Abstract
VEGFs activate 3 receptor tyrosine kinases, VEGFR-1, VEGFR-2, and VEGFR-3, promoting angiogenic and lymphangiogenic signaling. The extracellular receptor domain (ECD) consists of 7 Ig-homology domains; domains 2 and 3 (D23) represent the ligand-binding domain, whereas the function of D4-7 is unclear. Ligand binding promotes receptor dimerization and instigates transmembrane signaling and receptor kinase activation. In the present study, isothermal titration calorimetry showed that the Gibbs free energy of VEGF-A, VEGF-C, or VEGF-E binding to D23 or the full-length ECD of VEGFR-2 is dominated by favorable entropic contribution with enthalpic penalty. The free energy of VEGF binding to the ECD is 1.0-1.7 kcal/mol less favorable than for binding to D23. A model of the VEGF-E/VEGFR-2 ECD complex derived from small-angle scattering data provided evidence for homotypic interactions in D4-7. We also solved the crystal structures of complexes between VEGF-A or VEGF-E with D23, which revealed comparable binding surfaces and similar interactions between the ligands and the receptor, but showed variation in D23 twist angles. The energetically unfavorable homotypic interactions in D4-7 may be required for re-orientation of receptor monomers, and this mechanism might prevent ligand-independent activation of VEGFR-2 to evade the deleterious consequences for blood and lymph vessel homeostasis arising from inappropriate receptor activation.
Introduction
A plethora of growth factors, such as angiopoietins, VEGF family ligands, platelet-derived growth factors, fibroblast growth factors, and hepatocyte growth factors regulate blood and lymph vessel formation and homeostasis (reviewed in Cao1 ). VEGFs represent a large family of ligands: VEGF-A, VEGF-B, VEGF-C, VEGF-D, VEGF-E, and PlGF, which bind to and activate in a combinatorial fashion 3 type V receptor tyrosine kinases (RTKs), VEGFR-1, VEGFR-2, and VEGFR-3, which give rise to highly specific signal output. In mammals, VEGF-A signaling through VEGFR-2 is the major angiogenic signaling pathway, but VEGF-C plays essential, and in some cases complementary, roles in the activation of this receptor (reviewed in Grünewald et al2 ). The mechanism by which VEGFRs are activated is not understood in molecular detail, but clearly represents one of the many variations of RTK activation. In general, signaling by RTKs requires ligand-mediated dimerization with precise positioning of receptor subunits in active dimers. Dimeric ligand/receptor complexes subsequently initiate transmembrane signaling, resulting in the activation of the intracellular tyrosine kinase domains.3,4 Active VEGFRs instigate cell signaling and promote endothelial cell migration and proliferation, as well as vessel fenestration and permeabilization.5,6 The extracellular domain (ECD) of VEGFRs consists of 7 Ig-homology domains. The first 3 domains mediate ligand binding,7,8 whereas the membrane proximal domains are involved in ligand-induced receptor dimerization.7,9 Homotypic receptor interactions in ligand-bound dimers were subsequently identified in Ig-homology domains D4 and D7 using single-particle electron microscopy (EM) and small-angle X-ray scattering (SAXS).10,11 In addition, a crystal structure showed direct interactions mediated by the βE-F loop in the D7 dimers.12 The exact role of these contacts is unclear, but it is tempting to speculate that they fulfill a regulatory role in receptor activation, for example by properly positioning receptor monomers in active dimers.
To date, only partial structures of the VEGFR ECDs and kinase domains have been published, and we have crystal structures for all VEGFs.13-18 In addition, the structures of complexes between VEGF-A, VEGF-B, and PlGF with VEGFR-1 Ig domain 2 (D2)19-21 and of VEGFR-2 D23 in complex with VEGF-C are available.14 These structures show that high-affinity binding of VEGF to VEGFR-1 arises from interaction with D2, whereas VEGFR-2 binding requires both domains D2 and D3. These data agree well with a biochemical analysis showing that the loss of VEGFR-2 D3 results in a 1000-fold decrease in VEGF-A binding affinity.8
Based on structural EM and SAXS data10,11 and on a previously published comprehensive biochemical characterization,8 in the present study, we performed a thermodynamic and biophysical analysis of ligand binding to VEGFR-2. We show that D23 indeed represents the high-affinity ligand-binding site of VEGFR-2, whereas domains D4-7 reduce binding affinity by approximately 10-fold, suggesting that this domain plays a regulatory or proofreading role in receptor dimerization and activation. In an effort to understand ligand binding and subsequent receptor dimerization at the molecular level, we also performed a structural analysis. We determined the crystal structures of VEGF-A and VEGF-E in complex with D23 of VEGFR-2 and generated a model of the full-length receptor ECD bound to VEGF-E using SAXS solution scattering data. Based on these data, we propose a model for VEGFR-2 ligand binding and receptor activation that goes beyond simple dimerization.
