Abstract
Phytohemagglutin-stimulated child and adult leukocytes equally supported CCR5-dependent (R5) and CXCR4-dependent (X4) HIV-1 replication. In contrast, when phytohemagglutin-stimulated leukocytes from either healthy or congenitally immunodeficient children were cultured on feeder cells, they well supported R5, but not X4 HIV-1 replication, whereas both viruses equally spread in adult cells maintained in similar conditions. Both child and adult cells showed similar levels of proliferation and surface expression of CD4, CCR5, CXCR4, CD25, CD69, and HLA-DR. Lack of X4 HIV-1 replication in child versus adult cells was not caused by a differential expression of several known HIV-1 restriction factors. Similar levels of HIV DNA synthesis occurred in child cells infected with R5 and X4 viruses up to 48 hours after infection when R5 HIV-1 showed a significantly superior capacity to spread in culture than X4 virus. Cultured child cells well supported single round vescicular stomatitis virus-G pseudotyped virus replication, whereas superinfection of R5-infected cells with X4 HIV-1 (or vice versa) rescued the replication of this latter virus. Thus, child cells exposed to feeder cell culture represent a novel model system in which the superior capacity of R5 versus X4 viruses to spread can be investigated in primary, untransformed CD4+ cells.
Introduction
HIV type-1 (HIV-1) infects human beings of all ages and ethnicities. HIV-1 infects T lymphocytes and mononuclear phagocytes by engaging the primary receptor CD4 with its trimeric gp120 envelope (Env). Such an interaction leads to exposure of a second cryptic site of gp120 Env that becomes competent to bind to a chemokine coreceptor (CoR) molecule. Among other chemokine R, only CCR5 and CXCR4 are frequently used in vivo by HIV-1. Viruses using these CoRs are termed R5 and X4, respectively,1 and CoR use varies with different clinical phases with R5 viruses dominating the early stages and with CXCR4 use being mostly confined to subtype B viruses, although described also in subtype A and E.2 Furthermore, both in adults and children, infection starts as a monophyletic infection carried on by an R5 virus regardless of the viral phenotype predominant in the transmitter.3,4 In the case of subtype B infection (dominant in the North America, Europe, and Australia), an extension of CoR use to CXCR4, or the presence of so-called “dual mix” viral phenotypes, occurs in approximately 50% of persons resulting in dual-tropic R5×4 viruses; quite rarely, a true “switch” from R5 to X4 is observed.5 CXCR4 usage, however, is frequently associated with an accelerated disease progression.4
HIV-1 infection in children differs from that of adults by several parameters. First, although an acute mononucleosis-like illness is experienced in 50% to 70% of adults,6 the clinical outcomes of primary HIV infection are not as evident in infants.7 Second, quantification of plasma viremia during the first months of life frequently shows levels higher than those of acutely infected adults.8 The following decline in plasma virus in perinatally infected infants usually takes place over years rather than months in the absence of combination antiretroviral therapy (cART), therefore resulting in a higher systemic viral exposure during the first months of life.9 Third, although a clinically asymptomatic period follows primary HIV infection for many years in most adults (in the absence of cART), approximately 25% of infected children show a rapidly downhill course developing features characteristic of AIDS within the first year of life.10,11 In addition, the mortality rate of children who develop features of AIDS early in life is substantially higher than for those who become symptomatic later during childhood.12 However, as in adults, a minority of children do not show any signs or symptoms of AIDS by the age of 8 to 10 years.10,13
A link between HIV disease progression and CoR use in children has been reported5 with the emergence of CXCR4 using HIV-1 strains occurring in late stage of disease, as in adults.14 Phylogenetic analyses of gp120 Env sequences have demonstrated that CXCR4-using viruses in children did not derive from the maternal population of HIV-1 quasispecies but were consequent to the intrahost evolution of an originally transmitted homogeneous R5 HIV-1. Furthermore, although mothers harboring CXCR4-using viruses showed a significantly higher risk of transmitting the infection to their children than those infected by R5 viruses,15 in most cases the transmitted virus was an R5 strain.15,16 Thus, R5 HIV-1 both in adults17 and children is clearly better fit to be transmitted and spread in immunocompetent hosts than CXCR4-using viruses, although these latter may emerge once the immune system is severely compromised.
Despite a significant literature on HIV-infected infants, little is known about the in vitro susceptibility to R5 versus X4 HIV-1 infection of their cells. Therefore, we have investigated their susceptibility to infection and compared it with that of adult cells, by both conventional methods and by adopting a protocol optimized for maintaining untransformed activated cells of congenitally immunodeficient children in long-term culture.18,19 Although adult leukocytes supported both R5 and X4 replication in both culture conditions, when child CD4+ T cells were cultivated on feeder cells, they could efficiently support R5 but not X4 HIV-1 replication.
Methods
In vitro expansion of untransformed T-cell lines
Untransformed T-cell lines were established from Ficoll-Hypaque (Nycomed Pharma) purified PBMCs of 6 children (normal donors, age 1-11 years; median, 4 years) who underwent routine blood count for elective orthopedics or urological surgery and of 3 additional children affected by congenital adenosine deaminase deficiency severe combined immunodeficiency (ADA-SCID; age, 0.5-2 years; median, 1 year) before and after gene therapy (GT)20 whose blood was collected also for routine blood count. Cell lines from 5 adult normal donors (age > 18 years) were also obtained with the same protocol, as described.18 The ADA-SCID children were enrolled in a GT protocol approved by the San Raffaele Scientific Institute in Milano, Italy and by National Regulatory Authorities, and it is registered at www.clinicaltrials.gov (NCT00598481 and NCT00599781). Informed consent was obtained from adult donors or parents of pediatric donors in accordance with the Declaration of Helsinki, and the protocols were approved by the ethical committee of San Raffaele Scientific Institute in Milan, Italy.
Total leukocytes were stimulated with phytohemagglutinin (PHA, 1 μg/mL; Roche Diagnostics), recombinant human interleukin-2 (rhIL-2, 600 IU/mL; Novartis Italia) in the presence of an allogeneic feeder cell mix of PBMCs (1 × 106/mL, x-ray–irradiated at 60 Gy) and JY cells (105 cells/mL, x-ray–irradiated at 100 Gy). The cell cultures were maintained in complete medium (ie, IMEDM; Lonza) supplemented with YSSEL medium (10% volume/volume; Dyaclone), FBS (5% volume/volume; Lonza), and penicillin/streptomycin (100 U/mL; Bristol-Myers Squibb) for 3 days before separation from the feeder mix. Cells were then expanded in complete medium enriched with rhIL-2 and were restimulated by cocultivation with the feeder mix every 21 days. CD4+ T cells were purified 14 days after feeder stimulation by immunomagnetic technique using anti-CD4 Ab-coated microbeads (Miltenyi Biotec) according to the manufacturer's instructions (purity > 98%) and frozen for future experiments. After thawing, the cells were restimulated with PHA and IL-2 and infected after 6 days of culture (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
In a separate set of experiments, PBMCs from 8 immunocompetent children (age range, 3 months to 10 years; mean, 5.75 years; 5 males and 3 females) undergoing blood withdrawal after trauma (4 cases), upper respiratory tract infection (2 cases), epistaxis (1 case), or apnea (1 case) were obtained after Ficoll-Hypaque purification, washed, and stimulated with PHA for 72 hours. PHA blasts were then washed, resuspended in RPMI 1640 supplemented of 10% FBS, and immediately infected with R5 or X4 HIV-1 reference strains, as described in “HIV-1 infection of untransformed CD4+ T-cell lines and child PHA blasts.”
Flow cytometric analysis of cell surface determinants
Untransformed T-cell lines were washed with cold PBS plus 2% FBS to avoid nonspecific binding of Ab. The cells were then incubated with anti-CD4 (FITC), anti-CCR5 (PE), anti-CXCR4 (PE), anti-CD69 (PE), anti-CD25 (PE), and anti–HLA-DR (PE; BD Biosciences) mAb for 30 minutes at room temperature, washed with cold PBS-FBS (2%), and fixed with 2% paraformaldehyde. Flow cytometry was performed using a GALLIOS instrument (Beckman Coulter), and the results were analyzed with the FlowJo Version 8.4.3 software (TreeStar).
MTT cell proliferation assay
The cell proliferative capacity was assessed at the time of infection by the Cell Proliferation Kit I (Roche Diagnostics). The solubilized formazan product was spectrophotometrically quantified using a Microplate Reader 680 (Bio-Rad).
HIV-1 infection of untransformed CD4+ T-cell lines and child PHA blasts
CD4+ T-cell lines established from child and adult normal donors and from ADA-SCID patients were infected with prototypic laboratory-adapted, CCR5-dependent (R5) HIV-1BaL and CXCR4-dependent (X4) HIV-1LAI/IIIB subtype B HIV-1 strains. Infections were carried out at the multiplicities of infection (MOI) of 1 and 0.1. The isogenic X4 and R5 viruses, HIV-1NL4-3 and HIV-1NLAD8, derived from molecular infectious clones pNL4-3 and pNL(AD8), donated by Eric Freed (National Cancer Institute, National Institutes of Health), were also used. HIV-1NLAD8 was generated by replacing the 1.7-kb fragment containing env (encompassing both gp120 env and part of gp41 env) of HIV-1LAI/LAI/IIIB with the homologous portion of the R5 HIV-1ADA strain.21 After infection, culture supernatants were collected every 3 to 4 days for 5 weeks and stored at −20°C and later analyzed for virion-associated reverse transcriptase (RT) activity.22
VSV-G pseudotyped virus infection
The VSV-G–pseudotyped HIV-GFP virus was produced by cotransfection with a ratio of 1:7 of pMD2.G (a cytomegalovirus-driven expression plasmid that encodes the vesicular stomatitis virus [VSV] g envelope protein) together with pNLΔenvGFP that is pNL4-3 with an env-inactivating mutation and EGFP in place of nef.23
As control, cells were transduced with min-HIV-GFP, an HIV-1 vector expressing EGFP under the control of the spleen focus forming virus (SFFV) promoter, together with pMD2.G and psPax2, a packaging vector expressing HIV Gag-Pol23 at 1:3:4 ratios, respectively (supplemental Figure 2). Vector containing supernatants were harvested 48 hours after transfection, cleared by centrifugation, filtered (0.45 μm (MILLEX-HV PVDF; Millipore) and stored at −80°C.
R5 and X4 superinfection experiments
Untransformed CD4+ cells of 3 independent children were cocultured with feeder cells in the presence of PHA for 3 days and were then infected with either HIV-1BaL (R5) or HIV-1 LAI-IIIB (X4), as described in “HIV-1 infection of untransformed CD4+ T-cell lines and child PHA blasts.” For each donor, the kinetics of infection with the 2 viruses was followed as control; in addition, the supernatants of R5- and X4-infected cultures were cross-exchanged in different wells 24, 72 and 168 hours after the original infection. To avoid the potential transfer of contaminant cells (although not observed by optical microscopy), the supernatants from infected cultures were first centrifuged at 550g for 10 minutes before transfer to the counterinfected cell cultures. In essence, the supernatants from cells originally incubated with R5 HIV-1 were completely replaced with those collected from X4-infected cells (R5 super-infected with X4: R5s.i.w.X4) and vice versa (X4 super-infected with R5: X4s.i.w.R5). All the cultures were carried on for 2 weeks in parallel and their supernatants were collected and stored for RT activity evaluation.
To determine the coreceptor usage of the replicating virus in the different experimental conditions, supernatants of both control R5s.i.w.X4 and X4s.i.w.R5 cell cultures were collected at the peak of virus replication and were then tested for their capacity to infect U87 astrocytic cell lines stably transfected with and expressing human CD4 and either CCR5 or CXCR4.24
HIV-1 DNA quantification by real-time PCR
CD4+ T-cell lines were infected with DNase/RNasefree HIV-1BaL or HIV-1LAI/IIB (MOI = 0.1) and were then maintained in culture for 7 days. After infection, 2 × 106 cells were washed, resuspended in a lysis buffer, and digested at 65°C for 2 hours after 6, 24, 48, 96, and 168 hours after infection; proteinase K was then heat inactivated at 95°C for 15 minutes.25 An amount of lysate corresponding to 2.5 × 104 cells was amplified by real-time quantitative PCR reactions using primers and a probe recognizing the HIV-1 gag gene: forward primer, 5′-ACATCAAGCAGCCATGCAAAT-3′; reverse primer, 5′-ATCTGGCCTGGTGCAATAGG-3′; and probe, 5′-(FAM)CATCAATGAG GAAGCTGCAGGAATGGGATAGA(TAMRA)-3′. This primer/probe combination detects all forms of viral DNA synthesized after second-strand transfer mediated by RT. The number of HIV-1 DNA copies showed a linear distribution between 10 and 107 copies (r = 0.99) by an external curve and was normalized to that of human GAPDH. The primers and probe for GAPDH were: forward primer, 5′-ACCACAGTCCATGCATCACT-3′; reverse primer, 5′GGCCATCACGCCACAGITT-3′; and probe, 5′-(FAM) CCCAGAAGACTGTG GATGGCCCC (TAMRA)-3′. Thermal cycling conditions for real-time PCR were 50°C for 2 minutes and 95°C for 15 minutes followed by 40 cycles at 95°C for 15 seconds and 65°C for 1 minute.
Statistical analysis
Results are reported as mean values ± SEM. Student t test was applied to the RT activity levels measured at the day of the peak of virus replication (days 9-12 after infection) and to the percentage of CCR5+ and CXCR4+ child cells, before and after GT. To minimize interdonor variability, each experiment was performed in quintuplicate.
Results
R5, but not X4 HIV-1, replicates in untransformed CD4+ T-cell lines established from healthy children
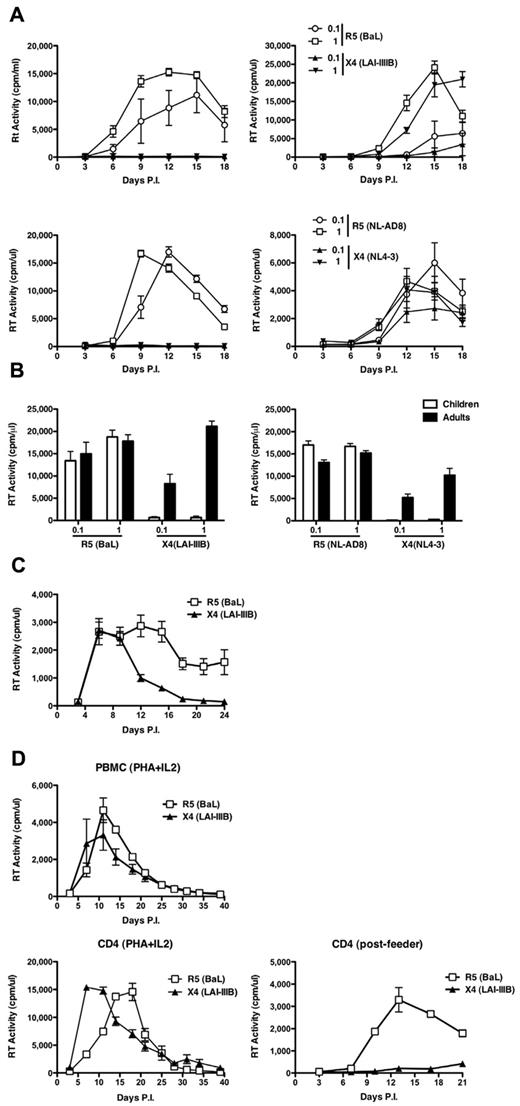
CD4+ T-cell lines were established from 6 pediatric and 5 adult healthy donors, respectively, and were then incubated with either R5 HIV-1BaL or X4 HIV-1LAI/IIIB strains (MOI, 0.1 and 1). Only R5 HIV-1 productively infected child CD4+ T cells at both MOI tested (Figure 1A top left panel). In contrast, both R5 and X4 viruses replicated with similar efficiency in adult CD4+ T-cell lines (Figure 1A top right panel). The same results were obtained when cell lines containing CD8+ cells or negatively selected for CD45RA+ cells or positively selected for CD45RO+ cells were established from the same donors (data not shown). The lack of virus replication of X4 strains in child cell lines was not the result of cytotoxicity or impaired cell proliferation (data not shown).
Discordant replication of R5 and X4 viruses in CD4+ T-cell lines established from children and adults. (A) Pediatric and adult cells were infected with 2 different MOI (0.1 and 1) of the laboratory-adapted R5 HIV-1BaL and X4 HIV-1IIIB (top panels) and isogenic viruses NL-AD8 (R5) and NL4-3 (X4; bottom panels). (Left panels) The results from a single infection representative of 6 independent experiments. (Right panels) A single infection of cell lines obtained from an adult, representative of 5 independent experiments. All experiments were performed in quintuplicate replicas. (B) Means ± SEM of the peak levels of replication observed in all aforementioned infections. R5 and X4 viruses showed comparable efficiency of replication in adult cell lines, but only R5 viruses spread in child cells. (C) Equal susceptibility of PHA blasts established from children's PBMCs (N = 7) to R5 (BaL) and X4 (LAI/IIIB) productive HIV-1 infection. (D) PBMCs from an independent child were infected by R5 or X4 HIV-1 as whole T-cell blasts (left panel), after enrichment in CD4+ T cells (middle panel), or after expansion and passage onto feeder cells (right panel). Both viruses replicated efficiently in the first 2 conditions, whereas only R5, but not X4, HIV-1 efficiently spread in CD4+ T cells cocultured for 3 days onto feeder cells.
Discordant replication of R5 and X4 viruses in CD4+ T-cell lines established from children and adults. (A) Pediatric and adult cells were infected with 2 different MOI (0.1 and 1) of the laboratory-adapted R5 HIV-1BaL and X4 HIV-1IIIB (top panels) and isogenic viruses NL-AD8 (R5) and NL4-3 (X4; bottom panels). (Left panels) The results from a single infection representative of 6 independent experiments. (Right panels) A single infection of cell lines obtained from an adult, representative of 5 independent experiments. All experiments were performed in quintuplicate replicas. (B) Means ± SEM of the peak levels of replication observed in all aforementioned infections. R5 and X4 viruses showed comparable efficiency of replication in adult cell lines, but only R5 viruses spread in child cells. (C) Equal susceptibility of PHA blasts established from children's PBMCs (N = 7) to R5 (BaL) and X4 (LAI/IIIB) productive HIV-1 infection. (D) PBMCs from an independent child were infected by R5 or X4 HIV-1 as whole T-cell blasts (left panel), after enrichment in CD4+ T cells (middle panel), or after expansion and passage onto feeder cells (right panel). Both viruses replicated efficiently in the first 2 conditions, whereas only R5, but not X4, HIV-1 efficiently spread in CD4+ T cells cocultured for 3 days onto feeder cells.
These results were confirmed with viruses derived from 2 isogenic infectious molecular clones (ie, HIV-1NL4-3 and HIV-1NLAD8) that differ only for their Env coding region.21 Only R5 HIV-1 fully replicated in child cells, whereas the X4 virus did not (Figure 1A bottom left panel) and both viruses showed similar profiles of spreading in adult cells (Figure 1A bottom right panel). This observation was highly consistent among all independent cultures tested (Figure 1B).
To understand whether the observed restriction to productive X4 HIV-1 infection was an intrinsic property of child cells or was consequent to the experimental protocol here adopted, PHA blasts from 7 independent HIV-seronegative children were infected with R5 and X4 viruses (HIV-1BaL and HIV-1LAI/IIIB). Indeed, child PHA blasts equally supported the replication of both viruses (Figure 1C), as independently reported,26 indicating that the observed X4 HIV-1 restriction was related to the protocol of activation and expansion of child cells. To further support this hypothesis, PBMCs from an independent child were isolated, stimulated for 3 days with PHA and IL-2 (PHA blasts), and either immediately infected with R5 or X4 HIV-1 strains or positively selected for CD4+ cells and then infected by both viruses. Finally, an aliquot of positively selected CD4+ cells was expanded for an additional 7 days, cultured for 3 days in the presence of feeder cells, and then infected with R5 or X4 HIV-1. Both PHA blasts and positively selected CD4+ cells (Figure 1D left and middle panels, respectively) were equally susceptible to both R5 and X4 infection, with the latter showing significantly higher levels of virus replication because of the removal of most CD8+ T cells. In contrast, when expanded CD4+ cells were first passaged onto feeder cells and then infected with both viruses, only the R5 virus efficiently replicated at levels comparable with those of PHA blasts, whereas X4 HIV-1 was significantly impaired (Figure 1D right panel).
Thus, maintenance of activated CD4+ T cells from children, but not from adults, in short-term culture in the presence of feeder cells seems to induce the emergence of a restriction for X4 HIV-1 replication.
Both adult and child CD4+ T-cell lines show similar levels of CD4, chemokine CoR, activation markers, and proliferative capacity
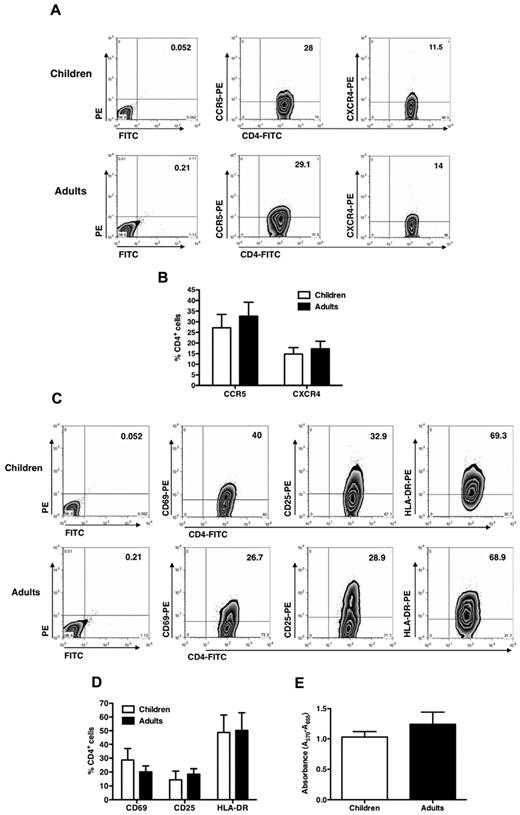
No significant differences between child and adult cells in terms of cell surface CD4, CCR5, or CXCR4 expression at the time of infection were observed (Figure 2A), although a lower percentage of CXCR4+ cells was noted in the cell lines from both children and adults compared with CCR5+ cells (Figure 2B). No secretion of CXCL12/stromal cell derived factor-1α, ligand of CXCR4, was detected in the supernatant of the cultures or of feeder cells (data not shown). Furthermore, child cells migrated similarly to those from adults in response to either CXCR4 or CCR5 ligands, indicating that these R were functional at the time of infection (supplemental Figure 2).
CD4, CCR5, and CXCR4 expression in CD4+ T-cell lines established from pediatric and adult donors. (A) T-cell lines were stained with anti-CD4–FITC, anti-CCR5–PE, and anti-CXCR4–PE mAb at the time of viral infection, and the percentage of positive cells was detected by cytofluorimetric analysis. The results from a single experiment on pediatric and adult cell lines representative of 6 and 5 different donors, respectively, are shown. No significant differences were seen between child and adult cell lines. (B) Percentage of CD4+ cells (mean ± SEM of 6 independent experiments made in quintuplicate) expressing CCR5 or CXCR4 in child and adult cell lines is reported without showing any significant difference. (C) T-cell activation marker expression of pediatric and adult CD4+ T-cell lines indicates no significant differences (one experiment representative of 6 pediatric and 5 adult cell lines is shown). (D) Mean expression levels of activation markers expressed by CD4+ T-cell lines are shown (n = 6 pediatric and 5 adult cell lines). (E) Proliferation of pediatric and adult CD4+ T-cell lines was assessed by the MTT assay at the time of infection. Mean ± SEM values of the proliferation (n = 6 pediatric and 5 adult cell lines) are shown and do not indicate differences in child versus adult cell lines.
CD4, CCR5, and CXCR4 expression in CD4+ T-cell lines established from pediatric and adult donors. (A) T-cell lines were stained with anti-CD4–FITC, anti-CCR5–PE, and anti-CXCR4–PE mAb at the time of viral infection, and the percentage of positive cells was detected by cytofluorimetric analysis. The results from a single experiment on pediatric and adult cell lines representative of 6 and 5 different donors, respectively, are shown. No significant differences were seen between child and adult cell lines. (B) Percentage of CD4+ cells (mean ± SEM of 6 independent experiments made in quintuplicate) expressing CCR5 or CXCR4 in child and adult cell lines is reported without showing any significant difference. (C) T-cell activation marker expression of pediatric and adult CD4+ T-cell lines indicates no significant differences (one experiment representative of 6 pediatric and 5 adult cell lines is shown). (D) Mean expression levels of activation markers expressed by CD4+ T-cell lines are shown (n = 6 pediatric and 5 adult cell lines). (E) Proliferation of pediatric and adult CD4+ T-cell lines was assessed by the MTT assay at the time of infection. Mean ± SEM values of the proliferation (n = 6 pediatric and 5 adult cell lines) are shown and do not indicate differences in child versus adult cell lines.
R5, but not X4 HIV-1, replicates in CD4+ T-cell lines from children with ADA-SCID before and after GT
To investigate whether cells with a congenitally altered intracellular signaling machinery were susceptible to R5 and X4 virus infection, untransformed T-cell lines established from 3 ADA-SCID patients were investigated. These cells were previously shown to possess severely compromised TCR/CD28-driven proliferation and cytokine production both at transcriptional and protein expression levels.18
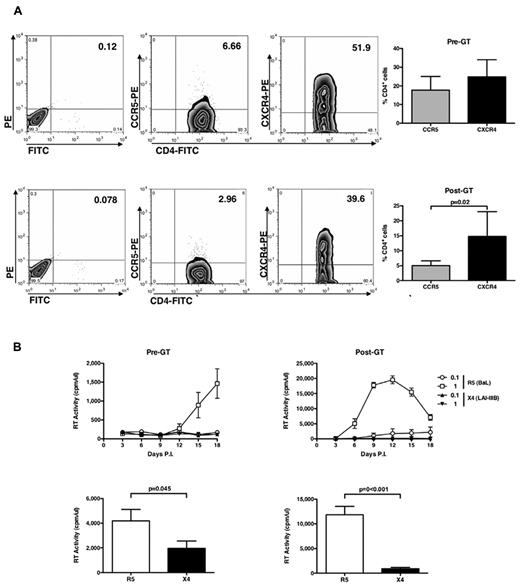
Before GT, even if the number of CD4+CXCR4+ cells were superior to that of CD4+CCR5+ cells (Figure 3A), R5 HIV-1 replicated in ADA-SCID CD4+ T cells, although at lower levels than observed in cells from healthy donors, whereas X4 HIV-1 did not (Figure 3B left panels). When infection was performed in child cells isolated 7 to 24 months after GT, a significantly higher capacity of supporting productive R5 HIV-1 infection was observed despite an even lower proportion of CD4+CCR5+ cells versus CD4+CXCR4+ cells compared with cells isolated before GT (Figure 3A bottom panel). The X4 virus still did not replicate (Figure 3B right panels), as confirmed also with isogenic HIV-1NL4-3 and HIV-1NLAD8 viruses (data not shown).
CD4, CCR5, and CXCR4 expression and HIV replication in untransformed CD4+ T-cell lines from ADA-SCID children. (A) CD4+ T-cell lines of 3 children affected by ADA-SCID that were established before and after GT were stained with anti-CD4–FITC, anti-CCR5–PE, and anti-CXCR4–PE mAb at the time of viral infection, and the percentage of positive cells was detected by cytofluorimetric analysis. (Left panels) The results from a single experiment on pediatric cell lines representative of the pattern observed with cells from 3 independent patients isolated before (top panel) and after (bottom panel) GT. (Right panels) Mean ± SEM values. No significant differences in terms of percentage of CD4+ T cells expressing the HIV-1 entry CoR was noted. (B) ADA-SCID CD4+ T-cell lines were infected with R5 and X4 HIV-1. Only R5 HIV-1 efficiently replicated in these cell lines before and after GT (which, however, significantly boosted the levels of virus replication). The results obtained with the cell lines established from a single child before and after GT are shown and are representative of those collected with the cell lines derived from 2 additional patients. (Bottom panels) Mean ± SEM of the peaks of virus replication in cell lines established before and after GT from the 3 patients.
CD4, CCR5, and CXCR4 expression and HIV replication in untransformed CD4+ T-cell lines from ADA-SCID children. (A) CD4+ T-cell lines of 3 children affected by ADA-SCID that were established before and after GT were stained with anti-CD4–FITC, anti-CCR5–PE, and anti-CXCR4–PE mAb at the time of viral infection, and the percentage of positive cells was detected by cytofluorimetric analysis. (Left panels) The results from a single experiment on pediatric cell lines representative of the pattern observed with cells from 3 independent patients isolated before (top panel) and after (bottom panel) GT. (Right panels) Mean ± SEM values. No significant differences in terms of percentage of CD4+ T cells expressing the HIV-1 entry CoR was noted. (B) ADA-SCID CD4+ T-cell lines were infected with R5 and X4 HIV-1. Only R5 HIV-1 efficiently replicated in these cell lines before and after GT (which, however, significantly boosted the levels of virus replication). The results obtained with the cell lines established from a single child before and after GT are shown and are representative of those collected with the cell lines derived from 2 additional patients. (Bottom panels) Mean ± SEM of the peaks of virus replication in cell lines established before and after GT from the 3 patients.
Thus, R5 HIV-1 shows a superior replicative capacity also in cell lines derived from ADA-SCID children, bearing a congenitally impaired signal transduction capacity.18
CD4+ T-cell lines established from both children and adult express similar levels of several host restriction factors
It has been recently demonstrated that R5 and X4 HIV-1 can differentially modulate the expression of host cell factors that can either enhance or impair virus replication.27 Therefore, we investigated the levels of mRNA expression of a comprehensive array of some of the best known restriction factors, including APOBEC-3G, -3F, -3A,28 TRIM5α, TRIM19, TRIM22 (with TRIM37 serving as a control),29 BST2/Tetherin,30 and SAMHD131,32 in PHA cells blasts, enriched CD4+ T cells, or CD4+ T cells maintained for 3 days on feeder cells. With the exception of APOBEC-3G and -3F, none of these restriction factors showed a significant pattern of association with the observed HIV-1 replication patterns in these different cell culture conditions (supplemental Figure 4), at least at the mRNA level. We therefore next investigated the expression of APOBEC3G protein at the time of infection, as well as up to 12 days after infection (data not shown) between child and adult CD4+ T-cell lines and observed that it was not different in the 2 cell types (supplemental Figure 5 left panel), Similarly, the levels of TRIM22 protein expression were similar in child and adult CD4+ T-cell lines before (supplemental Figure 5 right panel) and after infection (data not shown).
Similar early efficiency of R5 and X4 HIV-1 DNA synthesis in child and adult CD4+ T-cell lines
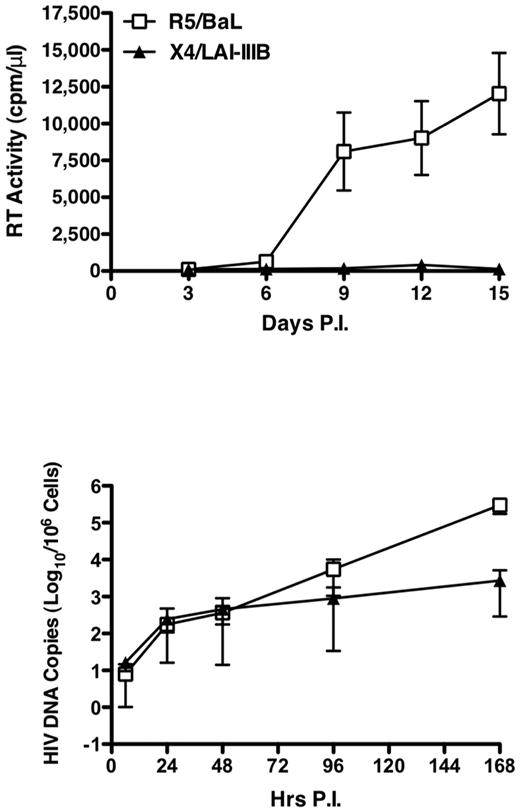
We next investigated whether X4 HIV-1 could undergo efficiently reverse transcription in cell lines established from 4 independent healthy children on infection with DNase-treated R5 (HIV-1BaL) and X4 (HIV-1LAI/IIIB) at the MOI of 0.1. Productive HIV-1 replication was exclusively observed on infection with the R5, but not with the X4, virus, as evaluated by RT activity in culture supernatants (Figure 4 top panel). In contrast, a comparable efficiency of HIV DNA synthesis during the first 48 hours of infection was observed with both viruses (Figure 4 bottom panel), although with some interdonor variability (supplemental Figure 6). Strikingly, a 100-fold increase in R5, but not X4, HIV-1 DNA synthesis occurred in child cell lines starting approximately 3 days after infection (Figure 4 bottom panel).
Discordant HIV DNA synthesis and R5 versus X4 virus replication in child CD4+ T-cell lines. Four independent CD4+ T-cell lines from pediatric donors were infected with R5 and X4DNase-treated viruses, and RT activity was detected in culture supernatant up to day 15 (top panel). The levels of HIV-1 gag DNA were measured between 6 and 168 hours after infection (bottom panel). Mean ± SEM of the 4 independent experiment are shown. Whereas only R5 virus replication was detected by RT activity, HIV DNA synthesis occurred with comparable efficiency after both R5 and X4 infection up to 48 hours after which R5 viruses showed a superior capacity to spread in culture.
Discordant HIV DNA synthesis and R5 versus X4 virus replication in child CD4+ T-cell lines. Four independent CD4+ T-cell lines from pediatric donors were infected with R5 and X4DNase-treated viruses, and RT activity was detected in culture supernatant up to day 15 (top panel). The levels of HIV-1 gag DNA were measured between 6 and 168 hours after infection (bottom panel). Mean ± SEM of the 4 independent experiment are shown. Whereas only R5 virus replication was detected by RT activity, HIV DNA synthesis occurred with comparable efficiency after both R5 and X4 infection up to 48 hours after which R5 viruses showed a superior capacity to spread in culture.
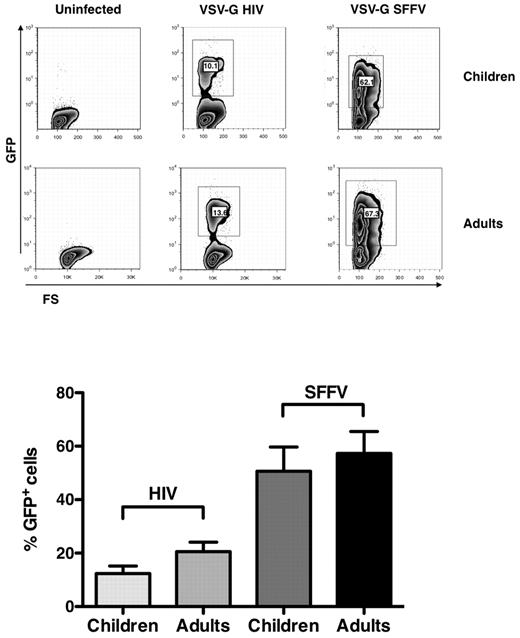
Similar efficiency of VSV-G pseudotyped HIV-1 in child and adult CD4+ T-cell lines
To define whether the replication of R5 and X4 viruses in child cells could be explained by their differential CoR use, these cells were infected by single-round pseudotyped viruses (ie, VSV-G HIV-1 and VSV-G SFFV), which do not share common regulatory elements with the HIV-1 LTR23 (supplemental Figure 2). CD4+ T-cell lines from 5 independent healthy children and adults were infected with the VSV-G pseudotyped viruses, and EGFP expression was evaluated 6 days after infection by cytofluorimetric analysis. The percentages of EGFP+ cell lines from children and adults were indeed comparable (Figure 5), although with an inferior efficiency in EGFP expression driven by the HIV-LTR versus that of the SFFV systems (Figure 5).
VSV-G HIV-1 and VSV-G SFFV infection of child and adult CD4+ T-cell lines. Child and adult cell lines were infected with the pseudotyped viruses, and EGFP expression was monitored by cytofluorimetric analysis 6 days after infection. (Top panel) Results of single experiment representative of 6 independently performed. (Bottom panel) Mean ± SEM. Child and adult cell lines showed a similar capacity of supporting both HIV-1 LTR and SFFV expression.
VSV-G HIV-1 and VSV-G SFFV infection of child and adult CD4+ T-cell lines. Child and adult cell lines were infected with the pseudotyped viruses, and EGFP expression was monitored by cytofluorimetric analysis 6 days after infection. (Top panel) Results of single experiment representative of 6 independently performed. (Bottom panel) Mean ± SEM. Child and adult cell lines showed a similar capacity of supporting both HIV-1 LTR and SFFV expression.
Overall, these results strongly indicate that after the first days of infection R5 HIV-1 possess a superior capacity of spreading than X4 virus in primary CD4+ T-cell lines established from children, while this restriction was not observed in cell lines of adults.
Rescue of X4 replication competence in child cell lines by R5 HIV-1 superinfection
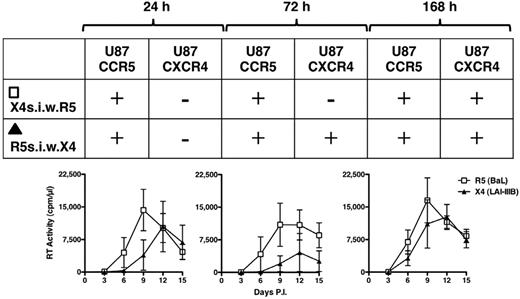
We finally explored whether the capacity of X4 HIV-1 to replicate in these untransformed cell lines was influenced in presence of an ongoing R5 infection. Vice versa, we tested whether the X4 abortive infection could interfere with the capacity of R5 HIV-1 to efficiently propagate in these cells. To this aim, child cell lines of 3 independent donors were infected either with R5 or X4 viruses after cultivation in the presence of feeder cells. The culture supernatants were collected 24, 72, and 168 hours after infection (supplemental Figure 7) and, after centrifugation to avoid the presence of contaminant cells, they were transferred onto the cells that were incubated with the other virus (ie, the supernatant from R5 infected cells completely replaced that of X4-infected cells and vice versa). Unlike what observed in the control X4 infection (supplemental Figure 7), virus replication was observed in all superinfected cultures although with variable kinetics and efficiency (Figure 6). To characterize the phenotype of the replicating virus in the different conditions, the supernatants collected at peak levels of RT activity (days 9-12) in the superinfected cultures were seeded onto U87-CD4 cells expressing either CCR5 or CXCR4. The results are summarized in Figure 6. After 24 hours of supernatant transfer, only R5 HIV-1 replicated in both cell culture conditions (ie, R5s.i.w.X4 and X4s.i.w.R5). However, when X4 infected cells were exposed to the R5 infectious supernatants collected 72 or 168 hours after infection, in addition to R5, also X4 HIV-1 replication was observed. Conversely, when R5 infected cells were incubated with X4 infectious supernatants collected 72 hours after infection the only replicating HIV-1 had an R5 phenotype, but when this last condition was repeated with X4 supernatant collected 168 hours after infection, both R5 and X4 HIV-1 replicated efficiently (Figure 6).
Superinfection by R5 HIV-1 infectious supernatant rescues X4 HIV-1 replicative capacity. After supernatant exchange from R5- and X4-infected cell cultures, the coreceptor use of the replicating viruses was tested at peak levels of virus replication onto U87-CD4-CCR5 and U87-CD4-CXCR4 cell lines. The outcome of these infections is represented in the embedded table showing a progressive rescue of X4 replicative capacity. X4s.i.w.R5 indicates X4-infected cells superinfected with R5 virus; and R5s.i.w.X4, R5-infected cells superinfected with X4 virus (n = 3).
Superinfection by R5 HIV-1 infectious supernatant rescues X4 HIV-1 replicative capacity. After supernatant exchange from R5- and X4-infected cell cultures, the coreceptor use of the replicating viruses was tested at peak levels of virus replication onto U87-CD4-CCR5 and U87-CD4-CXCR4 cell lines. The outcome of these infections is represented in the embedded table showing a progressive rescue of X4 replicative capacity. X4s.i.w.R5 indicates X4-infected cells superinfected with R5 virus; and R5s.i.w.X4, R5-infected cells superinfected with X4 virus (n = 3).
Overall, these findings strongly suggest that, at least in these experimental conditions, CCR5 usage is associated with a “permissivity signal” enabling also X4 viruses to (re-)gain replication competence, whereas X4 HIV-1 does not appear to impair significantly R5 virus replication.
Discussion
We here describe a restriction of HIV-1 replication occurring in untransformed CD4+ T-cell lines established from children (< 11 years old) versus those of adults (> 18 years old) that supported the productive infection of R5, but not X4 HIV-1, after short-term coculture with allogenic feeder cells. The restriction was not accounted for by differential levels of expression of CD4 or entry CoRs, activation state and proliferative capacity, or expression of several restriction factors in child versus adult cell lines, and it was confirmed in T-cell lines obtained from ADA-SCID children both before and after GT. HIV-1 DNA synthesis in the first 48 hours of both R5 and X4 virus infection occurred with similar efficiency, whereas R5 HIV-1 showed a superior spreading capacity after the initial cell-free virion infection. The discordant profile of virus replication was consequent to the differential engagement of CCR5 versus CXCR4 by HIV-1 gp120 Env because VSV-G mediated infection overcame HIV-1 restriction in child cells. Furthermore, superinfection of X4-infected cells with R5 infectious supernatant (or vice versa) rescued the replicative capacity of X4 HIV-1, whereas R5 virus production was not impaired.
Several studies have previously investigated the susceptibility of cord blood mononuclear cells to HIV-1 infection. Cord blood mononuclear cells showed an increased susceptibility to both R5 and X4 HIV-1 infection33 and LTR-driven transcription22,34 in respect to PBMCs independently of CoR engagement. However, infection of suboptimally activated CD4+ cord blood mononuclear cells resulted in the productive infection of R5, but not X4, virus.22,34 Remarkably, also in cord blood mononuclear cells, the restriction was dictated by gp120 Env CoR use and resulted in a comparable initial synthesis of HIV DNA of both viruses followed by a superior spreading capacity of R5 virus.22,34
In the present study, we adopted a cell culture system earlier optimized to support the survival and proliferation of T cells obtained from immunodeficient patients, such as ADA-SCID children,18,19 in whom the congenital defects impair cell survival and proliferative capacity. Only R5, but not X4 HIV-1, was able to establish a productive virus replication in these untransformed CD4+ T-cell lines established from healthy children, whereas both viruses efficiently replicated in those of healthy adults as well as when leukocytes from children were immediately infected after 3 days of PHA stimulation or even after expansion of CD4-enriched leukocytes. Before GT, R5 virus replication in ADA-SCID CD4+ T cells occurred at lower levels compared with cells from healthy children. After GT, however, the capacity of R5 virus to spread in culture achieved levels similar to those of healthy persons, further supporting previous results showing functional restoration after GT.18 In contrast, X4 virus replication was not observed either before or after GT.
In the present study, the observation that viral DNA synthesis in the first days after infection was overall comparable after R5 and X4 HIV-1 infection suggests an active contribution of CCR5-dependent signaling to viral replication, a hypothesis as supported by several studies.35 These include the observation that stimulation with R5 gp120 Env trimers triggered calcium fluxes in CD4+ T cells infected in vitro36 and virus replication in cultures of resting CD4+ T cells of infected persons.37 Of note, a signaling-deficient R5 HIV-1 was shown to successfully infect monocyte-derived macrophages but failed to replicate thereafter because of a block occurring at a postentry level.36 Thus, CCR5 engagement by HIV-1 triggers a signaling cascade that leads to productive viral replication, whereas gp120 Env/CXCR4 interaction seems to be devoid of such a function. This hypothesis is consistent with the observed comparable single-round replication efficiency in child and adult cell lines of VSV-G pseudotyped viruses causing virion/membrane fusion independently of CD4 and CoR as well as with the results of our superinfection experiments. Furthermore, the X4-related postentry block was not dependent on the up-regulation of several HIV-1 restriction factors, including A3A, A3F, A3G, TRIM5α, TRIM19, TRIM22, BST2/Tetherin, and SAMHD1. Overall, this study suggests that one of the features rendering R5 HIV-1 more fit than CXCR4-using viruses during interindividual transmission (including mother to child transmission38 also when the transmitting person harbors predominantly CXCR4-using viruses39 ) is a function of its superior capacity to spread in immunologically competent CD4+ cells after the initial cell-free infection.
Questions arise from our current observation, such as the definition of which functional differences emerge after short-term culture with feeder cells in adult versus child CD4+ T cells that result in their capacity to support both R5 versus X4 virus replication and when such differences are overcome as a function of the age. The observed increased efficiency of R5 virus replication in GT-reconstituted CD4+ cells from ADA-SCID versus their pre-GT levels suggest that the TCR-related signaling and proliferative defects curtails, although does not abolish, the efficiency of R5 virus in replicating in these cells, although it did not influence the inability of X4 virus to productively infect these cells. It should be underscored, however, that these differences do not appear to be “intrinsic” to child cells that, like adult cells, are fully permissive to X4 HIV-1 when infected after mitogen stimulation in the absence of feeder cells, as independently reported,26 or when they are superinfected by an R5 HIV-1, as here shown (Figure 6).
Nowadays more than 2 million children live with HIV-1 worldwide. The features of pediatric infection differ from those of the adults in different aspects.6,9,12 However, despite abundant epidemiologic information on pediatric HIV infection, very little was known about the in vitro susceptibility of child cells to HIV-1 infection.26 Thus, the development of a cellular system based on pediatric cells could be potentially useful to study the peculiarity of HIV infection in children and can help to elucidate the pathogenesis of pediatric AIDS. In this regard, as in adults,17 it is well established that HIV-1 infection of children from their mother (either via placenta, blood exchange during delivery, or by breastfeeding40,41 ) is started in most cases by an R5 monophyletic strain, regardless of the dominant quasispecies in the transmitting mother.8,14,15 However, transmission of CXCR4-using viruses has been also described and associated with rapid disease progression toward AIDS and death in the absence of cART.42 In this regard, it is conceivable that infection of the thymus early in prenatal or postnatal life43 may lead to a state of immunologic tolerance toward the virus that could significantly alter the host-virus interaction.
Our observation that the presence of an ongoing R5 HIV-1 spreading may rescue the replication of “dormant” X4 HIV-1 infection well fits with the observed emergence of R5×4 and “dual mix” phenotypes in the late stages of subtype B infection.1,2 Finally, it should also be underscored that mother-to-child HIV-1 transmission may occur through mucosal contact during delivery and it may involve infected cells lacking typical features of activation.44
In conclusion, we here describe, for the first time, that child CD4+ T cells are permissive to HIV-1 infection but bear an intrinsic potential to become restricted for X4 replication at a postentry level as revealed by their short-term cultivation on allogeneic feeder cells. This culture system may be suitable to unravel the biologic basis of the superior capacity of R5 viruses to spread compared with X4 HIVs, at least in children.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
S.A.M. performed this study as partial fulfillment of her PhD in Cellular and Molecular Biology, Vita-Salute San Raffaele University, Milan, Italy (jointly with the Open University of London, United Kingdom and the FP6 Europrise Network of Excellence).
This work was supported in part by the Telethon Foundation, Italy, Program of AIDS Research 2009-2010 of the Ministry of Health, Italy (grants 40H76 and 40H12; G.P. and E.V.), by Regione Lombardia (ID SAL-70 rif. 17165; G.P.), by CARIPLO Foundation (CAR-2230, G.P.), and by the Europrise Network of Excellence of the 6th Framework Program of the European Commission (S.A.M. and G.P.).
Authorship
Contribution: S.A.M. performed most of the experiments and wrote the first draft of the manuscript; I.B. prepared the primary cell lines and feeder cells from both immunocompetent and immunodeficient children and healthy adults; A.K.-R. prepared the VSV-G vectors and performed the single round infections; E.V. supervised the VSV-G experiment and wrote the manuscript; A.R. and A.P. provided the blood and clinical data of children for the PHA blast infection experiments; S.G. conducted and analyzed PHA blast infection experiments; A.A. supervised the establishment of the child and adult cell lines from both immunocompetent and immunodeficient children and healthy adults, interpreted the results, and wrote the manuscript; and G.P. supervised the whole project and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Guido Poli, P2/P3 Laboratories, DIBIT-1, Via Olgettina n 58, 20132, Milano, Italy; e-mail: poli.guido@hsr.it.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal