells. Bar graphs show mean ± SD; *P ≤ .05, (A-B) unpaired or (D-F) paired Student t test. All data are representative of at least 2 independent experiments.
We next considered the possibility that the decrease in Eri1−/− NK cells results from dysfunction in other hematopoietic cells. To test this hypothesis, we generated chimeras using a 1:1 mixture of Eri1+/+ (CD45.1+CD45.2+) and Eri1−/− (CD45.1−CD45.2+) E14.5 fetal liver cells. Mixed chimeras contained similar frequencies of BM Eri1+/+ and Eri1−/− Lin−Sca-1+c-Kit+ early hematopoietic precursors (Figure 1D); yet, we observed a significant reduction in all splenic Eri1−/− lymphocyte lineages, including but not limited to NK cells (Figure 1D-E). In contrast, Eri1+/+ and Eri1−/− splenic myeloid cell frequencies were similar. Because the presence of WT hematopoietic cells did not rescue the reduction in Eri1−/− NK cells, we conclude that Eri1 is required in a cell-intrinsic manner to maintain normal NK-cell numbers. Furthermore, competitive reconstitution unmasked a general defect in Eri1−/− lymphocyte homeostasis that was not apparent in Eri1−/− mice (Figure 1A-B). This may reflect defective peripheral turnover because of competition for limiting growth factors or reduced development from a common lymphocyte precursor.
Developing BM NK cells undergo ordered stages of maturation marked by the up-regulation or down-regulation of specific integrins and the acquisition of NK receptors.32 We examined 5 discrete populations of developing Eri1+/+ and Eri1−/− BM NK cells in mixed fetal liver chimeras (Figure 1F, supplemental Figure 2A). There were no differences in the relative frequencies of Eri1+/+ and Eri1−/− cells among early NK-cell precursors (stages I-III). However, Eri1−/− NK cells were significantly reduced starting at stage IV, which corresponds with a major proliferative phase, and at stage V, which is equivalent to mature CD11b+ NK cells. Although an underlying mechanism remains unclear, these data indicate that inefficient production of Eri1−/− NK cells in the BM contributes to their inability to populate peripheral compartments at normal frequencies.
Further analysis of developing WT BM NK populations revealed that stage III NK cells displayed high rates of cell death and apoptosis before they up-regulated CD49b and underwent a major proliferative burst at stage IV (supplemental Figure 2B-D). WT and Eri1−/− NK populations displayed similar frequencies of dead and apoptotic cells during all stages of BM development (supplemental Figure 2B-C). Eri1 deficiency also did not affect the frequency of BrdU-labeled cells after a 3- or 16-hour pulse (supplemental Figure 2D, data not shown). Rapid clearance of necrotic and apoptotic cells in vivo may preclude the detection of subtle differences in cell death, and differences in proliferation that occur outside the pulse window may not be detected by BrdU labeling. Nevertheless, we can exclude the possibility that major stage-specific differences in cell death and proliferation underlie the cell-intrinsic reduction of Eri1−/− BM NK cells.
Given the relatively large reduction in peripheral NK-cell numbers compared with the BM, we tested whether Eri1−/− NK cells have a homeostatic defect that exists independently of developmental impairment. WT and Eri1−/− NK cells incorporated BrdU provided in drinking water at the same rate, indicating normal NK-cell turnover at steady state in vivo (supplemental Figure 3A). Eri1−/− NK cells also expanded at the same rate as WT cells when transferred into lymphocyte-deficient Rag2−/−Il2rg−/− mice (supplemental Figure 3B). Constitutive signaling through the IL-15 receptor is essential for NK-cell development and survival.33 WT and Eri1−/− NK cells cultured in IL-15 ex vivo proliferated rapidly and underwent similar rates of cell death upon IL-15 withdrawal (supplemental Figure 3C-D). Thus, Eri1−/− NK cells are competent to signal through the IL-15 receptor and are not particularly sensitive to the proapoptotic effects of IL-15 withdrawal. Together, these data suggest that Eri1−/− NK cells undergo impaired production in the BM in the setting of normal peripheral turnover and IL-15–dependent proliferation.
Impaired maturation and Ly49 receptor expression in Eri1-deficient NK cells
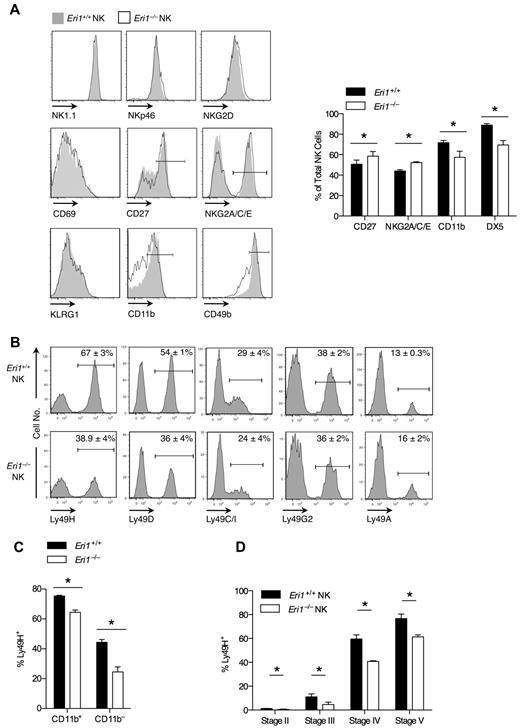
We examined the cell-surface phenotype of Eri1−/− and WT NK cells in mixed chimeras to determine whether Eri1 deficiency affects NK-cell maturation or activation status (Figure 2A). Eri1−/− NK cells expressed normal levels of the activating receptors NK1.1 and NKp46, which are used to identify the NK lineage. They also expressed normal levels of NKG2D and CD69, which are up-regulated acutely in activated cells, and KLRG1, which remains elevated on NK cells previously expanded by activation.34 A significantly higher frequency of Eri1−/− NK cells expressed the immature cell markers CD27 and NKG2A/C/E, and fewer Eri1−/− NK cells expressed CD49b and CD11b, which are up-regulated in the final stages of NK-cell maturation. Similar expression patterns were observed in Eri1−/− NK cells from unmixed chimeras (data not shown).
Impaired maturation and Ly49 receptor expression in Eri1−/− NK cells. B6 WT CD45.1+ lethally irradiated hosts were reconstituted with a 1:1 mixture of congenic WT (CD45.1+CD45.2+) and Eri1−/− (CD45.2+) fetal liver cells. (A-C) Splenic and (D) BM NK cells were analyzed by flow cytometry at 8-15 weeks. (A) Cell-surface expression of activating receptors (NKp46 and NK1.1), activation markers (NKG2D and CD69), and maturation markers (CD27, NKG2A/C/E, KLRG1, CD11b, and CD49b) on WT (CD45.1+CD45.2+) and Eri1−/− (CD45.1−CD45.2+) cells (left). Gated NK1.1+CD3ϵ− cells are shown for all stains, except for NK1.1, which shows gating on NKp46+TCRβ− cells. Summary of markers with significantly different expression on WT and Eri1−/− NK cells (right). (B) Splenic NK-cell activating and inhibitory Ly49 receptors. (C) Percentage of Ly49H+ cells among CD11b+ and CD11b− NK cells. (D) Percentage of Ly49H+ WT and Eri1−/− NK cells at various stages of NK-cell maturation in the BM (gating shown in supplemental Figure 2A). Bar graphs and flow cytometry plots show mean ± SD (N = 3 mice). *P ≤ .05; paired Student t test.
Impaired maturation and Ly49 receptor expression in Eri1−/− NK cells. B6 WT CD45.1+ lethally irradiated hosts were reconstituted with a 1:1 mixture of congenic WT (CD45.1+CD45.2+) and Eri1−/− (CD45.2+) fetal liver cells. (A-C) Splenic and (D) BM NK cells were analyzed by flow cytometry at 8-15 weeks. (A) Cell-surface expression of activating receptors (NKp46 and NK1.1), activation markers (NKG2D and CD69), and maturation markers (CD27, NKG2A/C/E, KLRG1, CD11b, and CD49b) on WT (CD45.1+CD45.2+) and Eri1−/− (CD45.1−CD45.2+) cells (left). Gated NK1.1+CD3ϵ− cells are shown for all stains, except for NK1.1, which shows gating on NKp46+TCRβ− cells. Summary of markers with significantly different expression on WT and Eri1−/− NK cells (right). (B) Splenic NK-cell activating and inhibitory Ly49 receptors. (C) Percentage of Ly49H+ cells among CD11b+ and CD11b− NK cells. (D) Percentage of Ly49H+ WT and Eri1−/− NK cells at various stages of NK-cell maturation in the BM (gating shown in supplemental Figure 2A). Bar graphs and flow cytometry plots show mean ± SD (N = 3 mice). *P ≤ .05; paired Student t test.
Eri1-deficient NK cells also displayed a skewed Ly49 receptor repertoire (Figure 2B). Surprisingly, Eri1−/− NK-cell populations displayed a specific reduction in the frequency of Ly49H- and Ly49D-expressing cells. In contrast to these activating receptors, the inhibitory receptors Ly49C/I, Ly49G2, and Ly49A were expressed normally. Within each Ly49+ subset, WT and Eri1−/− NK cells displayed no difference in the intensity of Ly49 receptor staining. Because activating Ly49 receptors are more frequently expressed on mature than immature NK cells, we considered the possibility that reduced Ly49H expression frequency could simply reflect the altered maturation status of Eri1−/− NK cells. However, Ly49H expression was decreased among both mature CD11b+ and immature CD11b−Eri1−/− NK cells (Figure 2C), indicating that this defect may occur independently of NK-cell maturation.
During their development, NK cells acquire Ly49 receptors in response to signals from the BM stroma.35,36 To determine when in NK-cell development Eri1−/− NK cells first show reduced Ly49H expression, we measured Ly49H+ NK-cell frequencies among stage II-V cells in the BM of mixed chimeras. Immature CD49b−αV+ NK cells acquire Ly49 receptors as they up-regulate c-Kit, which demarcates the transition from stage II to III cells (supplemental Figure 2A). Eri1−/− NK cells showed a marked reduction in Ly49H+ NK frequency at each developmental stage (Figure 2D). Furthermore, all measured Ly49 receptors, but not NKG2A/C/E receptors, were decreased in stage III Eri1−/− NK cells, including inhibitory receptors (Table 1). The inhibitory Ly49 repertoire normalizes in peripheral Eri1−/− NK cells, perhaps reflecting a selective growth advantage for NK cells with specific Ly49 repertoires. Together, these data indicate that peripheral Eri1−/− NK populations have an immature cell-surface phenotype and a skewed Ly49 repertoire with fewer Ly49 activating receptors.
Activating and inhibitory receptor repertoire on splenic and developing BM NK cells
| . | Eri1 . | II . | III . | IV . | V . | Spleen . |
|---|---|---|---|---|---|---|
| Ly49H | +/+ | 1.3 (0.1) | 11.0 (2.5) | 59.6 (3.4) | 76.7 (3.8) | 71.0 (3.5) |
| −/− | 0.5 (0.3) | 4.6 (2.0) | 40.8 (0.6) | 61.4 (1.7) | 50.1 (3.5) | |
| Ly49D | +/+ | 2.8 (0.5) | 13.6 (2.0) | 54.6 (2.9) | 55.4 (1.5) | 59.2 (1.0) |
| −/− | 2.3 (0.7) | 6.9 (1.8) | 47.7 (1.8) | 53.1 (1.6) | 48.4 (1.4) | |
| Ly49G2 | +/+ | 4.2 (1.6) | 15.9 (3.1) | 50.4 (5.6) | 49.2 (4.9) | 37.3 (2.7) |
| −/− | 4.2 (1.6) | 10.2 (2.4) | 56.6 (2.3) | 56.0 (5.2) | 38.1 (6.0) | |
| Ly49C/I | +/+ | 5.5 (0.7) | 19.0 (4.9) | 30.9 (1.2) | 34.6 (1.1) | 28.9 (3.5) |
| −/− | 3.5 (1.2) | 7.4 (1.2) | 26.0 (1.7) | 30.9 (1.9) | 24.4 (4.0) | |
| NKG2A/C/E | +/+ | 59.8 (0.7) | 60.1 (2.0) | 44.9 (0.7) | 45.5 (1.1) | 44.0 (1.3) |
| −/− | 60.7 (3.2) | 60.0 (1.8) | 55.9 (4.2) | 51.8 (2.8) | 52.2 (0.9) |
| . | Eri1 . | II . | III . | IV . | V . | Spleen . |
|---|---|---|---|---|---|---|
| Ly49H | +/+ | 1.3 (0.1) | 11.0 (2.5) | 59.6 (3.4) | 76.7 (3.8) | 71.0 (3.5) |
| −/− | 0.5 (0.3) | 4.6 (2.0) | 40.8 (0.6) | 61.4 (1.7) | 50.1 (3.5) | |
| Ly49D | +/+ | 2.8 (0.5) | 13.6 (2.0) | 54.6 (2.9) | 55.4 (1.5) | 59.2 (1.0) |
| −/− | 2.3 (0.7) | 6.9 (1.8) | 47.7 (1.8) | 53.1 (1.6) | 48.4 (1.4) | |
| Ly49G2 | +/+ | 4.2 (1.6) | 15.9 (3.1) | 50.4 (5.6) | 49.2 (4.9) | 37.3 (2.7) |
| −/− | 4.2 (1.6) | 10.2 (2.4) | 56.6 (2.3) | 56.0 (5.2) | 38.1 (6.0) | |
| Ly49C/I | +/+ | 5.5 (0.7) | 19.0 (4.9) | 30.9 (1.2) | 34.6 (1.1) | 28.9 (3.5) |
| −/− | 3.5 (1.2) | 7.4 (1.2) | 26.0 (1.7) | 30.9 (1.9) | 24.4 (4.0) | |
| NKG2A/C/E | +/+ | 59.8 (0.7) | 60.1 (2.0) | 44.9 (0.7) | 45.5 (1.1) | 44.0 (1.3) |
| −/− | 60.7 (3.2) | 60.0 (1.8) | 55.9 (4.2) | 51.8 (2.8) | 52.2 (0.9) |
WT CD45.1+ lethally irradiated hosts were reconstituted with a 1:1 mixture of fetal liver cells from congenic WT (CD45.1+CD45.2+) and Eri1−/− (CD45.2+) mice and analyzed 12 weeks later. Frequencies of NK-cell receptor expression on freshly isolated WT and Eri1−/− splenic NK cells (NK1.1+CD3ϵ−) and stage II-V developing BM NK cells (gating strategy shown in supplemental Figure 2A) are shown. Mean (SD) for 3 mice are listed.
Eri1 is dispensable for Ly49H-dependent NK cell–mediated cytotoxicity
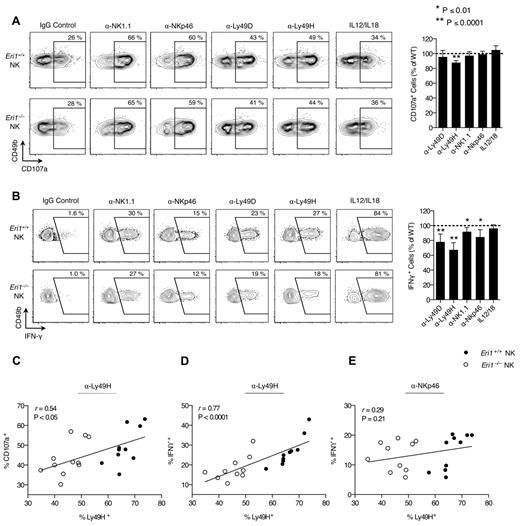
Activated NK cells degranulate and produce large amounts of IFN-γ. We measured IFN-γ production and the degranulation marker LAMP-1 (CD107a) in freshly isolated splenocytes activated with inflammatory cytokines or plate-bound Abs that ligate activating NK-cell receptors. WT and Eri1−/− cells from mixed chimeras were stimulated together, negating indirect feedback mechanisms on NK-cell activation. WT and Eri1-deficient NK cells showed similar LAMP-1 staining in all conditions tested except for ligation of Ly49H (Figure 3A). In contrast, Eri1−/− NK cells displayed modestly reduced IFN-γ expression upon cross-linking of all ITAM-associated receptors tested (Figure 3B). More significant decreases were observed with Ly49H and Ly49D cross-linking (35% and 20%, respectively). These results likely reflect the reduced frequency of Eri1−/− NK cells expressing these receptors rather than specific effects on Ly49 signaling. Indeed, starting Ly49H+ NK-cell frequency significantly correlated with both LAMP-1 and IFN-γ induction by Ly49H, but not NKp46 cross-linking (Figures 3C-E). In addition, a consistent ∼ 35% reduction in the frequency of IFN-γ–producing NK cells was observed when the Ly49H cross-linking Ab stimulus was titrated down over 2 orders of magnitude (data not shown).
Normal Ly49H-dependent NK-cell cytotoxicity in the absence of Eri1. B6 WT CD45.1+ lethally irradiated hosts were reconstituted with a 1:1 mixture of congenic WT (CD45.1+CD45.2+) and Eri1−/− (CD45.2+) fetal liver cells. (A) At 12 weeks, splenocytes were isolated and incubated in wells coated with IgG or mAbs against NK1.1, NKp46, Ly49D, or Ly49H in the presence of anti-CD107a mAb. Alternatively, NK cells were stimulated with IL-12 and IL-18. Frequency of degranulated WT (CD45.1+CD45.2+) and Eri1−/− (CD45.1−CD45.2+) CD107a+ NK cells (CD49b+TCRβ−) is shown. (B) Splenic NK cells stimulated as in panel A in the presence of brefeldin A and stained for intracellular IFN-γ. Flow cytometry plots show mean values for 10 mice. Summary graphs for Eri1−/− NK (right) show mean relative to WT ± SD (paired Student t test). (C-E) Regression of CD107a (C) or IFN-γ (D-E) expression on percentage of Ly49H+ NK cells after stimulation with indicated mAbs. The Pearson correlation coefficient (r) and significance test for nonzero correlation (P) are shown for each plot (N = 10 mice from 3 independent experiments).
Normal Ly49H-dependent NK-cell cytotoxicity in the absence of Eri1. B6 WT CD45.1+ lethally irradiated hosts were reconstituted with a 1:1 mixture of congenic WT (CD45.1+CD45.2+) and Eri1−/− (CD45.2+) fetal liver cells. (A) At 12 weeks, splenocytes were isolated and incubated in wells coated with IgG or mAbs against NK1.1, NKp46, Ly49D, or Ly49H in the presence of anti-CD107a mAb. Alternatively, NK cells were stimulated with IL-12 and IL-18. Frequency of degranulated WT (CD45.1+CD45.2+) and Eri1−/− (CD45.1−CD45.2+) CD107a+ NK cells (CD49b+TCRβ−) is shown. (B) Splenic NK cells stimulated as in panel A in the presence of brefeldin A and stained for intracellular IFN-γ. Flow cytometry plots show mean values for 10 mice. Summary graphs for Eri1−/− NK (right) show mean relative to WT ± SD (paired Student t test). (C-E) Regression of CD107a (C) or IFN-γ (D-E) expression on percentage of Ly49H+ NK cells after stimulation with indicated mAbs. The Pearson correlation coefficient (r) and significance test for nonzero correlation (P) are shown for each plot (N = 10 mice from 3 independent experiments).
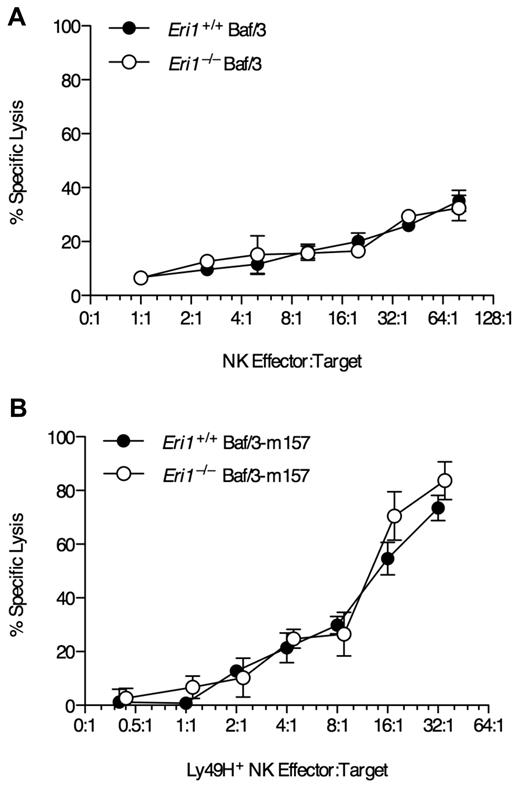
To test whether Eri1−/− NK cells can efficiently kill target cells in a Ly49H-dependent manner, we incubated freshly isolated NK cells ex vivo with Ba/F3 cells stably transduced with the MCMV Ly49H ligand m157. WT and Eri1−/− NK cells lysed the parental Ba/F3 line at similar rates (Figure 4A). Furthermore, WT and Eri1−/−Ly49H+ NK cells lysed Ba/F3-m157 targets at equal efficiency (Figure 4B). Together, these data indicate that Eri1−/− NK cells are competent to signal through NK-cell receptors and mediate normal Ly49H-dependent and -independent cytotoxicity. However, the reduced frequency of Ly49H+ cells in Eri1−/− NK-cell populations leads to proportional defects in Ly49H-dependent effector activities.
Normal NK-mediated killing in the absence of Eri1. Lethally irradiated CD45.1+ mice were reconstituted with CD45.2+ B6 WT or Eri1−/− fetal liver cells. Ten weeks later, splenic NK cells pooled from 3-4 mice were incubated with 51Cr-labeled target (A) Ba/F3 cells or (B) Ba/F3 cells stably expressing MCMV m157. Error bars indicate SD for triplicate measurements. Data are representative of 2 independent experiments.
Normal NK-mediated killing in the absence of Eri1. Lethally irradiated CD45.1+ mice were reconstituted with CD45.2+ B6 WT or Eri1−/− fetal liver cells. Ten weeks later, splenic NK cells pooled from 3-4 mice were incubated with 51Cr-labeled target (A) Ba/F3 cells or (B) Ba/F3 cells stably expressing MCMV m157. Error bars indicate SD for triplicate measurements. Data are representative of 2 independent experiments.
Eri1−/− NK cells expand less during MCMV infection and show poor control of viral load
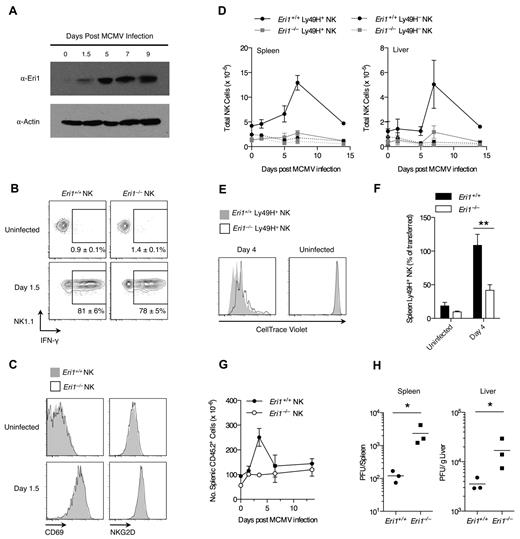
Ly49H+ NK cells are critical for controlling MCMV infection in B6 mice.37,38 Given the reduced Ly49H expression in Eri1−/− NK-cell populations, we investigated the in vivo activation of Eri1−/− NK cells by MCMV. Early in MCMV infection all NK cells are activated nonspecifically by proinflammatory cytokines such as IL-12, IL-18, and type I IFNs.39,40 Then, several days after infection, there is marked expansion of Ly49H+ NK cells driven directly by Ly49H ligation by the viral antigen m157.3,4 Eri1 is strongly up-regulated in Ly49H+ NK cells over the course of MCMV infection (Figure 5A), suggesting a potential role in NK-cell activation. To study Eri1−/− NK-cell activation in response to MCMV, we infected mixed chimeras, obviating discrepant cytokine environments or antigenic loads that could complicate experiments in separate WT and Eri1−/− mice.
Eri1 is required for Ly49H+ NK-cell expansion and control of viral titers in MCMV infection. (A) Anti-Eri1 mAb immunoblot of Ly49H+ NK cells pooled from 3-4 mice and sorted at various time points after MCMV infection. Data are representative of 2 independent experiments. (B-D) B6 WT CD45.1+ lethally irradiated hosts were reconstituted with a 1:1 mixture of congenic WT (CD45.1+ CD45.2+) and Eri1−/− (CD45.2+) fetal liver cells. At 8-15 weeks, chimeras were infected with MCMV. WT (CD45.1+CD45.2+) and Eri1−/− (CD45.1−CD45.2+) NK cells (NK1.1+ CD3ϵ−) were analyzed for (B) intracellular IFN-γ and (C) cell-surface CD69 and NKG2D expression. (D) Absolute numbers of WT and Eri1−/− Ly49H+ and Ly49H− NK cells in the spleen and liver at various time points after infection. Error bars in panels B and D indicate SD (N = 3 mice). (E-F) CD45.2+Eri1−/− splenic Ly49H+ NK cells from reconstituted fetal liver chimeras or B6 mice were mixed 1:1 with splenic Ly49H+ NK cells from CD45.1+CD45.2+ or CD45.1+ WT B6 mice. Mixed splenocytes were labeled with CellTrace Violet and transferred into B6 CD45.1+Ly49H−/− hosts. (E) CellTrace Violet dilution before and after MCMV infection. (F) Percentage of Ly49H+ NK cells transferred at day 0 (mean ± SEM, N = 13 infected and 4 uninfected mice from 4 independent experiments). **P ≤ .001, paired Student t test. (G-H) B6 WT CD45.1+ lethally irradiated hosts were reconstituted with CD45.2+ WT or Eri1−/− fetal liver cells and infected with MCMV at 12 weeks. (G) Absolute numbers of WT and Eri1−/− CD45.2+ splenocytes over the course of infection (mean ± SD, N = 3 mice). (H) Viral titers in the spleen and liver were determined at day 3.5 p.i. by plaque assays. Horizontal line indicates the mean of each group (N = 3 mice). *P ≤ .05; 2-tailed Mann-Whitney U test.
Eri1 is required for Ly49H+ NK-cell expansion and control of viral titers in MCMV infection. (A) Anti-Eri1 mAb immunoblot of Ly49H+ NK cells pooled from 3-4 mice and sorted at various time points after MCMV infection. Data are representative of 2 independent experiments. (B-D) B6 WT CD45.1+ lethally irradiated hosts were reconstituted with a 1:1 mixture of congenic WT (CD45.1+ CD45.2+) and Eri1−/− (CD45.2+) fetal liver cells. At 8-15 weeks, chimeras were infected with MCMV. WT (CD45.1+CD45.2+) and Eri1−/− (CD45.1−CD45.2+) NK cells (NK1.1+ CD3ϵ−) were analyzed for (B) intracellular IFN-γ and (C) cell-surface CD69 and NKG2D expression. (D) Absolute numbers of WT and Eri1−/− Ly49H+ and Ly49H− NK cells in the spleen and liver at various time points after infection. Error bars in panels B and D indicate SD (N = 3 mice). (E-F) CD45.2+Eri1−/− splenic Ly49H+ NK cells from reconstituted fetal liver chimeras or B6 mice were mixed 1:1 with splenic Ly49H+ NK cells from CD45.1+CD45.2+ or CD45.1+ WT B6 mice. Mixed splenocytes were labeled with CellTrace Violet and transferred into B6 CD45.1+Ly49H−/− hosts. (E) CellTrace Violet dilution before and after MCMV infection. (F) Percentage of Ly49H+ NK cells transferred at day 0 (mean ± SEM, N = 13 infected and 4 uninfected mice from 4 independent experiments). **P ≤ .001, paired Student t test. (G-H) B6 WT CD45.1+ lethally irradiated hosts were reconstituted with CD45.2+ WT or Eri1−/− fetal liver cells and infected with MCMV at 12 weeks. (G) Absolute numbers of WT and Eri1−/− CD45.2+ splenocytes over the course of infection (mean ± SD, N = 3 mice). (H) Viral titers in the spleen and liver were determined at day 3.5 p.i. by plaque assays. Horizontal line indicates the mean of each group (N = 3 mice). *P ≤ .05; 2-tailed Mann-Whitney U test.
Early in MCMV infection, both WT and Eri1−/− NK cells responded with robust IFN-γ production (Figure 5B) and up-regulation of NKG2D and CD69 (Figure 5C). These results are consistent with efficient IFN-γ production upon IL-12 and IL-18 stimulation ex vivo (Figure 3B). Later in MCMV infection, WT Ly49H+ NK cells showed robust expansion, peaking at day 7 in both spleen and liver (Figure 5D). In contrast, Eri1−/− Ly49H+ NK cells underwent inefficient expansion. As expected, Ly49H− NK cells showed little change in total cell number regardless of their genotype. To assess Eri1−/− NK-cell expansion independent of the reduced initial Ly49H+ cell frequency, we adoptively transferred equal numbers of CellTrace Violet–labeled WT and Eri1−/− Ly49H+ NK cells into Ly49H-deficient hosts. Four days after infection with MCMV, WT Ly49H+ NK cells had diluted the cell proliferation dye more and undergone greater expansion than Eri1−/− cells (Figure 5E-F). Thus, Eri1-deficient NK cells are activated normally in early MCMV infection yet undergo poor Ly49H-dependent proliferation as the infection progresses.
To determine whether Eri1 is required to control viral load, we infected unmixed chimeras with MCMV. As observed in mixed chimeras, Eri1−/− NK cells showed robust expression of IFN-γ and up-regulated the early activation markers CD69 and NKG2D yet displayed reduced expansion of Ly49H+ cells (data not shown). Like other mice with poor NK-cell expansion,6,23 Eri1−/− chimeras exhibited decreased splenomegaly and increased viral titers (Figure 5G-H). We conclude that Eri1 is required in a cell-intrinsic manner for the normal expansion of Ly49H+ NK cells and control of MCMV infection.
Reduced virus-specific T-cell responses in the absence of Eri1
Unlike NK cells, CD4 and CD8 lineage T cells were present at normal steady-state numbers and proportions in the thymus and peripheral lymphoid tissues of Eri1−/− fetal liver chimeras (data not shown, supplemental Figure 1A). The proportions of naive, memory, and regulatory T cells were also normal, and similar results were obtained in Eri1fl/fl;CD4-cre mice that lack Eri1 only in T cells (data not shown). However, given the more general defect in Eri1−/− lymphocyte development revealed in mixed fetal liver chimeras and the decreased expansion of Ag-specific Eri1−/− NK cells during MCMV infection, we examined MCMV-specific responses in Eri1−/− CD4+ and CD8+ T cells in mixed fetal liver chimeras.
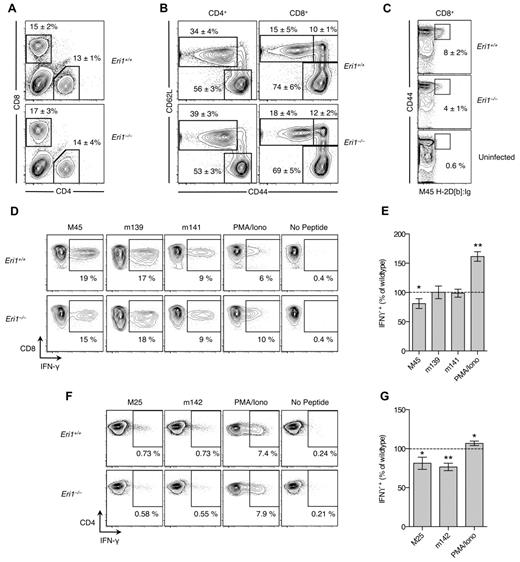
At day 8 p.i., T cells were evaluated by flow cytometry and by restimulation ex vivo with immunodominant MCMV peptides.25,41 As expected, infection increased the overall frequency of CD8+ T cells (Figure 6A) and the proportion of activated CD4+ and CD8+ T cells marked by high expression of CD44 and/or down-regulation of CD62L (Figure 6B). These changes occurred similarly in WT and Eri1−/− T cells (Figure 6A-B). However, fewer Eri1−/− CD8+ cells were specific for the H2-Db–restricted M45985-993 peptide epitope (Figure 6C). Accordingly, M45 induced fewer IFN-γ–producing Eri1−/− CD8+ cells, despite their increased response to PMA and ionomycin (Figure 6D-E). We also detected a reduced frequency of MCMV-specific IFN-γ–producing Eri1−/− CD4+ T cells despite robust IFN-γ production on PMA and ionomycin restimulation (Figure 6F-G). Together, these data indicate that, like NK cells, Eri1−/− T cells have diminished Ag-specific responses to MCMV infection. This defect is unlikely to contribute to poor MCMV control in Eri1−/− chimeras at day 3 p.i. (Figure 5H), when T cells, as well as B and NKT cells, are dispensable for viral clearance.42 However, decreased antiviral T-cell responses in the absence of Eri1 may have important functional consequences for the control of latent infection and the establishment of MCMV-specific immunologic memory.
Reduced MCMV-specific CD4+ and CD8+ T-cell frequency in the absence of Eri1. B6 WT CD45.1+ lethally irradiated hosts were reconstituted with a 1:1 mixture of congenic WT (CD45.1+ CD45.2+) and Eri1−/− (CD45.2+) fetal liver cells. At 8-15 weeks, chimeras were infected with MCMV and splenocytes were analyzed by flow cytometry at day 8 p.i. (A) Frequency of CD4+ and CD8+ cells among WT (CD45.1+ CD45.2+) or Eri1−/− (CD45.1− CD45.2+) splenocytes. (B) CD44 and CD62L expression on WT and Eri1−/− CD4+ and CD8+ cells. (C) Frequency of WT and Eri1−/− (day 8 p.i.) or WT control (uninfected) CD8+ cells labeled with M45-loaded H2-Db:Ig dimer. (D-G) Splenocytes from infected mice were restimulated with indicated peptides or PMA and ionomycin. Flow cytometric plots show the average percentage of gated (D) CD8+ or (F) CD4+ cells producing intracellular IFN-γ. Summary graphs for Eri1−/− splenic (E) CD8+ and (G) CD4+ T cells show mean IFN-γ expression relative to WT. Numbers indicate average values (A-C) ± SD or (E,G) SEM for 6 mice from 2 independent experiments. *P ≤ .05 **P ≤ .001; unpaired Student t test.
Reduced MCMV-specific CD4+ and CD8+ T-cell frequency in the absence of Eri1. B6 WT CD45.1+ lethally irradiated hosts were reconstituted with a 1:1 mixture of congenic WT (CD45.1+ CD45.2+) and Eri1−/− (CD45.2+) fetal liver cells. At 8-15 weeks, chimeras were infected with MCMV and splenocytes were analyzed by flow cytometry at day 8 p.i. (A) Frequency of CD4+ and CD8+ cells among WT (CD45.1+ CD45.2+) or Eri1−/− (CD45.1− CD45.2+) splenocytes. (B) CD44 and CD62L expression on WT and Eri1−/− CD4+ and CD8+ cells. (C) Frequency of WT and Eri1−/− (day 8 p.i.) or WT control (uninfected) CD8+ cells labeled with M45-loaded H2-Db:Ig dimer. (D-G) Splenocytes from infected mice were restimulated with indicated peptides or PMA and ionomycin. Flow cytometric plots show the average percentage of gated (D) CD8+ or (F) CD4+ cells producing intracellular IFN-γ. Summary graphs for Eri1−/− splenic (E) CD8+ and (G) CD4+ T cells show mean IFN-γ expression relative to WT. Numbers indicate average values (A-C) ± SD or (E,G) SEM for 6 mice from 2 independent experiments. *P ≤ .05 **P ≤ .001; unpaired Student t test.
Eri1 negatively regulates miRNA abundance in lymphocytes
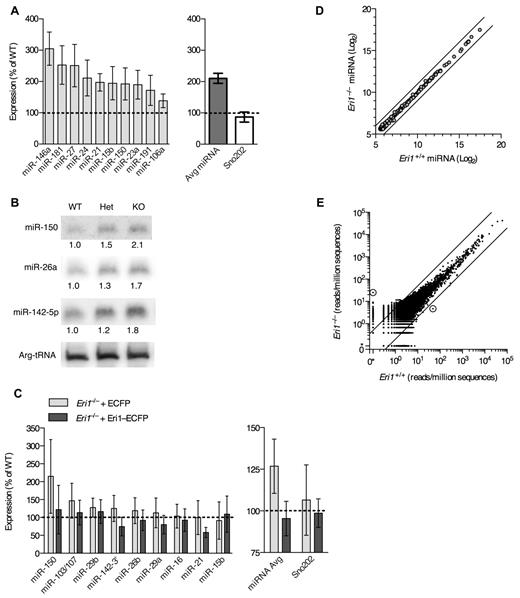
We next sought to identify important RNA targets for Eri1 in mouse lymphocytes. Eri1 inhibits RNAi in worms and mammalian cell lines and degrades siRNA duplexes in vitro.21,22 Given structural similarities between miRNA and siRNA duplexes, we conjectured that Eri1 regulates miRNA abundance in lymphocytes. Indeed, quantitative real-time PCR (qRT-PCR) revealed, on average, a 2-fold increase in miRNA expression in Eri1−/− NK cells compared with littermate controls (Figure 7A). Highly expressed miRNAs (eg, miR-150 and miR-21) and those expressed at 1-2 lower orders of magnitude (eg, miR-106a and miR-181) were affected.43 Other noncoding RNAs, including U6 snRNA, U7 snRNA, Arg-tRNA, and Sno202 were unaffected (Figure 7A, data not shown).
Eri1 negatively regulates miRNAs in lymphocytes. (A) qRT-PCR analysis of miRNA expression in ICR/B6 Eri1−/− NK (NK1.1+CD3ϵ−) cells purified by flow cytometry. (Left) miRNA levels in Eri1−/− NK cells shown relative to WT ( ). (Right) Sum of measurements from 10 miRNAs (
). (Right) Sum of measurements from 10 miRNAs ( ) and Sno202 (□). Data were normalized to U6 snRNA. Graphs indicate mean ± SEM (N = 6, ICR/B6 littermates). (B) Northern blot analysis of miRNAs from Eri1 WT (WT, Eri1+/+;CD4-cre), heterozygous (Het, Eri1fl/+;CD4-cre), and knockout (KO, Eri1fl/fl;CD4-cre) CD4+ T cells. Values indicate miRNA-specific signals quantified by phosphoimager, normalized to Arg-tRNA, and expressed relative to WT T cells. (C Left) qRT-PCR analysis of miRNA expression in Eri1-deficient (Eri1fl/fl;CD4-cre) T cells transduced with retroviruses encoding Thy1.1 and ECFP (
) and Sno202 (□). Data were normalized to U6 snRNA. Graphs indicate mean ± SEM (N = 6, ICR/B6 littermates). (B) Northern blot analysis of miRNAs from Eri1 WT (WT, Eri1+/+;CD4-cre), heterozygous (Het, Eri1fl/+;CD4-cre), and knockout (KO, Eri1fl/fl;CD4-cre) CD4+ T cells. Values indicate miRNA-specific signals quantified by phosphoimager, normalized to Arg-tRNA, and expressed relative to WT T cells. (C Left) qRT-PCR analysis of miRNA expression in Eri1-deficient (Eri1fl/fl;CD4-cre) T cells transduced with retroviruses encoding Thy1.1 and ECFP ( ) or Thy1.1 and Eri1-ECFP (
) or Thy1.1 and Eri1-ECFP ( ). Total RNA was prepared from transduced Thy1.1+ T cells purified by FACS. Data were normalized to U6 snRNA and expressed relative to miRNA measured in WT (Eri1+/+;CD4-cre) T cells transduced with ECFP retrovirus. (Right) Average of all miRNAs measured and Sno202 control. Columns show mean ± SEM (4 independent experiments). (D) Microarray comparison of miRNA expression patterns in WT and Eri1-deficient CD4+ T cells. Circles show average log2 hybridization fluorescence intensity values for quantile-normalized data from 3 independent T-cell samples. Black diagonal lines show 2-fold intensity differences. (E) Small RNA read counts from WT and Eri1−/− T-cell sequencing libraries. Dots show read counts at independent genomic loci with reads normalized to total genomic sequences in each library. Black lines indicate 5-fold expression differences. Circled dots show loci with > 90% posterior probability of a 5-fold expression difference between libraries. The location of these loci and gene origin of the most frequently cloned RNA from that locus are (left) Chr13:98860450-98860650, Rps18 pseudogene and (right) Chr8:73490090-73490290, Ell.
). Total RNA was prepared from transduced Thy1.1+ T cells purified by FACS. Data were normalized to U6 snRNA and expressed relative to miRNA measured in WT (Eri1+/+;CD4-cre) T cells transduced with ECFP retrovirus. (Right) Average of all miRNAs measured and Sno202 control. Columns show mean ± SEM (4 independent experiments). (D) Microarray comparison of miRNA expression patterns in WT and Eri1-deficient CD4+ T cells. Circles show average log2 hybridization fluorescence intensity values for quantile-normalized data from 3 independent T-cell samples. Black diagonal lines show 2-fold intensity differences. (E) Small RNA read counts from WT and Eri1−/− T-cell sequencing libraries. Dots show read counts at independent genomic loci with reads normalized to total genomic sequences in each library. Black lines indicate 5-fold expression differences. Circled dots show loci with > 90% posterior probability of a 5-fold expression difference between libraries. The location of these loci and gene origin of the most frequently cloned RNA from that locus are (left) Chr13:98860450-98860650, Rps18 pseudogene and (right) Chr8:73490090-73490290, Ell.
Eri1 negatively regulates miRNAs in lymphocytes. (A) qRT-PCR analysis of miRNA expression in ICR/B6 Eri1−/− NK (NK1.1+CD3ϵ−) cells purified by flow cytometry. (Left) miRNA levels in Eri1−/− NK cells shown relative to WT ( ). (Right) Sum of measurements from 10 miRNAs (
). (Right) Sum of measurements from 10 miRNAs ( ) and Sno202 (□). Data were normalized to U6 snRNA. Graphs indicate mean ± SEM (N = 6, ICR/B6 littermates). (B) Northern blot analysis of miRNAs from Eri1 WT (WT, Eri1+/+;CD4-cre), heterozygous (Het, Eri1fl/+;CD4-cre), and knockout (KO, Eri1fl/fl;CD4-cre) CD4+ T cells. Values indicate miRNA-specific signals quantified by phosphoimager, normalized to Arg-tRNA, and expressed relative to WT T cells. (C Left) qRT-PCR analysis of miRNA expression in Eri1-deficient (Eri1fl/fl;CD4-cre) T cells transduced with retroviruses encoding Thy1.1 and ECFP (
) and Sno202 (□). Data were normalized to U6 snRNA. Graphs indicate mean ± SEM (N = 6, ICR/B6 littermates). (B) Northern blot analysis of miRNAs from Eri1 WT (WT, Eri1+/+;CD4-cre), heterozygous (Het, Eri1fl/+;CD4-cre), and knockout (KO, Eri1fl/fl;CD4-cre) CD4+ T cells. Values indicate miRNA-specific signals quantified by phosphoimager, normalized to Arg-tRNA, and expressed relative to WT T cells. (C Left) qRT-PCR analysis of miRNA expression in Eri1-deficient (Eri1fl/fl;CD4-cre) T cells transduced with retroviruses encoding Thy1.1 and ECFP ( ) or Thy1.1 and Eri1-ECFP (
) or Thy1.1 and Eri1-ECFP ( ). Total RNA was prepared from transduced Thy1.1+ T cells purified by FACS. Data were normalized to U6 snRNA and expressed relative to miRNA measured in WT (Eri1+/+;CD4-cre) T cells transduced with ECFP retrovirus. (Right) Average of all miRNAs measured and Sno202 control. Columns show mean ± SEM (4 independent experiments). (D) Microarray comparison of miRNA expression patterns in WT and Eri1-deficient CD4+ T cells. Circles show average log2 hybridization fluorescence intensity values for quantile-normalized data from 3 independent T-cell samples. Black diagonal lines show 2-fold intensity differences. (E) Small RNA read counts from WT and Eri1−/− T-cell sequencing libraries. Dots show read counts at independent genomic loci with reads normalized to total genomic sequences in each library. Black lines indicate 5-fold expression differences. Circled dots show loci with > 90% posterior probability of a 5-fold expression difference between libraries. The location of these loci and gene origin of the most frequently cloned RNA from that locus are (left) Chr13:98860450-98860650, Rps18 pseudogene and (right) Chr8:73490090-73490290, Ell.
). Total RNA was prepared from transduced Thy1.1+ T cells purified by FACS. Data were normalized to U6 snRNA and expressed relative to miRNA measured in WT (Eri1+/+;CD4-cre) T cells transduced with ECFP retrovirus. (Right) Average of all miRNAs measured and Sno202 control. Columns show mean ± SEM (4 independent experiments). (D) Microarray comparison of miRNA expression patterns in WT and Eri1-deficient CD4+ T cells. Circles show average log2 hybridization fluorescence intensity values for quantile-normalized data from 3 independent T-cell samples. Black diagonal lines show 2-fold intensity differences. (E) Small RNA read counts from WT and Eri1−/− T-cell sequencing libraries. Dots show read counts at independent genomic loci with reads normalized to total genomic sequences in each library. Black lines indicate 5-fold expression differences. Circled dots show loci with > 90% posterior probability of a 5-fold expression difference between libraries. The location of these loci and gene origin of the most frequently cloned RNA from that locus are (left) Chr13:98860450-98860650, Rps18 pseudogene and (right) Chr8:73490090-73490290, Ell.
Eri1 also regulated miRNA abundance in CD4+ T cells. Northern blot analyses showed a modest, consistent increase in the expression of several miRNAs in Eri1-deficient T cells (Figure 7B). Because we could transduce primary Eri1−/− T cells more easily than NK cells, we used these lymphocytes to rescue miRNA levels by ectopic expression of Eri1. Transducing Eri1-deficient T cells with a retrovirus encoding an Eri1-ECFP fusion protein15 reduced miRNA expression to WT levels (Figure 7C). Thus, altered miRNA abundance in Eri1-deficient lymphocytes was not the indirect result of impaired lymphocyte development.
To test whether Eri1 preferentially affects some miRNAs, we performed microarray analysis of 3 independent Eri1-deficient and matched WT control T-cell samples (Figure 7D). Despite a high degree of reproducibility between samples (Pearson correlation coefficient r ≥ 0.99), this analysis revealed no differentially expressed miRNAs in Eri1-deficient cells when using a false discovery rate of 5%. Note that array data were quantile normalized to compare expression of each miRNA relative to all other miRNAs, so these experiments do not detect global changes in miRNA expression. The high degree of similarity in miRNA expression patterns between WT and Eri1-deficient T cells indicated that Eri1 globally regulates the homeostasis of all miRNAs without any discernible sequence specificity.
In C elegans, ERI-1 interacts with Dicer to form a complex that is required to generate some classes of endo-siRNAs.19,20 To determine whether Eri1 is required for the biogenesis of any noncanonical classes of small RNAs in mouse lymphocytes, we used deep sequencing to broadly profile the small RNA transcriptome of WT and Eri1−/− T cells (Figure 7E, supplemental Figure 4). All sequences were mapped to the mouse genome and assigned to 390 000 empirically determined genomic loci as described previously.29 This analysis revealed a high degree of similarity between WT and Eri1−/− small RNA libraries (r > 0.97; Figure 7E). Only 2 genomic loci had a > 90% probability of a 5-fold or greater expression difference between the 2 libraries. A 5-fold cutoff for significance was established based on the observation that C elegans deficient in components of the ERI-1–Dicer complex show at least a 5-fold decrease in specific classes of endo-siRNAs.44 Of the 2 loci differentially expressed in Eri1-deficient T cells, one (chromosome 13) could be accounted for by a single nucleotide polymorphism present in the Eri1−/− but not the WT sample. The other locus (chromosome 8) contained a 28-nt RNA that mapped to a nonconserved intronic region of Ell. Using qRT-PCR, we could not confirm that this RNA was differentially expressed in Eri1−/− T cells (data not shown). Together, our small RNA profiling data show that Eri1 negatively regulates miRNA abundance in a sequence-independent manner and that Eri1 is not required for the biogenesis of any abundant classes of small RNAs in T cells.
Discussion
Eri1 is a highly conserved exoribonuclease that has been recruited into small RNA regulatory pathways in evolutionarily diverse organisms. Our findings establish that mammalian Eri1 modulates global miRNA abundance, a novel regulatory activity that may be important for proper lymphocyte development and effector function. While Eri1−/− mice had normal numbers of B, T, and NKT cells, all Eri1−/− lymphocyte lineages were reduced in mixed fetal liver chimeras. Thus, in the absence of competition, the homeostatic control of peripheral B- and T-cell numbers in Eri1−/− mice likely masks a defect in their development. In contrast, steady-state NK-cell populations were reduced to half their normal number. This observation may reflect differences in the regulation of NK-cell versus B- and T-cell homeostasis, or an NK cell–specific dependence on Eri1 activity. The remaining Eri1−/− NK cells exhibited an immature phenotype and skewed Ly49 repertoire marked by reduced Ly49H+ cells. Furthermore, these Eri1−/− Ly49H+ NK cells failed to expand efficiently during MCMV infection. Eri1−/− CD4+ and CD8+ T cells also displayed diminished Ag-specific MCMV responses, suggesting that Eri1 may generally enhance lymphocyte-mediated antiviral immunity.
Immature BM CD49b−αV+ NK cells acquire Ly49 receptors in a developmentally regulated fashion that correlates with c-Kit up-regulation.32 Ly49 induction requires direct contact with the BM stroma and is altered in the setting of signaling pathway defects.35,36,45,46 Similar to NK cells deficient in PI3K subunits or phospholipase Cγ2, Eri1−/− BM NK cells showed delayed acquisition of multiple Ly49 receptors. However, unlike these mutants, which show mirrored peripheral Ly49 reduction, Eri1−/− NK splenocytes have a normal Ly49 inhibitory repertoire and a selective reduction in Ly49D and Ly49H activating receptors. We hypothesize that the specific reduction in activating receptor repertoires results from delayed acquisition of all Ly49 receptors in the BM followed by the selective outgrowth of NK cells bearing specific inhibitory receptors.
Ly49A, Ly49C, Ly49I, Ly49G2, and Ly49D all recognize class I MHC, and MHC may in turn shape their expression on NK cells.47 In contrast, the MCMV protein m157 is the only known ligand of Ly49H. The observed decrease in Ly49H- and Ly49D-expressing Eri1−/− NK cells from mixed chimeras indicates that a cell-intrinsic mechanism underlies this defect rather than aberrant Ly49 ligand distribution. The adapter molecules DAP10 and DAP12 stabilize activating Ly49 receptors on the cell surface, and loss of either of these proteins leads to reduced Ly49D and Ly49H expression.24,48 Yet, receptor destabilization is an unlikely mechanism for Ly49 repertoire skewing on Eri1−/− NK cells, as they showed no change in receptor density at the cell surface. Another possibility is that Ly49 activating receptors are direct miRNA targets that become down-regulated by miRNA derepression in Eri1−/− NK cells. This is also unlikely given that miRNA depletion from mature NK cells does not alter Ly49 expression.8 A more probable scenario is that Eri1 alters Ly49H and Ly49D acquisition indirectly through effects on Ly49 transcriptional regulators. Such a mechanism has been proposed for aberrant Ly49A acquisition observed in miR-150 mutant mice.9 Further study of Eri1−/− NK cells may provide new insight into the BM signals that drive Ly49 expression in developing NK cells.
The best predictor of effective antiviral immunity to MCMV in B6 mice is the expansion of pathogen-specific NK cells.3,37,38,49 Eri1−/− NK cells displayed a reduced Ly49H+ repertoire and poor Ly49H-dependent expansion in response to viral infection, with a corresponding deficiency in viral clearance during the acute phase of infection. Although we observed a decreased frequency of MCMV-specific Eri1−/− T cells in mixed fetal liver chimeras, T, B, and NKT cells are dispensable for viral clearance during early acute infection.42 Therefore, we speculate that NK-cell dysfunction is the primary reason Eri1−/− chimeras poorly controlled MCMV viral load. However, we cannot rule out the possibility that other hematopoietic populations may contribute to diminished viral clearance. Further study is required to fully define the extent of Eri1 regulation of immune responses.
Reduced NK-cell expansion was evident when equal numbers of WT and Eri1−/− Ly49H+ NK cells were transferred to Ly49H-deficient hosts, suggesting an NK cell–intrinsic defect. Eri1−/− NK cells also displayed a modest defect in IFN-γ production on cross-linking of ITAM-associated receptors in vitro, suggesting a general defect in activating receptor signal transduction that may contribute to poor viral control. Yet, on a per-cell basis, Eri1−/− and WT Ly49H+ NK cells surprisingly showed equal Ly49H-dependent degranulation and target cell killing. Thus, Eri1−/− NK cells are not generally hyporesponsive to Ly49H ligation or unable to mediate effector functions. In addition, reduced NK-cell expansion in MCMV infection does not reflect a general proliferation defect, as we observed normal IL-15–driven expansion in vitro and in vivo. Further research is needed to identify the affected pathways that mediate poor expansion of Eri1−/− Ly49H+ NK cells during MCMV infection.
Although we do not yet understand how Eri1 modulates mature miRNA abundance, we speculate that it acts by direct enzymatic degradation of precursor or mature miRNAs, as is observed for the small RNA exonucleases SDN1 in Arabidopsis and XRN-2 in C elegans.50,51 More generally, our data suggest that Eri1-dependent regulation of endogenous small RNAs in mammalian somatic cells is distinctly different from that observed in S pombe or C elegans, where Eri1 regulates endo-siRNA abundance. Deep sequencing analysis of small RNAs in WT and Eri1-deficient T cells revealed no small RNA species that were as dependent on Eri1 as some classes of endo-siRNAs are in C elegans. Thus, miRNAs are likely the major small RNA target for Eri1 in somatic mammalian cells. This activity may not be restricted to mammals, as one previous report found that eri-1 mutant C elegans expressed increased levels of mature miR-238.20
We cannot exclude the possibility that other Eri1 substrates, such as ribosomal RNA, may mediate some of the phenotypes observed in Eri1−/− lymphocytes. Interestingly, Eri1−/− mice share some phenotypic similarities with humans who have Diamond-Blackfan anemia (DBA),15 a heterogeneous disease most commonly attributed to Rps19 mutations.52 Lymphocyte deficiency is a common feature of many ribosomopathies including DBA, Shwachman-Diamond syndrome, and dyskeratosis congenita.53 Of note, we were unable to detect NK-cell homeostasis defects in Rps19 mutant mice (Dsk3) or Rps20 mutants (Dsk4; data not shown).54 Studies are currently under way to determine whether NK-cell deficiency occurs in other mouse strains with mutations in ribosome-associated proteins.
These results imply that miRNAs as a class are negatively regulated in lymphocytes and, by extension, so is miRNA-mediated gene silencing. Although Eri1 is broadly expressed because of its constitutive role in 5.8S rRNA maturation, it is enriched in lymphoid organs and is strongly up-regulated in activated lymphocytes. Thus, Eri1-mediated repression of miRNAs may lead to cell-type–specific defects. In this capacity, Eri1 comprises a growing class of factors that modulate miRNA expression at a global level.14 Many of these factors, including Eri1, are attractive therapeutic targets whose inhibition could enhance miRNA- or siRNA-mediated gene repression.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ariya Lapan, Laura Smith, and Eric Yanni for technical assistance; Chris Eisley, Rebecca Barbeau, Andrea Barczak, and David Erle (SABRE Functional Genomics Core) for expert assistance with microarray experiments; Jon Woo (University of California, San Francisco [UCSF] Genomics Core Facility) for assistance with small RNA sequencing; Natalie Bezman for critical comments on the manuscript; and Joseph Sun, Sandra Lopez-Verges, Maelig Morvan, and Yosuke Kamimura for helpful discussions.
This work was supported by the Burroughs Wellcome Fund (CABS 1006173), the National Institutes of Health (NIH; HL109102 [K.M.A.], and AI089828, AI70788 and AI068129 [L.L.L.]), the German Research Foundation (DFG HE 3359/3-1), and the UCSF/NIH Medical Scientist Training Program (M.F.T.).
L.L.L. is an American Cancer Society Professor.
National Institutes of Health
Authorship
Contribution: M.F.T. and K.M.A. performed research, analyzed data, and wrote the manuscript; S.A.-W. and M.P. performed some of the T-cell experiments, including miRNA qPCR for transduced T cells (S.A.-W.); J.E.B., M.R., and P.W. assisted with small RNA deep sequencing analysis and statistics; and K.M.A. and M.F.T. designed research with critical input from L.L.L. and V.H.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: K. Mark Ansel, PhD, Department of Microbiology and Immunology, University of California San Francisco, 513 Parnassus Ave, Box 0414, San Francisco, CA 94143-0414; e-mail: mark.ansel@ucsf.edu.