Methods
Protein expression and purification
Human VEGF-A121, VEGF-A165, and VEGF-C and pox virus VEGF-E NZ2 with an aminoterminal hexahistidine tag were cloned into the pPICZAlphaA vector (Invitrogen), and the proteins were expressed in Pichia pastoris, as described previously.22 Culture supernatants were concentrated and dialyzed against 50mM Tris-HCl (pH 8.0) and 300mM NaCl and the proteins further purified by immobilized metal affinity chromatography (IMAC) using a Ni2+-charged chelating Sepharose column (GE Healthcare), and eluted with an imidazole gradient from 0-500mM. The proteins were deglycosylated with Endoglycosidase F1 (Endo F1) and further purified by size-exclusion chromatography (SEC) on Superdex 200 (GE Healthcare) equilibrated with 50mM HEPES (pH 7.5), 150mM NaCl, and 5% (vol/vol) glycerol. VEGFR-2 recombinant proteins were expressed as secreted proteins in Sf21 insect cells. The ECD of VEGFR-2, consisting of Ig-homology domains 1-7, residues 1-764, with a hexahistidine tag at the carboxy terminus, was cloned into the pFASTBAC (Invitrogen) baculovirus transfer vector. Recombinant baculovirus was produced in Sf21 insect cells in serum-free Insect-Express medium (Lonza) at 27°C. For protein expression, Sf21 insect cells at a density of 106 cells/mL were infected with recombinant baculovirus at 27°C. The supernatant was collected 5 days after infection, concentrated, and dialyzed against 50mM Tris-HCl (pH 8.0) and 300mM NaCl. The protein was first purified by IMAC and eluted with an imidazole gradient from 0-500mM. The relevant fractions were pooled, dialyzed against 20mM Tris-HCl (pH 8.0), and applied to Source 15Q (GE Healthcare) ion exchange chromatography. Protein was eluted with a NaCl gradient from 0-500mM. The samples were finally purified by SEC on Superdex 200 equilibrated with 50mM HEPES (pH 7.5), 150mM NaCl, and 5% (vol/vol) glycerol. The ligand-binding domain of VEGFR-2, D23, consisting of Ig-homology domains 2 and 3, residues 120-326, with an Fc tag at the carboxy terminus, used for isothermal titration microcalorimetry (ITC) was produced and purified as described previously.14
Isothermal titration microcalorimetry
All proteins were applied to SEC on a Superdex 200 (GE Healthcare) column equilibrated with PBS before being used for ITC. The relevant fractions were pooled, concentrated, and adjusted to PBS by dialysis. VEGF-A121, VEGF-A165, VEGF-C, and VEGF-E ITC titrations to the full-length VEGFR-2 ECD or subdomains D23 were carried out at 4°C using an iTC200 calorimeter (MicroCal; GE Healthcare). VEGFR-2 constructs were used in the calorimeter cell at a concentration of 10-30μM and the VEGF ligands in the syringe at 150-250μM. To increase the signal-to-noise ratio, the following parameters were used: injection speed of 0.5 μL/s, data filter of 1 s, and typically 15 injections per titration. Overtitration was performed where needed to obtain a precise estimate of unspecific heat. The data were concatenated using the ConCat program (MicroCal) and processed and analyzed using Origin Version 7.0 software (OriginLab) supplemented with the ITC plug-in provided by the instrument manufacturer. The concentration of native protein was estimated as described previously,23,24 taking into account scattering of protein particles.
Small-angle X-ray scattering
The SAXS experiments were conducted at the cSAXS beamline at the Swiss Light Source (SLS), as described previously.11 Briefly, the proteins and protein complexes were purified as described in “Protein expression and purification,” concentrated to 1-5 mg/mL, and measured in 1-mm quartz capillaries (Hilgenberg). The beam energy was 12.4 keV, and the scattering patterns were collected using a Pilatus 2M detector placed 2.15 m downstream of the sample. Two hundred frames of 0.5 seconds for each protein and the corresponding buffer were collected, integrated, checked for radiation damage, and averaged. After background subtraction, data were analyzed using the ATSAS Version 2.3 program package.25 The distance distribution functions were calculated from the scattering curves using GNOM (Figure 3A) and ab initio shape reconstructions were done with DAMMIF. Twenty independent reconstructions were aligned, averaged, and filtered by DAMAVER to produce the model shown in Figure 3B. Rigid-body homology modeling of solution-scattering data was conducted using SASREF (Figure 3C). For the VEGF-E/VEGFR-2 ECD complex, the structure of the ligand-binding domain D23 with VEGF-E was taken as one rigid body; domains 1, 4, 5, and 6 were derived from c-KIT (2E9W)26 domains 1, 4, 5, and 6, respectively, and domain 7 from the VEGFR-2 structure of the D7 dimer (3KVQ).12 The resulting 10 rigid bodies were held together by linker regions of maximum 7 Å lengths, but otherwise allowed to move and rotate with respect to each other. The independent SASREF runs converged. The models (Figure 3C) are very similar to the ab initio shape reconstructions and were superimposed to the average envelope (Figure 3B) by SUPCOMB13 (Figure 3D). It should be emphasized that the ab initio and rigid-body modeling were done completely independently.
Crystallization, structure determination, and analysis
Purified VEGFR-2 ECD and VEGF-A121 or VEGF-E were mixed in a 1:1.5 molar ratio. The complexes were purified by SEC on Superdex 200 equilibrated with 50mM HEPES (pH 7.5), 150mM NaCl, and 5% (vol/vol) glycerol. The complexes were concentrated to 4-6 mg/mL. In situ limited proteolysis was performed by the addition of α-chymotrypsin in a 1:100 (w/w) ratio to complexes right before crystallization. Crystallization conditions were screened using the sitting drop vapor-diffusion technique at 20°C by mixing the reservoir solution and the protein in a 1:1 drop ratio. Crystals of the VEGF-A complex grew in 0.1M HEPES (pH 7.5), 0.1M NaCl, and 1.6M ammonium sulfate within 1-3 weeks at 20°C. Crystals of the VEGF-E complex grew in 0.1M HEPES (pH 6.5) and 0.8M ammonium sulfate within 2-3 weeks at 20°C. Limited proteolysis led to crystallization of the ligand-binding domain of VEGFR-2 (Ig-homology domains 2 and 3) in complex with VEGF-A or VEGF-E, respectively. Both complexes crystallized in space group P6522 with very similar unit cell parameters (hexagonal cell constants were a = b = 81.0 Å, c = 332.1 Å, and a = b = 79.9 Å, c = 340.0 Å for the VEGF-A and VEGF-E complex, respectively). Complete diffraction datasets were collected at 100 K from crystals flash-frozen in liquid nitrogen at the beamline XS06A at the SLS. The data were processed with XDS27 and the CCP4 suite of programs28 (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
The structures were solved by molecular replacement using Phaser29 and further optimized using Phenix30 for automated refinement and Coot31 for interactive modeling. First, the structure of the VEGF-E complex was solved using as search models the receptor D23 structure from 2X1X and the VEGF-E monomer structure from 1GNN (chain A). The asymmetric unit was found to contain one VEGFR and one VEGF-E ligand chain (solvent content, 70%) such that the dimeric complex is generated by application of a crystallographic 2-fold rotation. Initial electron density maps calculated after a first round of Phenix refinement showed that the model needed manual corrections, including some deletions and insertions at the chain termini and in some loop regions. Most notably, loop Ala83-Asn89 of VEGF-E (not present in the search model from 2GNN) showed strong difference density and could be added to the model. Many solvent-exposed side chains lacked electron density and were truncated. Strong difference electron density, continuous with that of asparagine side chains, was observed at several putative glycosylation sites of the receptor chain (Asn158, Asn245, and Asn318) and of VEGF-A at Asn75. After convergence of the refinement procedure, a final cycle with translational libration screw refinement with the 2 receptor domains and the VEGF-E chain defined as rigid units was carried out. The resulting model was used to solve the structure of the VEGF-A complex by molecular replacement. VEGF-E was then substituted with VEGF-A (initial model from 2VPF, chain A) and refinement proceeded as described for the VEGF-E complex except that only little manual remodeling was required. The final model of the VEGF-A complex comprised residues 13-107 of VEGF-A and residues 132-263, 272-277, and 283-329 of the receptor; the final model of the VEGF-E complex comprised residues 14-107 of VEGF-E and residues 131-263, 269-275, and 283-329 of the receptor. Because of poor side chain densities, we cannot exclude a registry error in chain segments 272-277 and 269-275, respectively. The final R-factors were 0.244/0.301 and 0.237/0.276 for Rwork/Rfree for the VEGF-A and VEGF-E complex, respectively. Detailed refinement statistics for both structures are given in supplemental Table 1.
All figures were generated with PyMol (http://www.pymol.org/). Surface interface areas of the complexes were determined using the PISA server (Protein Interfaces, Surfaces, and Assemblies at the European Bioinformatics Institute, http://www.ebi.ac.uk/pdbe/prot_int/pistart.html, authored by E. Krissinel and K. Henrick). The structure-based ΔG values were determined as described previously.32-35 The coordinates for the VEGF-A and VEGF-E complex structures were deposited at The Protein Databank (www.pdb.org) as 3V2A and 3V6B, respectively.
Results
VEGF family proteins bind to 3 type V RTKs, VEGFR-1, VEGFR-2, and VEGFR-3, and interact with coreceptors and extracellular matrix components. This hampers a thorough analysis of the specific roles played by individual Ig domains in VEGFR binding and activation in live cells. We first investigated the function of the membrane proximal Ig-homology domains 4-7 in ligand binding and receptor dimerization using recombinant proteins encompassing either the core of the ligand-binding domain, D23, or the full-length ECD comprising Ig-homology domains 1-7. Recombinant receptor ECD constructs were produced in insect cells and purified by IMAC and gel filtration. The ligand proteins were expressed in P pastoris and purified as described previously.22 Monodispersity and functionality of recombinant ligand and receptor proteins was determined by incubating the proteins at various molar ratios, followed by gel filtration and multi-angle laser scattering analysis. All ligand/receptor complexes had the expected molecular weights and consisted of 2 receptor molecules and 1 VEGF dimer (supplemental Figure 1).
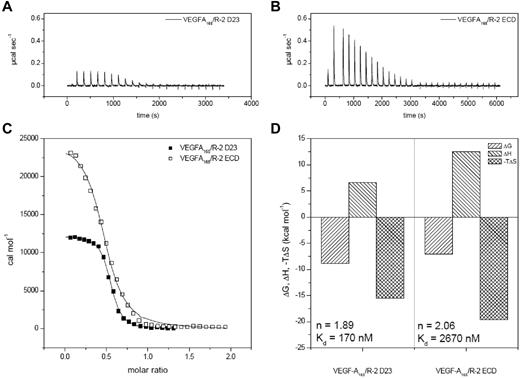
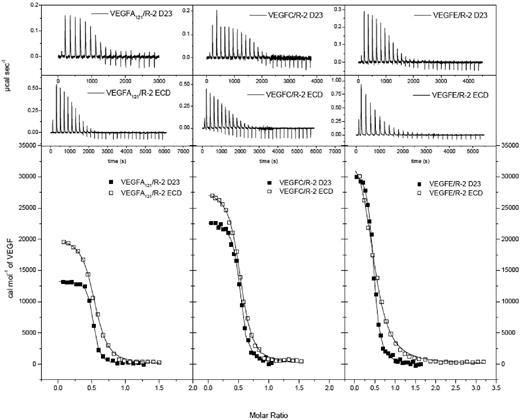
The thermodynamic parameters of 3 VEGF ligands interacting with D23 and D1-7 were determined by ITC (Figure 1, Figure 2, and supplemental Figure 2). Binding constants varied between 0.093 and 0.28μM for D23 and between 1.12 and 5.99μM for the full-length ECD construct. VEGF-A121 showed the highest affinity and VEGF-E the lowest (Table 1). These differences resulted from both enthalpic and entropic contributions to ligand/receptor interaction. Surprisingly, ITC analysis of ligand binding to the full-length ECD revealed that the presence of the membrane proximal Ig-homology domains D4-7 significantly reduced the binding affinity for all complexes (Figures 1-2, Table 1). The ratios of the ligand-binding affinities for D23 and the full-length ECD differed by up to 3.5-fold, but the ΔΔ values (ΔΔG, ΔΔH, and −TΔΔS) showed the same thermodynamic footprints. The positive value for ΔΔG was in all cases dominated by an unfavorable enthalpic change (supplemental Figure 2A-C and Table 1). This finding is somewhat counterintuitive and shows that the homotypic interactions in D4-7 observed in our EM and SAXS analysis are endothermic. Therefore, VEGF binding is driven by direct ligand/receptor interactions in D23, whereas the energetically unfavorable homotypic receptor interactions in D4-7 may ensure proper positioning of receptor monomers in active dimers.
Thermodynamic analysis of VEGF-A165 binding to VEGFR-2 D23 and the full-length ECD. (A-B) Raw titration data of VEGFR-2 domains 2 and 3 and full-length ECD with VEGF-A165, respectively. (C) Integrated and concentration normalized isothermograms from panels A and B. Solid lines represent the best fit according to the “One Set of Sites” model. (D) Thermodynamic analysis of the interaction of VEGFR-2 with VEGF. All reactions are driven by entropy and inhibited by enthalpy. The stoichiometry and binding constants are displayed in the corresponding parts of the panel. R-2 indicates VEGFR-2.
Thermodynamic analysis of VEGF-A165 binding to VEGFR-2 D23 and the full-length ECD. (A-B) Raw titration data of VEGFR-2 domains 2 and 3 and full-length ECD with VEGF-A165, respectively. (C) Integrated and concentration normalized isothermograms from panels A and B. Solid lines represent the best fit according to the “One Set of Sites” model. (D) Thermodynamic analysis of the interaction of VEGFR-2 with VEGF. All reactions are driven by entropy and inhibited by enthalpy. The stoichiometry and binding constants are displayed in the corresponding parts of the panel. R-2 indicates VEGFR-2.
Thermodynamic analysis of VEGFR-2 D23 and full-length ECD binding to VEGF ligands. Top 2 rows show raw titration data of VEGFR-2 D23 and full-length ECD, respectively, with VEGF-A121, VEGF-C, and VEGF-E. Bottom row shows integrated and concentration normalized isothermograms. Solid lines represent the best fit according to the “One Set of Sites” model.
Thermodynamic analysis of VEGFR-2 D23 and full-length ECD binding to VEGF ligands. Top 2 rows show raw titration data of VEGFR-2 D23 and full-length ECD, respectively, with VEGF-A121, VEGF-C, and VEGF-E. Bottom row shows integrated and concentration normalized isothermograms. Solid lines represent the best fit according to the “One Set of Sites” model.
Thermodynamic parameters of VEGF/VEGFR-2 interactions determined by ITC*
| . | N . | Kd, μM . | ΔG, kcal/mol . | ΔH, kcal/mol . | −TΔS, kcal/mol . | c† . |
|---|---|---|---|---|---|---|
| VEGF-A121/VEGFR-2 D23 | 2.01 | 0.093 | −8.9 | 6.6 | −15.5 | 281 |
| VEGF-A165/VEGFR-2 D23 | 1.89 | 0.17 | −8.6 | 6.5 | −15.1 | 150 |
| VEGF-C/VEGFR-2 D23 | 1.91 | 0.20 | −8.5 | 12.0 | −20.5 | 131 |
| VEGF-E/VEGFR-2 D23 | 2.21 | 0.28 | −8.3 | 13.2 | −21.5 | 91 |
| VEGF-A121/VEGFR-2 ECD | 1.97 | 1.12 | −7.5 | 10.4 | −17.9 | 57 |
| VEGF-A165/VEGFR-2 ECD | 2.06 | 2.67 | −7.1 | 12.5 | −19.6 | 24 |
| VEGF-C/VEGFR-2 ECD | 1.9 | 1.21 | −7.5 | 14.8 | −22.3 | 53 |
| VEGF-E/VEGFR-2 ECD | 2.09 | 5.99 | −6.6 | 17.7 | −24.3 | 11 |
| ΔΔG, kcal/mol | ΔΔH, kcal/mol | −TΔΔS, kcal/mol | ||||
| VEGF-A121‡ | 1.4 | 3.8 | −2.4 | |||
| VEGF-A165‡ | 1.5 | 6.0 | −4.5 | |||
| VEGF-C‡ | 1.0 | 2.8 | −1.8 | |||
| VEGF-E‡ | 1.7 | 4.5 | −2.8 | |||
| . | N . | Kd, μM . | ΔG, kcal/mol . | ΔH, kcal/mol . | −TΔS, kcal/mol . | c† . |
|---|---|---|---|---|---|---|
| VEGF-A121/VEGFR-2 D23 | 2.01 | 0.093 | −8.9 | 6.6 | −15.5 | 281 |
| VEGF-A165/VEGFR-2 D23 | 1.89 | 0.17 | −8.6 | 6.5 | −15.1 | 150 |
| VEGF-C/VEGFR-2 D23 | 1.91 | 0.20 | −8.5 | 12.0 | −20.5 | 131 |
| VEGF-E/VEGFR-2 D23 | 2.21 | 0.28 | −8.3 | 13.2 | −21.5 | 91 |
| VEGF-A121/VEGFR-2 ECD | 1.97 | 1.12 | −7.5 | 10.4 | −17.9 | 57 |
| VEGF-A165/VEGFR-2 ECD | 2.06 | 2.67 | −7.1 | 12.5 | −19.6 | 24 |
| VEGF-C/VEGFR-2 ECD | 1.9 | 1.21 | −7.5 | 14.8 | −22.3 | 53 |
| VEGF-E/VEGFR-2 ECD | 2.09 | 5.99 | −6.6 | 17.7 | −24.3 | 11 |
| ΔΔG, kcal/mol | ΔΔH, kcal/mol | −TΔΔS, kcal/mol | ||||
| VEGF-A121‡ | 1.4 | 3.8 | −2.4 | |||
| VEGF-A165‡ | 1.5 | 6.0 | −4.5 | |||
| VEGF-C‡ | 1.0 | 2.8 | −1.8 | |||
| VEGF-E‡ | 1.7 | 4.5 | −2.8 | |||
N indicates the stoichiometry of ligand/receptor complexes.
Error estimation: N, ± 0.01; Kd, ± 5%; ΔG, ΔH, and −TΔS, ± 0.5 kcal mol−1.
The c value is calculated as: c = N*[R2]/Kd.
ΔΔ values represent the difference between the ΔG/ΔH/−TΔS values between the full-length ECD and the D23 ligand complexes.
We also studied the structural details of receptor binding of the 3 ligands. Using single-particle EM analysis, we showed previously that ligand-induced VEGFR-2 dimerization leads not only to contacts in D23, where the ligands directly interact with the receptor, but also in the membrane proximal Ig-homology domains, presumably in D4 and D7. Mutagenesis of D7 confirmed that these interactions are structurally and functionally relevant.10,12 To verify that the apparent homotypic D4-D4 and D7-D7 interactions were not an artifact of sample preparation, we analyzed ligand/receptor complexes in solution by SAXS (Figure 3A and Kisko et al11 ). The ab initio molecular envelope (Figure 3B) and the rigid-body–modeled molecular shape (Figure 3C) of the full-length VEGFR-2 ECD bound to VEGF-E look similar to the single-particle EM model and the published SAXS-derived model of the VEGF-C/VEGFR-2 ECD complex.10,11 The ligand-binding site is clearly visible and the membrane-proximal domains apparently interact with each other. The structural models derived from the solution-scattering data therefore confirm the homotypic interactions between receptor Ig domains D4-7 suggested by the negative-stain EM analysis.10
SAXS analysis of the VEGF-E/VEGFR-2 ECD complex. (A) Scattering intensity and distance distribution function showing scattering data (blue dots), DAMMIF fit (red line), and SASREF fit (black dashed line). (B) Ab initio shape reconstruction of the VEGF-E/VEGFR-2 ECD complex. The mesh represents the average structure of 20 independent shape reconstructions. (C) Rigid-body homology modeling of the SAXS data to deduce the possible arrangement of the individual domains. VEGF-E is shown in blue; VEGFR-2 D23 is shown in dark gray; domains D1, D4, D5, and D6 modeled by c-KIT domains D1, D4, D5, and D5 (pdb 2E9W), respectively, are shown in pink; and the D7 dimer of VEGFR-2 is shown in purple (pdb 3KVQ). (D) Overlay of the independent ab initio and rigid-body homology models derived from SAXS data.
SAXS analysis of the VEGF-E/VEGFR-2 ECD complex. (A) Scattering intensity and distance distribution function showing scattering data (blue dots), DAMMIF fit (red line), and SASREF fit (black dashed line). (B) Ab initio shape reconstruction of the VEGF-E/VEGFR-2 ECD complex. The mesh represents the average structure of 20 independent shape reconstructions. (C) Rigid-body homology modeling of the SAXS data to deduce the possible arrangement of the individual domains. VEGF-E is shown in blue; VEGFR-2 D23 is shown in dark gray; domains D1, D4, D5, and D6 modeled by c-KIT domains D1, D4, D5, and D5 (pdb 2E9W), respectively, are shown in pink; and the D7 dimer of VEGFR-2 is shown in purple (pdb 3KVQ). (D) Overlay of the independent ab initio and rigid-body homology models derived from SAXS data.
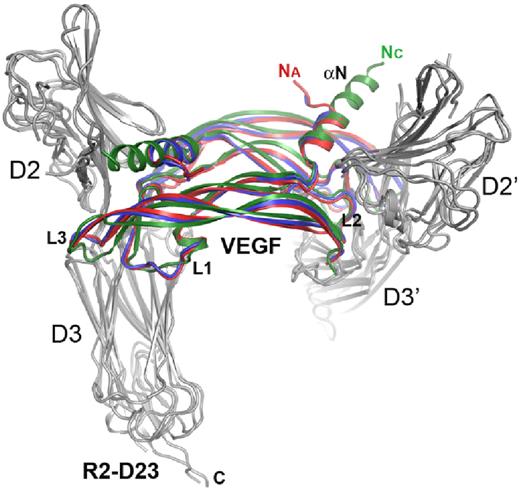
To further analyze binding of VEGF-A and VEGF-E to VEGFR-2, we determined the structures of the ligand/receptor complexes by X-ray crystallography. Preformed complexes between VEGF-A and VEGF-E, respectively, and the full-length receptor ECD were purified by gel filtration and crystallized by in situ limited proteolysis. The crystallographic data are summarized in supplemental Table 1. Crystals contained complexes between D23 and a ligand dimer and were compared with our previously published structure of the VEGF-C receptor complex.14 The overall architecture of the 3 structures was very similar, revealing a symmetrical 2:2 complex and providing the structural basis of high-affinity ligand binding by the 2 receptor domains (Figure 4). VEGF-A and VEGF-E form antiparallel homodimers with symmetrical receptor binding sites at opposite poles. D2 and D3 of the 2 chains of VEGFR-2 in the complex are arranged in a left-handed helical conformation, with the twist angles varying for the 3 complexes. With approximately 6 degrees, the twist angle difference between the VEGF-A and VEGF-E complexes was noticeably smaller than the 16 and 25 degrees observed between the VEGF-E and VEGF-C and the VEGF-A and VEGF-C complexes, respectively.
Comparison of ligand/VEGFR-2 D23 complex structures. Ribbon diagram of the superposition of VEGF-A (red), VEGF-C (green, pdb 2X1X), and VEGF-E (blue) complexes with VEGFR-2 D23 (gray). Na and Nc amino termini of VEGF-A and -C, respectively, the VEGF loops L1-L3, and the aminoterminal helix (αN) are labeled where applicable.
Comparison of ligand/VEGFR-2 D23 complex structures. Ribbon diagram of the superposition of VEGF-A (red), VEGF-C (green, pdb 2X1X), and VEGF-E (blue) complexes with VEGFR-2 D23 (gray). Na and Nc amino termini of VEGF-A and -C, respectively, the VEGF loops L1-L3, and the aminoterminal helix (αN) are labeled where applicable.
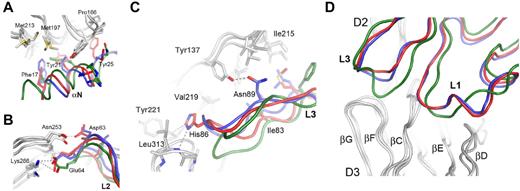
The VEGFR-2–binding epitopes of VEGF family monomers are composed of 3 connecting loops (L1-L3) and an N-terminal α-helix (αN). Similar to the VEGF-C receptor complex structure, the aminoterminal helix of VEGF-A and VEGF-E interacts with VEGFR-2 D2, whereas the D2/D3 linker reaches into the groove separating the 2 VEGF monomers. The interface between the aminoterminal helices of VEGF-A and VEGF-E and D2 of VEGFR-2 result from mostly hydrophobic interactions, including Phe17, Tyr21, and Tyr25 of VEGF-A (Figure 5A). The ligands lack sequence identity but show surprising structurally and chemically similar complementary interfaces. L2 contributes conserved polar interactions between Asp63 and Glu64 (VEGF-A numbering) and Asn253 and Lys286 of VEGFR-2, respectively (Figure 5B). L3 contributes both conserved and similar hydrophobic interactions, as well as polar interactions mediated by His86 of VEGF-A and Asn89 of VEGF-E to receptor binding (Figure 5C). L3 of VEGF-E shows the most significant structural differences between the free and the receptor-bound ligand structure. It is highly flexible in unbound VEGF-E and adopts an extended β-sheet conformation after receptor binding.15 Finally, L1 of all VEGFs interacts solely with D3 (Figure 5D). Our results provide the structural basis for the interaction of VEGF family proteins with VEGFR-2 and define subtle but distinct differences in their receptor-binding mode.
Binding interfaces between the aminoterminal helices (αN) and loops L1-L3 of the VEGFs and VEGFR-2 D2 and D3, representing the major ligand-binding domain. Key residues are highlighted and labeled using VEGFR-2 and VEGF-A numbering, except for Asn89 of VEGF-E. VEGF-A is shown in red, VEGF-C is shown in green, VEGF-E is shown in blue, and VEGFR-2 D23 is shown in gray. Hydrogen bonds and salt bridges are shown by gray dashed lines. In panel A, the view is centered on the aminoterminal helices (αN) of VEGFs, and in panel B, on the contacts between L2 of VEGF with D3 of the receptor. (C) Interaction of loop L3 of VEGFs with VEGFR-2 D23. Hydrogen bonds and hydrophobic interactions for VEGF-A His86 and VEGF-E Asn89 are shown by gray dashed lines. (D) The binding interfaces between loops L1 and L3 of the VEGFs with VEGFR-2.
Binding interfaces between the aminoterminal helices (αN) and loops L1-L3 of the VEGFs and VEGFR-2 D2 and D3, representing the major ligand-binding domain. Key residues are highlighted and labeled using VEGFR-2 and VEGF-A numbering, except for Asn89 of VEGF-E. VEGF-A is shown in red, VEGF-C is shown in green, VEGF-E is shown in blue, and VEGFR-2 D23 is shown in gray. Hydrogen bonds and salt bridges are shown by gray dashed lines. In panel A, the view is centered on the aminoterminal helices (αN) of VEGFs, and in panel B, on the contacts between L2 of VEGF with D3 of the receptor. (C) Interaction of loop L3 of VEGFs with VEGFR-2 D23. Hydrogen bonds and hydrophobic interactions for VEGF-A His86 and VEGF-E Asn89 are shown by gray dashed lines. (D) The binding interfaces between loops L1 and L3 of the VEGFs with VEGFR-2.
To date, no differences in receptor signaling for the 3 ligands VEGF-A, VEGF-C, and VEGF-E have been described. However, the binding affinities of the 3 ligands differ significantly, both in cell-binding assays and in solution, as shown here by ITC. Ligand binding to D23 or D1-7 also differs significantly, with VEGF-A showing the highest affinity and VEGF-E the lowest. For a full comparison we analyzed the buried interface surface area and the number of ionic interactions for the 3 VEGFR-2 D23 complexes based on our crystallographic data using the PISA server (supplemental Figure 3).35 To better describe the interactions, we have previously assigned 2 binding sites, site 1 and 2, corresponding to VEGF monomer A (αN, L2) and B (L1, L3) interactions with the receptor.14 The total buried interface area in site 1 for the VEGF-A, VEGF-C, and VEGF-E complexes would be 930, 1530, and 1110 Å,2 respectively. However, the site 1 comparison is not valid because VEGFR-2 residues 276-282 in the VEGF-A and VEGF-E complexes, comprising part of the site 1 interface in the VEGF-C complex, were omitted from the final models. The buried surface area in site 2 for the VEGF-A, VEGF-C, and VEGF-E complexes are 1310, 1250, and 1160 Å,2 respectively. According to the PISA interface analysis, the number of hydrogen bonds and salt bridges at the VEGFR-2 interfaces is comparable: 11 for VEGF-A, 13 for VEGF-C, and the largest, 14, for VEGF-E (supplemental Figure 3). The largest difference in the number of ionic interactions is in site 2, where VEGF-E has 6 hydrogen bonds and VEGF-A and VEGF-C only 2. Although methods for calculating the free energy for complex formation of proteins based on structural data are still unreliable, the estimates determined by the PISA server show that the VEGF-E complex is thermodynamically less favorable. The calculated ΔG values for the VEGF-A and VEGF-C complexes were −24.7 and −18.4 kcal/mol, respectively, and for the VEGF-E complex, −6.2 kcal/mol, which is in agreement with the lower affinity of the latter complex.
Discussion
The thermodynamic analysis of receptor binding of VEGFs shown here reveals a surprising finding: the Gibbs free energy of ligand binding to the full-length VEGFR-2 ECD (D1-7) compared with binding to D23 was less negative by 1.0-1.7 kcal/mol for all ligands (Table 1), and therefore the affinity constants are lower by 6- to 21-fold. Ligand binding to VEGFR-2 and the subsequent receptor dimerization apparently depend on 2 types of interactions: (1) direct ligand interaction with D23, which is in all cases thermodynamically highly favorable despite differences in the binding mode of each ligand; and (2) an unfavorable endothermic component arising from interactions between subdomains 4 through 7. Homotypic interactions in D4-7 are clearly visible in the single particles analyzed by negative-stain EM and in the structural models derived from SAXS data. We thus infer that in the full-length receptor ECD complexes, the symmetric optimal ligand interactions with D23, as observed in the ligand/D23 complexes, can only be maintained at the expense of an endothermic contribution originating from the limited structural flexibility of each receptor monomer.
Because no high-resolution crystallographic information for full-length ECD complexes is available at present, we analyzed these structures by SAXS. Solution-scattering data were used to build structural models using a software suite developed by Svergun et al.25 Our models of the receptor complexes in solution are in good agreement with published data for VEGFR-210-13 and the related type III RTKs c-Kit,26 PDGFR-β,36,37 Fms,38 and Flt3.39 Except for Flt3, all receptors form homotypic contacts in membrane proximal Ig-homology domains, which are distinct from the contacts mediated by the ligand-binding domain. These interactions were also shown to be functionally relevant and, based on these results, we propose the following order for the mechanism of receptor activation: (1) the ligand-bound ECD dimerizes the receptor, (2) homotypic receptor contacts between receptor monomers resulting from dimerization reorient the intracellular kinase domains, leading to (3) activation of the receptor kinase and (4) phosphorylation of specific sites in the intracellular receptor domain, resulting in (5) the activation of ligand- and receptor-specific downstream signaling pathways.
Our structural analysis completes the analysis of VEGFR-2 binding to its major ligands VEGF-A, VEGF-C, and VEGF-E. VEGFR-2 signaling is the predominant pathway in physiologic and pathologic angiogenesis and is an important drug target. VEGF-A binding to VEGFR-2 was originally studied in a large alanine mutant screen by identifying single mutations that affected receptor binding.40 The 5 amino acids most important for binding were Ile46 from strand β2, Ile83 from strand β4, Glu64 from loop L2, Phe17 from the N-terminal helix, and Gln79 from strand β5. The VEGF-A/VEGFR-2 D23 complex structure presented in the present study allows the definition of specific roles for these residues. Glu64 of VEGF-A interacts with Lys286 in D3 of VEGFR-2 (Figure 5B), and Phe17 is part of the hydrophobic interface formed with D2 (Figure 5A). Ile46 and Ile83 are adjacent residues in the structure and pack against Val217 in the D2/D3 linker region. The other important residues identified in the alanine mutant screen were Ile43 from loop L1 and Lys84 and Pro85 from strand β5. Ile43 packs against Phe258 and Phe288 in D3 of VEGFR-2. Pro85 packs against Gly255 in D3, allowing the tip of L3 (His86) to interact with VEGFR-2. Gln79 and Lys84 of VEGF-A seem to have structural rather than VEGFR-2 interacting roles. Consistent with our complex structures, VEGF-A binding to VEGFR-2 was reported previously to be strongly dependent on D3.8 Asn253 and Lys286 in D3 are involved in hydrophilic interactions with residues in loop L2 conserved in VEGF-A, VEGF-C, and VEGF-E (Figure 5B). The structures also reveal multiple ligand-specific interactions by loops L3 and L1 of VEGF with D3 of VEGFR-2. L1 and L3 show the largest differences, both in conformation and in sequence, of all of the structural elements of VEGFR-2 specific ligands. Our data also clearly show that ligand structure is modulated by receptor binding; for example, L3 of VEGF-E, which is flexible in the unbound form,15 adopts an extended β-sheet conformation apparently required for optimal interaction with D2 and D3. Combining the structural information now available for all 3 complexes with the analysis of the VEGF-A alanine mutant screen will be helpful in designing small-receptor–inhibitory polypeptides with therapeutic potential.
The binding affinities of the 3 VEGFs for D23 and D1-7 of VEGFR-2 differ by a factor of 3 and 5, respectively, reflecting the differences in affinity observed in vivo in cell-binding assays. The calculation of binding affinities of interacting proteins from structural information is based on rather empirical energy functions and therefore such results must be interpreted with caution. Analysis of the surfaces of the 3 complexes by PISA shows that the buried surface area in site 2 is significantly smaller for the VEGF-E complex. In addition, structure-based calculation of the ΔG values for the 3 complexes shows the same trend as the experimental thermodynamic data.
VEGF-A, VEGF-C, VEGF-E, and the VEGF-C homolog VEGF-D are all potent VEGFR-2 ligands and induce VEGFR-2–mediated angiogenic signaling in many experimental conditions.34,41 It remains unclear how subtle differences in the binding affinities and the interfaces described herein affect biologic signal output. The crystal structures of the VEGFR-2 D23 complexes reveal essentially identical binding sites (Figure 5) and conserved hydrophobic and hydrophilic interactions. In addition, thermodynamic analysis of the ligand-binding domains 2 and 3 by ITC showed similar thermodynamic footprints, with large entropic contributions presumably arising from multiple hydrophobic interactions arising during complex formation. Based on a published structure for VEGF-D, one might propose a similar role for these hydrophilic interactions and the large hydrophobic surface in receptor binding for this ligand.13 The 3 structures presented herein also reveal significant differences, most significantly variation in the D23 twist angles, which might control ligand-specific signal output by modulating, for example, the kinetics of receptor activation, signaling, and trafficking. In agreement with this hypothesis, it was shown previously that, in some cases, ligand-specific receptor signaling is determined by coreceptor recruitment, in particular of neuropilins.42-44 Neuropilins bind VEGF-A and VEGF-E through specific sequences in the carboxy terminus.45,46 It will be interesting in future studies to determine whether coreceptor assembly alters the overall structure of VEGFR-2 dimers, for example, by affecting the structure of receptor interfaces and thus the exact positioning of the intracellular kinase domains relative to each other.
VEGFR-2, unless expressed at extremely high levels, does not spontaneously dimerize in the absence of ligand, presumably because of repulsion between the extracellular domains.47 It has also been shown in another system, fibroblast growth factor receptor 3, that the ECD prevents ligand-independent receptor dimerization, thereby avoiding constitutive receptor signaling.48 The ITC data presented herein for VEGFR-2 specify the distinct roles of individual extracellular subdomains in ligand-mediated dimerization; ECD subdomains 2 and 3 form the high-affinity ligand-binding site, whereas the energetically unfavorable homotypic interactions in domains 4-7 reduce the overall binding affinity. Tao et al showed previously that deletion of D4-7 led to constitutive ligand-independent receptor activation.49 In agreement with our thermodynamic data, the investigators in that study proposed that structural motifs in D4-7 block spontaneous VEGFR-2 dimerization, kinase activation, and downstream signaling. We propose that this represents a proofreading mechanism safeguarding endothelial cells against ligand-independent receptor activation. Such a mechanism is of pivotal importance in tissue homeostasis in that it averts the deleterious consequences of aberrant vascularization documented in many diseases.50 This novel and surprising finding opens new possibilities for developing anti-angiogenic drugs interfering with VEGFR dimerization and activation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Swiss National Science Foundation and Oncosuisse for continuous support of our work; the staff at the crystallography and cSAXS beamlines at the Swiss Light Source for technical support; and Dr J. H. Missimer for critical data evaluation.
This work was funded by the Swiss National Science Foundation (grant 31003A-130463) and by Oncosuisse (grant OC2 01200-08-2007).
Authorship
Contribution: M.S.B., S.B., and K.K. designed and performed the experiments, analyzed the data, and wrote the manuscript; T.S. prepared the recombinant protein; V.-M.L. and F.K.W. analyzed and processed the structural data and wrote the manuscript; K.A. assisted in manuscript preparation; and K.B.-H. designed the experiments, supervised the project, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kurt Ballmer-Hofer, Paul Scherrer Institut, Department of Biomolecular Research, Molecular Cell Biology, CH-5232 Villigen PSI, Switzerland; e-mail: kurt.ballmer@psi.ch.
References
Author notes
M.S.B., S.B., and K.K. contributed equally to this work.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal