Abstract
Diagnosis and classification of VWD is aided by molecular analysis of the VWF gene. Because VWF polymorphisms have not been fully characterized, we performed VWF laboratory testing and gene sequencing of 184 healthy controls with a negative bleeding history. The controls included 66 (35.9%) African Americans (AAs). We identified 21 new sequence variations, 13 (62%) of which occurred exclusively in AAs and 2 (G967D, T2666M) that were found in 10%-15% of the AA samples, suggesting they are polymorphisms. We identified 14 sequence variations reported previously as VWF mutations, the majority of which were type 1 mutations. These controls had VWF Ag levels within the normal range, suggesting that these sequence variations might not always reduce plasma VWF levels. Eleven mutations were found in AAs, and the frequency of M740I, H817Q, and R2185Q was 15%-18%. Ten AA controls had the 2N mutation H817Q; 1 was homozygous. The average factor VIII level in this group was 99 IU/dL, suggesting that this variation may confer little or no clinical symptoms. This study emphasizes the importance of sequencing healthy controls to understand ethnic-specific sequence variations so that asymptomatic sequence variations are not misidentified as mutations in other ethnic or racial groups.
Introduction
VWF is a large, multimeric plasma glycoprotein that binds to damaged vessel walls and mediates platelet adhesions to the vascular endothelium at sites of injury.1 In addition, VWF binds and stabilizes factor VIII (FVIII) in the circulation. Defects in VWF associated with bleeding symptoms may cause VWD, a common inherited bleeding disorder with a reported incidence ranging from 0.01%-1%.2,3 VWD is classified by quantitative (types 1 and 3) and qualitative (type 2) defects in VWF.1 The defects observed in type 2 VWD include defects in multimerization, platelet interactions, or FVIII binding that may be identified by laboratory tests that assess the functional characteristics of VWF. Molecular analysis of the VWF gene is also useful in the diagnosis and classification of VWD. Many type 2 mutations are localized to specific functional domains in exons 18-25 (type 2N), exon 28 (types 2A, 2B, and 2M), and exons 11-16, 24-26, and 51-52 (type 2A), whereas the type 1 and type 3 VWD mutations have been identified in or near exons throughout the VWF gene.4 Extensive sequence variation has been identified in VWF by studies carried out in the European Union, the United Kingdom, and Canada.5,6 These studies concentrated on sequencing the VWF gene in patients with type 1 VWD, but did not sequence healthy controls or examine sequence variations in different ethnic groups.
One of the characteristics used to distinguish benign from disease-causing mutations is the allele frequency. Sequence variations present at > 1% in a population are typically considered to be polymorphisms. Our knowledge of single nucleotide polymorphisms (SNPs) has been enhanced by the HapMap project, but our knowledge of rare SNPs or SNPs specific to a population or ethnic group is still incomplete. Numerous studies have shown a higher level of nucleotide diversity in Africans relative to non-Africans,7 but the diversity has not been characterized in all genes, including VWF. Sequence variations may be rare in one population but frequent in another. For example, the β-hemoglobin G6V mutation causing sickle cell disease is rare in the white population, but is found in 8%-10% of AAs.8 This situation is complicated by the definition that a polymorphism is a sequence variation found in the population at > 1% frequency. It is not uncommon when sequencing genes responsible for rare disorders to identify sequence variations of unknown significance. The significance of unknown variations can be a particular problem when studying a disorder—such as VWD—that some predict to be present in 1% of the population.2,9 Given the disease frequency, it is more difficult to distinguish disease-causing alleles from benign polymorphisms. The significance of the variation can sometimes be resolved by functional analysis when possible, by family studies, or by the identification of the variation in several individuals without the disorder. Sequencing of healthy controls in the ethnic groups being studied is crucial to the understanding of new sequence variations, particularly when the racial/ethnic admixture can result in the rare identification of a sequence variation in an person with slightly reduced VWF, leading to the assumption that this sequence alteration is causative.
The T.S. Zimmerman Program for the Molecular and Clinical Biology of VWD (ZPMCB-VWD) is a multinational National Institutes of Health Program Project established to further the study of VWD in the United States and to contrast these studies with the studies initiated previously in the European Union, United Kingdom, and Canada. As part of this study, we sought further insight into the polymorphisms and sequence variations more frequently identified in AAs by performing DNA sequence analysis of VWF coding regions and laboratory testing on healthy controls enrolled in the study. Our data indicate that the sequence variations common in the AA population are rare in whites and may have been misidentified as VWF mutations.
Methods
Patient population
Subjects with no previous diagnosis of a bleeding disorder were enrolled in the ZPMCB-VWD as healthy controls. Exclusion criteria included a previous diagnosis of VWD and known pregnancy. Subjects from the general population were recruited at the 7 primary centers in Atlanta, Detroit, Houston, Indianapolis, Milwaukee, New Orleans, and Pittsburgh. All healthy controls were 18 years of age or older. Race was determined by self-identification. A bleeding questionnaire was administered to all control subjects. Questions included those used in the European Molecular and Clinical Markers for the Diagnosis and Management of type 1 VWD study (MCMDM-1VWD),10 as well as a set of additional questions specific to the ZPMCB-VWD. Bleeding scores herein were calculated using the MCMDM-1VWD scoring system. Laboratory testing and VWF gene sequencing was completed on all healthy controls included in this study. This study is a report on the first 184 healthy controls, including 118 whites and 66 AAs. Only controls with completed laboratory testing and gene sequencing results were included in this analysis.
Blood was collected and the plasma was sent to a central reference laboratory at BloodCenter of Wisconsin for VWF Ag (VWF:Ag), VWF ristocetin cofactor (VWF:RCo), FVIII activity, VWF collagen binding (VWF:CB), VWF propeptide (VWFpp), ABO blood type, and multimer analysis. A blood sample was also collected to prepare DNA for VWF full gene sequencing. The study was approved by the Human Research Review Board at each institution and informed consent was obtained from all subjects in accordance with the Declaration of Helsinki.
DNA sequence analysis
DNA was isolated using the PureGene method (QIAGEN). PCR and VWF gene sequencing was performed at Partners Healthcare Center for Genetics and Genomics, Harvard Medical School (Boston, MA). Primers were designed to specifically amplify the entire coding sequence of VWF, including intron-exon boundaries, 3.5 kb upstream of exon 1, and 0.8 kb downstream of the C-terminal stop codon while excluding the VWF pseudogene sequence that includes exons 23-34. The primer sequences are available on request. Bidirectional VWF gene sequencing was carried out using Applied Biosystems Bigdye Terminator Version 3.1 cycle sequencing chemistry software. Sequence data were compared with the VWF reference sequence NC_000012.1.
VWF testing
VWF:Ag was determined by ELISA using 2 different mAbs (105.4 and AVW-1) for capture and a rabbit polyclonal Ab for detection.11 VWF:RCo was measured using formalin-fixed platelets and standard concentrations of ristocetin on a BCS (Dade-Behring). VWF multimers were assessed by 0.65% agarose multimer gels.12 FVIII activity was measured using a 1-stage clotting assay. VWF:CB was measured using type III human placental collagen (Southern Biotechnology Associates) for capture and a rabbit polyclonal Ab for detection of VWF binding. The assay of VWFpp was done as described previously.13
Results
As part of ZPMCB-VWD, in the present study, we performed full VWF laboratory testing and gene sequencing of 184 healthy controls to better characterize the sequence variations throughout the VWF gene and in different ethnic groups. The study included 118 whites and 66 AAs. The healthy controls were individuals with a negative bleeding history. Laboratory values included VWF:Ag, VWF:RCo, FVIII levels, VWF:CB, and VWFpp levels and were all in the normal range (Table 1) except for 3 samples, 1 with a low VWF:Ag (36 IU/dL) and 3 with a slightly low VWF:RCo (46-51 IU/dL). The sample with low VWF:Ag also had a decreased VWF:RCo (46 IU/dL) but a normal VWF:RCo/VWF:Ag ratio.
VWF laboratory data for healthy controls
| . | VWF:Ag . | VWF:RCo . | FVIII . | VWF:CB . | VWFpp . |
|---|---|---|---|---|---|
| Laboratory normal range | 50-230 | 54-279 | 60-195 | 59-249 | 60-190 |
| All controls, (n = 184) | 109 (36-285) | 116 (46-301) | 106 (53-218) | 127 (50-332) | 111 (47-296) |
| African Americans, (n = 66) | 123 (66-285) | 118 (51-290) | 118 (63-198) | 146 (71-332) | 116 (59-201) |
| Whites, (n = 118) | 101 (36-235) | 114 (46-301) | 100 (53-218) | 117 (50-325) | 108 (47-296) |
| . | VWF:Ag . | VWF:RCo . | FVIII . | VWF:CB . | VWFpp . |
|---|---|---|---|---|---|
| Laboratory normal range | 50-230 | 54-279 | 60-195 | 59-249 | 60-190 |
| All controls, (n = 184) | 109 (36-285) | 116 (46-301) | 106 (53-218) | 127 (50-332) | 111 (47-296) |
| African Americans, (n = 66) | 123 (66-285) | 118 (51-290) | 118 (63-198) | 146 (71-332) | 116 (59-201) |
| Whites, (n = 118) | 101 (36-235) | 114 (46-301) | 100 (53-218) | 117 (50-325) | 108 (47-296) |
Log-transformed mean laboratory values and ranges (in parentheses) for all controls, African Americans, and whites. The laboratory normal range is the log-transformed mean ± 2 SD. All values are in IU/dL.
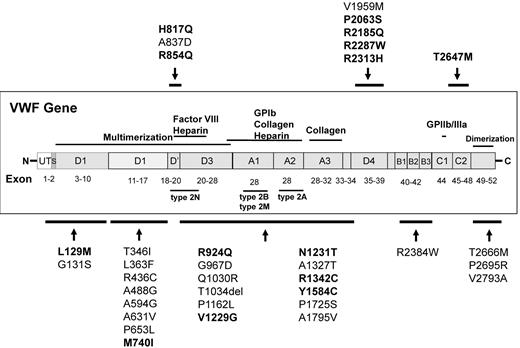
VWF DNA sequence analysis of healthy controls revealed 35 sequence variations that were either unreported or reported previously as a VWF mutation (Figure 1). We identified 21 new sequence variations in healthy controls (Table 2). These sequence variations were observed throughout the 52 exons of the gene. Most of the variations were SNPs, but we also observed a dinucleotide change (3485_3486CA > TG) and a 3-bp deletion (3101_3103delCCA). Thirteen (62%) of 21 sequence variations were found exclusively in AAs. Two of the sequence variations (2900G > A (G967D) and 7997C > T (T2666M)) were found in 10%-15% of the AA samples, suggesting that they were polymorphisms. Overall, 80.4% of the new sequence variations were observed in AAs and 19.6% were observed in whites. One of the new sequence variations, A631V, was observed in a white subject, the one healthy control with abnormal VWF:Ag and VWF:RCo.
Novel sequence variations and reported VWF mutations identified in healthy controls. The approximate location of VWF sequence variations is indicated. The sequence variations reported previously as VWF mutations are in bold.
Novel sequence variations and reported VWF mutations identified in healthy controls. The approximate location of VWF sequence variations is indicated. The sequence variations reported previously as VWF mutations are in bold.
New sequence variations identified in healthy controls
| Exon . | Nucleotide . | Amino acid . | Observations, n . | ||
|---|---|---|---|---|---|
| Total . | African Americans . | Whites . | |||
| 5 | 391G > A | G131S | 5 | 5 | 0 |
| 9 | 1037C > T | T346I | 1 | 1 | 0 |
| 9 | 1087C > T | L363F | 1 | 0 | 1 |
| 12 | 1306C > T | R436C | 1 | 1 | 0 |
| 13 | 1463C > G | A488G | 1 | 1 | 0 |
| 15 | 1781C > G | A594G | 1 | 0 | 1 |
| 15 | 1892C > T | A631V | 1 | 0 | 1 |
| 16 | 1958C > T | P653L | 1 | 0 | 1 |
| 19 | 2510C > A | A837D | 2 | 2 | 0 |
| 22 | 2900G > A | G967D | 7 | 7 | 0 |
| 23 | 3089A > G | Q1030R | 1 | 1 | 0 |
| 23 | 3101_3103delCCA | T1034del | 2 | 2 | 0 |
| 26 | 3485_3486CA > TG | P1162L | 1 | 1 | 0 |
| 28 | 3979G > A | A1327T | 1 | 0 | 1 |
| 30 | 5173C > T | P1725S | 2 | 2 | 0 |
| 31 | 5384C > T | A1795V | 1 | 1 | 0 |
| 35 | 5875G > A | V1959M | 1 | 0 | 1 |
| 42 | 7150C > T | R2384W | 2 | 2 | 0 |
| 49 | 7997C > T | T2666M | 12 | 10 | 2 |
| 49 | 8084C > G | P2695R | 1 | 0 | 1 |
| 52 | 8378T > C | V2793A | 1 | 1 | 0 |
| Total | 46 | 37 | 9 | ||
| Percentage of total | 80.4% | 19.6% | |||
| Exon . | Nucleotide . | Amino acid . | Observations, n . | ||
|---|---|---|---|---|---|
| Total . | African Americans . | Whites . | |||
| 5 | 391G > A | G131S | 5 | 5 | 0 |
| 9 | 1037C > T | T346I | 1 | 1 | 0 |
| 9 | 1087C > T | L363F | 1 | 0 | 1 |
| 12 | 1306C > T | R436C | 1 | 1 | 0 |
| 13 | 1463C > G | A488G | 1 | 1 | 0 |
| 15 | 1781C > G | A594G | 1 | 0 | 1 |
| 15 | 1892C > T | A631V | 1 | 0 | 1 |
| 16 | 1958C > T | P653L | 1 | 0 | 1 |
| 19 | 2510C > A | A837D | 2 | 2 | 0 |
| 22 | 2900G > A | G967D | 7 | 7 | 0 |
| 23 | 3089A > G | Q1030R | 1 | 1 | 0 |
| 23 | 3101_3103delCCA | T1034del | 2 | 2 | 0 |
| 26 | 3485_3486CA > TG | P1162L | 1 | 1 | 0 |
| 28 | 3979G > A | A1327T | 1 | 0 | 1 |
| 30 | 5173C > T | P1725S | 2 | 2 | 0 |
| 31 | 5384C > T | A1795V | 1 | 1 | 0 |
| 35 | 5875G > A | V1959M | 1 | 0 | 1 |
| 42 | 7150C > T | R2384W | 2 | 2 | 0 |
| 49 | 7997C > T | T2666M | 12 | 10 | 2 |
| 49 | 8084C > G | P2695R | 1 | 0 | 1 |
| 52 | 8378T > C | V2793A | 1 | 1 | 0 |
| Total | 46 | 37 | 9 | ||
| Percentage of total | 80.4% | 19.6% | |||
Twenty-one new sequence variations were identified. The variations were mainly observed in African Americans (80.4%) versus whites (19.6%). Thirteen of 21 variations were only observed in African Americans (shaded). G131S, G967D, and T2666M were present in 5 or more individuals. All these controls had normal laboratory values except for the sample with A631V, which had low VWF:Ag (36 IU/dL) and decreased VWF:RCo (46 IU/dL) but a normal VWF:RCo/VWF:Ag ratio.
We identified 14 sequence variations in the healthy controls that had been reported previously as VWF mutations in the ISTH-SSC VWF database (http://www.VWF.group.shef.ac.uk/; Table 3). The majority of these sequence variations were reported as type 1 VWD mutations. Similar to what was observed for new sequence variations in healthy controls, the reported VWD mutations were mainly observed in AAs. Twelve (86%) of the 14 mutations were found in AAs and 10 (71%) were exclusively observed in this group. The frequency of 3 of mutations (M740I, H817Q, and R2185Q) was 15%-18% in AAs. Ten controls had the reported type 2N H817Q mutation and 1 of the 10 was homozygous. Four mutations were observed in whites (R854Q, R924Q, P2063S, and R2185Q). R854Q and P2063S occurred exclusively in whites.
Reported VWF mutations identified in healthy controls (HC)
| Nucleotide . | Amino Acid . | VWD type . | Observations, n . | Frequency, % . | Average laboratory values, IU/dL . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HC . | AA . | W . | HC . | AA . | W . | VWF:Ag . | VWF:RCo . | FVIII . | VWFpp . | VWF:CB . | |||
| 385C > A | L129M | 1 | 1 | 1 | 0 | 0.5 | 1.5 | 0.0 | 233 | 225 | 143 | 165 | 266 |
| 2220G > A | M740I | 1/2M | 12 | 12 | 0 | 6.3 | 18.2 | 0.0 | 112 (69-231) | 93 (57-248) | 102 (63-151) | 114 (88-167) | 134 (71-265) |
| 2451T > A | H817Q | 2N | 10 | 10 | 0 | 5.2 | 15.2 | 0.0 | 106 (69-231) | 91 (57-248) | 99 (63-151) | 115 (88-167) | 129 (71-265) |
| 2561G > A | R854Q | 2N | 2 | 0 | 2 | 1.0 | 0.0 | 1.7 | 122 (117-128) | 150 (145-155) | 92 (74-115) | 139 (134-145) | 115 (105-126) |
| 2771G > A | R924Q | 1/2N | 4 | 1 | 3 | 2.1 | 1.5 | 2.5 | 95 (82-114) | 135 (107-169) | 95 (78-18) | 96 (87-107) | 108 (93-138) |
| 3686T > G | V1229G | 1 | 3 | 3 | 0 | 1.6 | 4.5 | 0.0 | 157 (87-210) | 150 (97-193) | 109 (72-152) | 145 (114-171) | 152 (103-208) |
| 3692A > C | N1231T | 1 | 2 | 2 | 0 | 1.0 | 3.0 | 0.0 | 128 (87-187) | 137 (97-193) | 93 (72-119) | 140 (114-171) | 146 (103-208) |
| 4024C > T | R1342C | 1 | 1 | 1 | 0 | 0.5 | 1.5 | 0.0 | 90 | 51 | 95 | 95 | 98 |
| 4751A > G | Y1584C | 1 | 1 | 1 | 0 | 0.5 | 1.5 | 0.0 | 61 | 47 | 95 | 65 | 69 |
| 6187C > T | P2063S | 1 | 2 | 0 | 2 | 1.0 | 0.0 | 1.7 | 94 (74-119) | 84 (74-96) | 102 (75-138) | 114 (103-127) | 81 (65-102) |
| 6554G > A | R2185Q | 1 | 15 | 12 | 3 | 7.9 | 18.2 | 2.5 | 118 (69-231) | 120 (67-248) | 120 (89-198) | 113 (93-167) | 142 (96-265) |
| 6859C > T | R2287W | 1/2A | 2 | 2 | 0 | 1.0 | 3.0 | 0.0 | 103 (99-107) | 83 (71-98) | 124 (98-156) | 133 (128-138) | 116 (95-141) |
| 6938G > A | R2313H | 1 | 1 | 1 | 0 | 0.5 | 1.5 | 0.0 | 173 | 220 | 162 | 184 | 227 |
| 7940C > T | T2647M | 1 | 1 | 1 | 0 | 0.5 | 1.5 | 0.0 | 83 | 114 | 97 | 90 | 120 |
| Nucleotide . | Amino Acid . | VWD type . | Observations, n . | Frequency, % . | Average laboratory values, IU/dL . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HC . | AA . | W . | HC . | AA . | W . | VWF:Ag . | VWF:RCo . | FVIII . | VWFpp . | VWF:CB . | |||
| 385C > A | L129M | 1 | 1 | 1 | 0 | 0.5 | 1.5 | 0.0 | 233 | 225 | 143 | 165 | 266 |
| 2220G > A | M740I | 1/2M | 12 | 12 | 0 | 6.3 | 18.2 | 0.0 | 112 (69-231) | 93 (57-248) | 102 (63-151) | 114 (88-167) | 134 (71-265) |
| 2451T > A | H817Q | 2N | 10 | 10 | 0 | 5.2 | 15.2 | 0.0 | 106 (69-231) | 91 (57-248) | 99 (63-151) | 115 (88-167) | 129 (71-265) |
| 2561G > A | R854Q | 2N | 2 | 0 | 2 | 1.0 | 0.0 | 1.7 | 122 (117-128) | 150 (145-155) | 92 (74-115) | 139 (134-145) | 115 (105-126) |
| 2771G > A | R924Q | 1/2N | 4 | 1 | 3 | 2.1 | 1.5 | 2.5 | 95 (82-114) | 135 (107-169) | 95 (78-18) | 96 (87-107) | 108 (93-138) |
| 3686T > G | V1229G | 1 | 3 | 3 | 0 | 1.6 | 4.5 | 0.0 | 157 (87-210) | 150 (97-193) | 109 (72-152) | 145 (114-171) | 152 (103-208) |
| 3692A > C | N1231T | 1 | 2 | 2 | 0 | 1.0 | 3.0 | 0.0 | 128 (87-187) | 137 (97-193) | 93 (72-119) | 140 (114-171) | 146 (103-208) |
| 4024C > T | R1342C | 1 | 1 | 1 | 0 | 0.5 | 1.5 | 0.0 | 90 | 51 | 95 | 95 | 98 |
| 4751A > G | Y1584C | 1 | 1 | 1 | 0 | 0.5 | 1.5 | 0.0 | 61 | 47 | 95 | 65 | 69 |
| 6187C > T | P2063S | 1 | 2 | 0 | 2 | 1.0 | 0.0 | 1.7 | 94 (74-119) | 84 (74-96) | 102 (75-138) | 114 (103-127) | 81 (65-102) |
| 6554G > A | R2185Q | 1 | 15 | 12 | 3 | 7.9 | 18.2 | 2.5 | 118 (69-231) | 120 (67-248) | 120 (89-198) | 113 (93-167) | 142 (96-265) |
| 6859C > T | R2287W | 1/2A | 2 | 2 | 0 | 1.0 | 3.0 | 0.0 | 103 (99-107) | 83 (71-98) | 124 (98-156) | 133 (128-138) | 116 (95-141) |
| 6938G > A | R2313H | 1 | 1 | 1 | 0 | 0.5 | 1.5 | 0.0 | 173 | 220 | 162 | 184 | 227 |
| 7940C > T | T2647M | 1 | 1 | 1 | 0 | 0.5 | 1.5 | 0.0 | 83 | 114 | 97 | 90 | 120 |
The reported mutations were frequently identified in African Americans (AA). M701I, H817Q and R2185 were found in greater than 15% of African-American healthy controls. R854Q and P2063S were only seen in Whites (W). Laboratory values are reported as a log transformed average (range) if there is more than one individual. Slightly abnormal values were only seen for Y1584C (low VWF:RCo) and R1342C (low VWF:RCo/VWF:Ag).
In healthy control individuals with the reported VWF mutations, there was a negative bleeding history. The average laboratory values for VWF:Ag, VWF:RCo, FVIII, VWFpp, and VWF:CB are shown in Table 3. The controls with reported type 1 mutations had VWF:Ag levels within the normal range and the average level for these controls (115 IU/dL) was higher than all healthy controls (109 IU/dL). For the most frequently observed mutations, M740I and R2185Q, the average VWF Ag level was greater than 130 IU/dL. Two of the healthy controls with reported VWF mutations had abnormal VWF:RCo values. The sample from a type O individual containing the Y1584C mutation had a VWF:Ag of 61 IU/dL and a VWF:RCo of 47 IU/dL. The VWF:Ag was also reduced in this sample, so the VWF:RCo/VWF:Ag ratio was normal. These data are consistent with Y1584C being a VWD type 1 mutation. The healthy control sample containing the R1342C mutation had a normal VWF:Ag level (90 IU/dL) but a decreased VWF:RCo (51 IU/dL). The abnormal VWF:RCo/VWF:Ag ratio of 0.59 suggests that this sequence variation may cause a type 2 defect. However, this sample was also heterozygous for D1472H, which can disrupt ristocetin binding and cause an artificially low VWF:RCo activity that may not result in a functional defect.14 The presence of D1472H would explain the low VWF:RCo/VWF:Ag in this subject.
Ten AA subjects (15%) had the reported type 2N H817Q mutation; one individual was homozygous for H817Q. The average FVIII level in this group was 99 IU/dL. The H817Q homozygous individual had a FVIII level of 151 IU/dL and an absence of bleeding symptoms, suggesting that the H817Q sequence variation may confer few or no clinical symptoms.
Overall, we identified new sequence variations or reported VWF mutations in 63 (34%) of 184 healthy controls, specifically in 43 (65%) of 66 AAs and 20 (17%) of 118 whites. More than one sequence variations was observed in 27 (63%) of the 43 AAs and in 2 (10%) of 20 whites.
Discussion
Molecular testing for VWF sequence variations can be useful in the diagnosis of VWD.15 Targeted mutation analysis is commonly used to detect type 2 mutations because they are localized to specific functional domains in exons 18-25 (type 2N), exon 28 (types 2A, 2B, and 2M), and exons 11-16, 24-26, and 51-52 (type 2A). Therefore, more is known about the sequence variations and their frequency in these VWF exons. In contrast, mutations causing type 1 and type 3 VWD have been reported in exons throughout this large gene. Furthermore, less is known about the sequence variations present in the less frequently studied exons of the VWF gene. Recently, full gene sequence analysis was used to study large groups of patients with type 1 VWD in the United Kingdom, European Union, and Canada.5,6 These groups identified many new sequence variations in the VWF gene. These studies, however, concentrated on sequencing the VWF gene in type 1 VWD patients, but did not sequence healthy controls or examine sequence variations in different ethnic groups.
When full gene analysis is newly introduced in the diagnosis of a genetic disorder, it is not uncommon to ascertain new sequence variations of unknown significance in patients. The inability to determine whether the sequence variation is disease causing or benign is because of the incomplete knowledge of gene, protein function, and ethnic-specific mutations in the general population. It is not unusual for some sequence variations to be common in one population and rare in another. As an example, there are distinct differences between the frequency of sequence variations found in the genes for blood group Ags in whites and AAs.16 An example in the VWF gene was reported by Flood et al, who observed that AAs in the ZPMCB-VWD were overrepresented in the subgroup of VWD patients diagnosed with type 2M VWD,14 subsequently demonstrating that a common VWF polymorphism in AAs near the ristocetin-binding site D1472H affects the measurement of VWF function by ristocetin cofactor but does not appear to result in abnormal in vivo VWF function. This variation was present in 61% of AAs and 20% of white controls.
In contrast to previous studies, our study included full laboratory testing and sequencing of healthy control samples and provides further insight into the sequence variations in the VWF gene in both white and AAs. We identified 2 groups of sequence variations in the healthy controls, novel sequence variations and sequence variations reported previously as VWF mutations.
We report 21 new sequence variations in the VWF gene. The majority (80%) of the new variations were found in AAs, likely because of the predominance of white subjects in previous studies. Interestingly, all but one of the sequence variations was found exclusively in either AAs or whites. Only the most common variation, T2666M, was found in both ethnic groups, but 10 of 12 instances of T2666M were in AAs. G131S, G967D, and T2666M were present in 5-10 AA individuals, accounting for a frequency of 8%-15% in this group. The frequency of these variations is well above the level designated for polymorphisms (1%), yet these variations were not described previously. With the exception of the sample with A631V, all controls had normal laboratory values, suggesting that these variations are not disease-causing mutations. A631V was observed in a one white healthy control with a reduced VWF:Ag (36 IU/dL) and VWF:RCo (46 IU/dL) but a normal ratio. Additional studies are needed to confirm that this sequence variation is causative. The identification of these novel sequence variations is important: if they were detected in a VWD patient, they could be misinterpreted as VWD-causing mutations.
Two of the healthy controls with a reported VWF mutation had abnormal VWF laboratory results. One person with the Y1584C had an abnormal VWF:RCo of 47 IU/dL, but the VWF:Ag was also decreased (61 IU/dL). The resulting VWF:RCo/VWF:Ag ratio was normal. The low VWF:Ag observed is consistent with Y1584C being a type 1 mutation.5,17 One individual with R1342C had a low VWF:RCo (51 IU/dL) and a slightly abnormal VWF:RCo/VWF:Ag ratio of 0.59, but this individual was also heterozygous for D1472H, which would explain the low VWF:RCo and VWF:RCo/VWF:Ag in this sample.14 VWF:CB (98 IU/dL) and the multimers were also normal in this individual. The one index case with R1342C in the European study had a VWF:Ag of 20 and a VWF:RCo of 3.6 These laboratory values for the Y1584C and R1342C represent data from a single healthy control, so it is not clear whether the abnormality is specific to the mutation or specific to the sample tested.
In addition to these 2 potential VWF mutations, we identified 12 other sequence variations reported previously as VWF mutations in 57 of the healthy controls. The majority of these sequence variations were reported as type I VWD mutations (ISTH-SSC VWF database, http://www.VWF.group.shef.ac.uk/). All but 3 of the mutations were found in AAs and the frequency of 3 of mutations (M740I, H817Q, and R2185Q) was 15%-18% suggesting these are not disease-causing mutations but rather polymorphisms frequently found in AAs. M740I was reported previously in 3 index cases in the European study but was co-inherited with R1205H6 but none of the individuals with M740I in our study also had R1205H. R2185Q was reported previously in one index case and the investigators were uncertain whether this allele was pathogenic.5 H817Q was not reported in either the European or the Canadian study. Ten AA subjects in our study had the reported type 2N H817Q mutation and one was homozygous. The FVIII levels were normal in heterozygous and homozygous controls with H817Q. Because we have not identified an increased frequency of 2N VWD in AAs and the FVIII levels are normal, this suggests that the sequence variation may confer little or no clinical symptoms. This mutation has been reported together with R782W in one subject with a type 2N phenotype.18 The subject was not AA.
Finally, the V1229G and N1231T variations have been reported to occur on the same haplotype. This haplotype has been reported in type 1 VWD patients, but the pathogenicity was uncertain because this haplotype was also present in unaffected family members.5 We found V1229G and N1231T exclusively in AAs, with one example of V1229G in the absence of N1231T; the VWF:Ag was greater than 140 IU/dL in this subject. Our VWF laboratory data and the frequency of these variations in healthy controls would support the hypothesis that these sequence variations are benign and not disease causing. However, because many of these variations were observed in 1-2 healthy controls or index cases, expression studies and /or more extensive analysis of healthy controls may be necessary to confirm these conclusions.
The role of molecular testing in the diagnosis of type 1 VWD has been debated.15,19 VWF mutations are found in only 60%-65% of more severe type 1 cases (< 25 IU/dL) and in less than 50% of cases when the Ag levels are > 25 IU/dL. The highly polymorphic nature of VWF makes it more difficult to distinguish neutral variants from pathogenic mutations, especially for type 1 VWD, possibly leading to misclassification of a sequence variant. Our data from healthy controls suggests some reported type 1 mutations may be misclassified. Sequencing results from type 1 patients need careful analysis to confirm that the sequence variants detected are not new or ethnic-specific sequence variations that not associated with VWD. Molecular analysis of type 1 cases will have the best clinical utility in patients with a well-characterized type 1 VWD phenotype with very low Ag levels.
Our data indicate that the sequence variation in the VWF gene is extensive and that unique sequence variations are frequently detected in AAs. We found that 65% of AA healthy controls had at least one new sequence variation or a previously reported VWF mutation and 40% had more than one. Although this study was limited to whites and AAs, our results suggest that it is critical to perform similar studies in other ethnic groups. Some of the sequence variations in AAs appear to be associated with one another. For example, all H817Q subjects in our study were either homozygous (n = 4) or heterozygous (n = 5) for D1472H. In addition, M740I is frequently found in subjects with H817Q. This suggests there may be distinct haplotypes in the AA population. In conclusion, this study emphasizes the importance of sequencing large numbers of healthy controls to understand ethnic-specific sequence variations so that polymorphisms are not misidentified as mutations.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all of the subjects, physicians, and staff involved in the ZPMCB-VWD study.
This work was supported by a National Institutes of Health program project grant (L081588) and by a Clinical and Translational Science Institute Award (1-UL1-RR031973).
National Institutes of Health
Authorship
Contribution: D.B.B., P.A.C., and R.R.M. designed the research project; D.B.B., P.A.C., V.H.F, J.C.G., K.D.F., S.L.H., A.D.S., T.C.A., C.L., W.K.H., J.M.L., and M.V.R. analyzed the results; D.B.B and R.R.M. wrote the manuscript; and all authors had full access to the data and edited the final paper.
Conflict-of-interest disclosure: J.C.G. is consultant to Baxter, CSL Behring and Octapharma. K.D.F. is consultant to CSL Behring and Octapharma. R.M.M is consultant to Gen-Probe, Baxter, CSL Behring, and Bayer. T.C.A is on the advisory board for CSL Behring. C.L. receives speaker fees from CSL Behring, Grifols, and Baxter and receives research funding and is on the advisory board for CSL Behring, Baxter, Bayer, Novo Nordisk, and Pfizer. The remaining authors declare no competing financial interests.
The current affiliation for W.K.W. is National Heart, Lung, and Blood Institute, Bethesda, MD.
Correspondence: Daniel B. Bellissimo, PhD, BloodCenter of Wisconsin, PO Box 2178, 638 N 18th St, Milwaukee, WI 53201-2178; e-mail: dan.bellissimo@bcw.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal