Abstract
Normal human erythroid cell maturation requests the transcription factor GATA-1 and a transient activation of caspase-3, with GATA-1 being protected from caspase-3–mediated cleavage by interaction with the chaperone heat shock protein 70 (Hsp70) in the nucleus. Erythroid cell dysplasia observed in early myelodysplastic syndromes (MDS) involves impairment of differentiation and excess of apoptosis with a burst of caspase activation. Analysis of gene expression in MDS erythroblasts obtained by ex vivo cultures demonstrates the down-regulation of a set of GATA-1 transcriptional target genes, including GYPA that encodes glycophorin A (GPA), and the up-regulation of members of the HSP70 family. GATA-1 protein expression is decreased in MDS erythroblasts, but restores in the presence of a pan-caspase inhibitor. Expression of a mutated GATA-1 that cannot be cleaved by caspase-3 rescues the transcription of GATA-1 targets, and the erythroid differentiation, but does not improve survival. Hsp70 fails to protect GATA-1 from caspases because the protein does not accumulate in the nucleus with active caspase-3. Expression of a nucleus-targeted mutant of Hsp70 protects GATA-1 and rescues MDS erythroid cell differentiation. Alteration of Hsp70 cytosolic-nuclear shuttling is a major feature of MDS that favors GATA-1 cleavage and differentiation impairment, but not apoptosis, in dysplastic erythroblasts.
Introduction
Myelodysplastic syndromes (MDS) are clonal disorders of the HSC characterized by ineffective hematopoiesis leading to peripheral blood cytopenias, mainly anemia. Erythroid cell dysplasia is observed in ∼ 50% of early stage MDS, including refractory anemia (RA), refractory cytopenia with (RCMD) or without multilineage dysplasia, 5q− syndrome, refractory anemia with ring sideroblasts (RARS or RCMD-RS), and RA with excess of blasts (RAEB) with < 10% medullary blasts (RAEB 1).1 Dyserythropoiesis associates with a defective maturation of erythroid precursors and an excessive apoptosis.2-4 The initial erythropoietic cell damage differs among MDS subtypes, for example, 5q− syndrome involves RPS14 haploinsufficiency, defective ribosomal biogenesis, and p53 activation,5,6 while RARS includes aberrant iron accumulation and mitochondrial stress.7
Commitment of the BM progenitor cells is achieved through expression of lineage-specific transcription factors. GATA-1 is a zinc finger hematopoietic transcription factor expressed in erythroid cells, megakaryocytes, mast cells, and eosinophils.8 This transcription factor is required for the commitment of multipotent hematopoietic progenitor to the erythroid and megakaryocytic lineages. Mutant embryonic stem cells deleted of GATA-1 fail to give rise to mature RBCs in vitro.9 Deletion of GATA-1 gene in mice results in a lethal phenotype between embryonic day (E) 10.5 and E11.5 because of severe anemia. GATA-1− embryos at E9.5 contain erythroid cells arrested at an early proerythroblast-like stage of their differentiation,10 thus demonstrating that GATA-1 is also required for the terminal erythroid cell maturation.
Regulation of GATA-1 expression takes place at the transcriptional, translational, and posttranslational level. Exposure of erythroblasts to death-receptor ligands or cytokine starvation results in caspase-mediated degradation of GATA-1.11 Caspase-3 is also transiently activated during erythropoietin (Epo)–driven normal erythroid differentiation at the transition from basophilic erythroblasts to polychromatophilic erythroblasts and cleaves a series of proteins such as poly(ADP-ribose) polymerase, acinus, or lamin B, but not GATA-1.12,13 GATA-1 cleavage does not occur in Epo-stimulated cells because of a protective interaction with the ATP-dependent chaperone, heat shock protein 70 (Hsp70) that migrates in the nucleus at the onset of caspase activation.14 The present study was designed to determine whether the GATA-1/Hsp70 interplay was disabled in MDS erythroblasts. We demonstrate a caspase-dependent decrease of GATA-1 expression because of a default in the nuclear localization of Hsp70.
Methods
Patient and control cells
A total of 51 samples including low-grade MDS patients with dyserythropoiesis (RA, n = 11; undefined MDS [MDS-U], n = 2; RCMD, n = 5; RARS/RCMD-RS, n = 7; RAEB1, n = 16) or without dyserythropoiesis (n = 10) were collected at diagnosis (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Normal CD34+ cells purified from BM (n = 14) or peripheral blood cytapheresis (n = 6) were used as controls (supplemental Table 1). The study was approved by the local ethics committee from Cochin Hospital and informed consent was obtained from all patients and volunteers in accordance with the Declaration of Helsinki.
Erythroid cell culture and flow cytometry
CD34+ progenitors were isolated from BM mononuclear cell fraction on MidiMacs system (Miltenyi Biotec) and cultured as described.3 In some experiments, erythroblasts were incubated with 100μM Q-VD-OPH (BioVision Research Products) or 20μM MG132 (Sigma-Aldrich). Cell differentiation was assessed on May-Grünwald Giemsa–stained cytospins or flow cytometry with FITC-coupled CD71 and PE-coupled glycophorin A (GPA) labeling (Beckman Coulter). Apoptosis was measured using PE-coupled annexin V (Alexis Biochemicals) and 7-amino-actinomycin D (7-AAD; Beckman Coulter), or annexin V and hexamethylindodicarbo-cyanine iodide (HIDC; Sigma Aldrich) and caspase-3-like activity was measured using the CaspGLOW Fluorescein Active Caspase-3 Staining Kit (BioVision Research Products). Analyses were performed on a FC500 apparatus (Beckman Coulter).
Microarray experiments and RT-qPCR
Five MDS with dyserythropoiesis (MDS nos. 15, 23, 36, 38, 46) and 4 control samples obtained from the laboratory of P.V. were cultured in erythroid conditions and processed for RNA extraction and hybridization to GeneChip Human Genome U133A gene chips (Affymetrix), as described.15 Image data were quantified with Microarray Software Suite Version 5.0 (Affymetrix). Data from 10820_at labeled probes were normalized to median of 3 replicates by GC-Robust MultiChip Analysis and significance analysis of microarrays used for marker selection was performed in the software R. Neighborhood analysis was performed using the published methods.16 Genes differentially expressed (ANOVA test; P < .01) were identified by hierarchic clustering using the GeneSpring Version 6.0 software (Silicon Genetics). Microarray data have been deposited on the Genome Expression Omnibus (GEO) database of the NCBI Web site (http://www.ncbi.nlm.nih.gov/geo/ accession number GSE34268). Expression level of specific genes was validated by RT-qPCR on the training set and a validation set of 4 MDS with dyserythropoiesis (MDS nos. 1, 4, 9, 10) and 3 controls (CTL nos. 2, 6, 11) on Light Cycler apparatus (Roche Diagnostics). A standard curve and an internal calibrator were prepared from U937 cell line. The specific fluorescence threshold (Cq) compared with the calibrator, normalized to reference genes GAPDH or hypoxanthine-guanine phosphoribosyltransferase (HPRT) and expressed as normalized relative quantity (nRQ) was calculated using the Relquant software using the ΔCq method.
Immunoblot
Cells were lysed on ice in 0.2% NP40, 20mM HEPES pH7.9, 10mM KCl, 1mM EDTA, 10% glycerol, phosphatase, and protease inhibitors and centrifuged at 10 000g for recovering cytosolic fractions. Nuclear fractions were extracted from the pellets in 350mM NaCl, 20% glycerol; 20mM HEPES pH 7.9, 10mM KCl, 1mM EDTA. Immunoblotting was performed using Abs to GATA-1 (C20), Hsc70 (Santa Cruz Biotechnology), Hsp70 (Stressgen) or actin (Sigma-Aldrich). Chemiluminescence was quantified with MultiGauge Version 3.0 Fujifilm software.
Immunofluorescence
Cytospins were fixed in acetone and permeabilized with 0.2% Triton X-100 in PBS pH7.4.14 After washing, cells were sequentially incubated with anti–GATA-1 N6 (Santa Cruz Biotechnology), anti–rat-Cyanine (Cy)3, rabbit anti-Hsp70 and anti–rabbit FITC (Jackson ImmunoResearch Laboratories) Abs. Nuclei were stained with diaminido phenyl indol (DAPI; Sigma-Aldrich). Alternatively, cells were sequentially labeled with anti-cleaved caspase mouse Ab (SantaCruz Biotechnology), goat anti–mouse-cyanine 5 Ab (Jackson ImmunoResearch Laboratories), rabbit anti-Hsp70 Ab (Stressgen Bioreagents) and donkey anti–rabbit-FITC Ab (Jackson ImmunoResearch Laboratories). Images were obtained on a Leica DMB microscope and analyzed using Metamorph software (Molecular Devices). Signals were quantified using NIH ImageJ software. In some experiments, anti-HA Abs (Sigma-Aldrich) were used and slides were examined with a confocal laser microscope (LSM 510; Carl Zeiss).
Generation of Hsp70 mutant
Construction of mutant Hsp70 S400A was done by directed mutagenesis following the instruction of the supplier (Stratagen's QuickChange II Site Directed Mutagenesis Kit; Agilent Technologies). The pcDNA3 Hsp70 wild-type (wt) plasmid was used as a template for the annealing of the following mutagenic oligonucleotides: reverse primer (5′-TG GAC GTG GCT CCC CTG GCG CTG GG-3′), forward primer (5′-CC CAG CGC CAG GGG AGC CAC GTC CA-3′). Normal erythroblasts were transfected with plasmids using the Amaxa protocol according to the manufacturer's instructions.
Retrovirus and lentivirus production and MDS CD34+ cell transduction
wtGATA-1 and the uncleavable mutant μGATA-1 cloned in the PINCO retroviral vector upstream a CMV promoter-Green Fluorescent Protein (GFP) cassette (a generous gift from R. De Maria, Istituto Superiore di Sanita, Rome, Italy) were used to produce vector particles by cotransfection using Lipofectamine 2000 (Invitrogen) of 293EBNA cells with the vector plasmid, an encapsidation plasmid (gag-pol) lacking all accessory HIV-1 proteins, and an expression plasmid (pHCMV-G) encoding the vesicular stomatitis virus (VSVg) envelope. wtBcl-2, wtHsp70, and the nuclear mutant Hsp70 S400A were cloned in the pTripΔU3EF1α lentiviral vector upstream a IRESECMV–Green Fluorescent Protein (GFP) cassette. Infectious vector particles were produced in 293T cells by Lipofectamine 2000 cotransfection with the encapsidation plasmid p8.9 and the expression plasmid pHCMV-G. CD34+ cells were transduced twice with viral supernatants at day 1 and 2 after purification. The empty vectors were used as controls. On day 3, cells were washed twice and seeded in medium for erythroid culture. Transduction efficacy was measured as the percentage of GFP+ cells. Apoptosis and differentiation markers were always quantified in the GFP+ population.
Results
Defective erythroid cell maturation of MDS erythroblasts
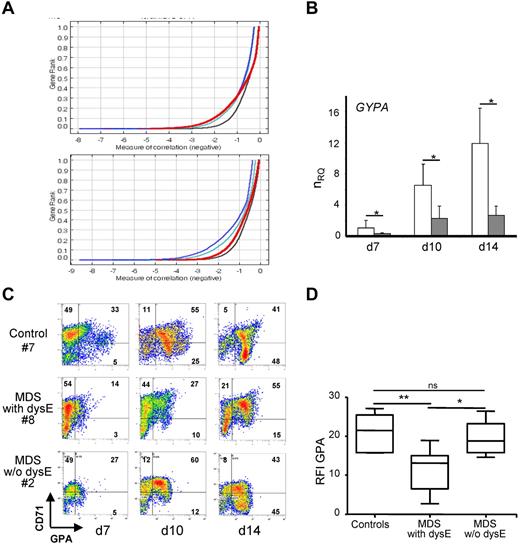
To investigate the pathophysiology of dyserythropoiesis in MDS, we compare gene expression profile of erythroblasts obtained by liquid culture of CD34+ cells sorted from MDS with morphologic erythroid cell dysplasia (n = 5) and control (n = 4) BM. RNA is extracted from erythroblasts harvested at 3 different days of culture and hybridized with hU133A gene chips. By neighborhood analysis, we identify a MDS dyserythropoiesis signature of 1612 genes in which a set of 265 down-regulated genes could be used to make a prediction of MDS disease (Figure 1A). These down-regulated genes include a series of known GATA-1 target genes involved in erythroid differentiation such as GYPA, ALAD, ACYP1, CA2, UROS that encode GPA, aminolevulinate δ dehydratase, acylphosphatase erythrocyte, carbonic anhydrase II, and uroporphyrinogen III synthetase, respectively (supplemental Table 2a).17
Deregulation of erythroid marker expression in MDS erythroblasts. (A) Gene expression profiling. RNA from MDS with dyserythropoiesis (dysE; n = 5) versus normal (n = 4) cultured erythroblasts are hybridized on Affymetrix gene chip U133A. Neighborhood analysis of data from day 7 shows the probe sets predictive of MDS (red line) versus normal together with curves showing the 50% (black line), 5% (green line), and 1% (blue line) significance levels representing randomly permuted class distinctions. An under-expressed probe set (n = 265) predicts MDS (top panel) with a less than 5 in 100 chances of error, whereas no overexpressed probe set can make the distinction between MDS and normal (bottom panel). (B) RT-qPCR. Quantification of GYPA transcripts in the training and validation sets (9 MDS:  ; 7 controls: □), pooled and expressed as mean normalized relative quantity (nRQ) ± SEM (SE) to GAPDH. (C-D) Erythroid differentiation. (C) Representative dual fluorescence histograms of MDS with dysE (n = 30), control (n = 19), and MDS w/o dysE (n = 11) erythroblasts labeled with Abs to CD71 (FL1) and GPA (FL2). Percentages are indicated. (D) Ratio of median fluorescence intensity (RFI) of GPA to isotype per cell at day 14 expressed as box plots: lines within boxes represent the medians (Student t test for P values; *P < .05; **P < .01; ns indicates not significant).
; 7 controls: □), pooled and expressed as mean normalized relative quantity (nRQ) ± SEM (SE) to GAPDH. (C-D) Erythroid differentiation. (C) Representative dual fluorescence histograms of MDS with dysE (n = 30), control (n = 19), and MDS w/o dysE (n = 11) erythroblasts labeled with Abs to CD71 (FL1) and GPA (FL2). Percentages are indicated. (D) Ratio of median fluorescence intensity (RFI) of GPA to isotype per cell at day 14 expressed as box plots: lines within boxes represent the medians (Student t test for P values; *P < .05; **P < .01; ns indicates not significant).
Deregulation of erythroid marker expression in MDS erythroblasts. (A) Gene expression profiling. RNA from MDS with dyserythropoiesis (dysE; n = 5) versus normal (n = 4) cultured erythroblasts are hybridized on Affymetrix gene chip U133A. Neighborhood analysis of data from day 7 shows the probe sets predictive of MDS (red line) versus normal together with curves showing the 50% (black line), 5% (green line), and 1% (blue line) significance levels representing randomly permuted class distinctions. An under-expressed probe set (n = 265) predicts MDS (top panel) with a less than 5 in 100 chances of error, whereas no overexpressed probe set can make the distinction between MDS and normal (bottom panel). (B) RT-qPCR. Quantification of GYPA transcripts in the training and validation sets (9 MDS:  ; 7 controls: □), pooled and expressed as mean normalized relative quantity (nRQ) ± SEM (SE) to GAPDH. (C-D) Erythroid differentiation. (C) Representative dual fluorescence histograms of MDS with dysE (n = 30), control (n = 19), and MDS w/o dysE (n = 11) erythroblasts labeled with Abs to CD71 (FL1) and GPA (FL2). Percentages are indicated. (D) Ratio of median fluorescence intensity (RFI) of GPA to isotype per cell at day 14 expressed as box plots: lines within boxes represent the medians (Student t test for P values; *P < .05; **P < .01; ns indicates not significant).
; 7 controls: □), pooled and expressed as mean normalized relative quantity (nRQ) ± SEM (SE) to GAPDH. (C-D) Erythroid differentiation. (C) Representative dual fluorescence histograms of MDS with dysE (n = 30), control (n = 19), and MDS w/o dysE (n = 11) erythroblasts labeled with Abs to CD71 (FL1) and GPA (FL2). Percentages are indicated. (D) Ratio of median fluorescence intensity (RFI) of GPA to isotype per cell at day 14 expressed as box plots: lines within boxes represent the medians (Student t test for P values; *P < .05; **P < .01; ns indicates not significant).
In the training set of 5 MDS and 4 controls and in a validation set of 4 MDS and 3 control samples of cultured erythroblasts, we use RT-qPCR to confirm that GYPA transcript is decreased in day 7, 10, and 14 MDS erythroblasts compared with the controls (Figure 1B). By flow cytometry, dual fluorescence analysis of CD71 and GPA markers during the culture demonstrates that erythroid maturation is delayed with persistence of CD71 and low expression of GPA in MDS compared with control or MDS without morphologic erythroid cell dysplasia (Figure 1C). The ratio of median fluorescence intensity (RFI) of GPA per cell is significantly lower in MDS dyserythropoietic erythroblasts (11.2 ± 4.1) compared with controls (21.1 ± 4.4; P < .001) or MDS without dyserythropoiesis (19 ± 5.0; P < .05). This further indicates that low GPA is a marker of MDS dyserythropoiesis (Figure 1D). Additional GATA-1 target genes whose expression is decreased in MDS include HBB gene that encodes β-globin and PBGDe encoding the erythroid-specific porphobilinogen deaminase (supplemental Figure 1). Altogether, these results suggest that GATA-1 may experience functional inactivation in mature dysplastic erythroblasts.
Caspase-mediated cleavage of GATA-1 in MDS erythroblasts
When measured by RT-qPCR, the level of GATA1 mRNA is equivalent in MDS and control erythroblasts, ruling out a defect in GATA1 gene transcription (P = .350; Figure 2A). MDS erythroblasts exhibit an increase in apoptotic cell death compared with normal cells (Figure 2B), which correlates with a burst of caspase activation that is detectable by flow cytometry and can be blocked by the pan-caspase inhibitor Q-VD-OPH (100μM from day 8 of culture, Figure 2C). Immunoblot analysis of nuclear extracts demonstrates that GATA-1 expression is lower in 7 of 11 MDS with dyserythropoiesis compared with control erythroblasts (n = 8) at day 14 of the liquid culture and the quantification shows that the difference was significant (Figure 2D; P < .05). The expression of GATA-1 both at day 12 and day 14 is rescued by cell treatment with Q-VD-OPH, but not by cell treatment with MG132, a proteasome and cathepsin inhibitor (Figure 2E). Although GATA-1 may be degraded through proteasome,18 we observe that a caspase-mediated cleavage, rather than a proteasome/lysosome-dependent degradation, is responsible for GATA-1 decrease in MDS erythroblasts.
A caspase-mediated cleavage of GATA-1 in MDS erythroblasts. (A) GATA-1 gene expression. RT-qPCR for GATA1 transcripts in MDS versus control erythroblasts. Normalized relative quantity (nRQ) to HPRT. Median values indicated as horizontal bars. (B) Apoptosis. Flow cytometry for annexin V+/7-AAD− cells in MDS (n = 30; ■) and control (n = 22; ○) erythroblasts. Mean percentages ± SE. (C) Caspase-3 activity. Flow cytometry for caspase-3 like activity in MDS (n = 8; ■), pan-caspase inhibitor Q-VD-OPH (100μM)–treated MDS (n = 1;  ) and control (n = 4; ○) erythroblasts. Results as mean fluorescence intensity. (D) GATA-1 protein expression. Nuclear fraction proteins blotted with the indicated Abs (top panel). Quantification of GATA-1 by densitometry expressed as a ratio to Hsc70, normalized to the ratio of GATA-1 to Hsc70 in UT-7 control cell line for each blot (bottom panel) in MDS with (n = 11) or without (n = 1) dyserythropoiesis and 8 controls. (E) Inhibition of caspases rescues GATA-1 expression. Q-VD-OPH (100μM) or MG132 (20μM) is added on MDS and control cultures. Immunoblot for GATA-1 in whole-cell lysates. Actin or Hsc70 are used as loading controls. Student t test for P value; *P < .05; **P < .01; ns indicates not significant.
) and control (n = 4; ○) erythroblasts. Results as mean fluorescence intensity. (D) GATA-1 protein expression. Nuclear fraction proteins blotted with the indicated Abs (top panel). Quantification of GATA-1 by densitometry expressed as a ratio to Hsc70, normalized to the ratio of GATA-1 to Hsc70 in UT-7 control cell line for each blot (bottom panel) in MDS with (n = 11) or without (n = 1) dyserythropoiesis and 8 controls. (E) Inhibition of caspases rescues GATA-1 expression. Q-VD-OPH (100μM) or MG132 (20μM) is added on MDS and control cultures. Immunoblot for GATA-1 in whole-cell lysates. Actin or Hsc70 are used as loading controls. Student t test for P value; *P < .05; **P < .01; ns indicates not significant.
A caspase-mediated cleavage of GATA-1 in MDS erythroblasts. (A) GATA-1 gene expression. RT-qPCR for GATA1 transcripts in MDS versus control erythroblasts. Normalized relative quantity (nRQ) to HPRT. Median values indicated as horizontal bars. (B) Apoptosis. Flow cytometry for annexin V+/7-AAD− cells in MDS (n = 30; ■) and control (n = 22; ○) erythroblasts. Mean percentages ± SE. (C) Caspase-3 activity. Flow cytometry for caspase-3 like activity in MDS (n = 8; ■), pan-caspase inhibitor Q-VD-OPH (100μM)–treated MDS (n = 1;  ) and control (n = 4; ○) erythroblasts. Results as mean fluorescence intensity. (D) GATA-1 protein expression. Nuclear fraction proteins blotted with the indicated Abs (top panel). Quantification of GATA-1 by densitometry expressed as a ratio to Hsc70, normalized to the ratio of GATA-1 to Hsc70 in UT-7 control cell line for each blot (bottom panel) in MDS with (n = 11) or without (n = 1) dyserythropoiesis and 8 controls. (E) Inhibition of caspases rescues GATA-1 expression. Q-VD-OPH (100μM) or MG132 (20μM) is added on MDS and control cultures. Immunoblot for GATA-1 in whole-cell lysates. Actin or Hsc70 are used as loading controls. Student t test for P value; *P < .05; **P < .01; ns indicates not significant.
) and control (n = 4; ○) erythroblasts. Results as mean fluorescence intensity. (D) GATA-1 protein expression. Nuclear fraction proteins blotted with the indicated Abs (top panel). Quantification of GATA-1 by densitometry expressed as a ratio to Hsc70, normalized to the ratio of GATA-1 to Hsc70 in UT-7 control cell line for each blot (bottom panel) in MDS with (n = 11) or without (n = 1) dyserythropoiesis and 8 controls. (E) Inhibition of caspases rescues GATA-1 expression. Q-VD-OPH (100μM) or MG132 (20μM) is added on MDS and control cultures. Immunoblot for GATA-1 in whole-cell lysates. Actin or Hsc70 are used as loading controls. Student t test for P value; *P < .05; **P < .01; ns indicates not significant.
Expression of uncleavable mutant of GATA-1 rescues MDS erythroid differentiation
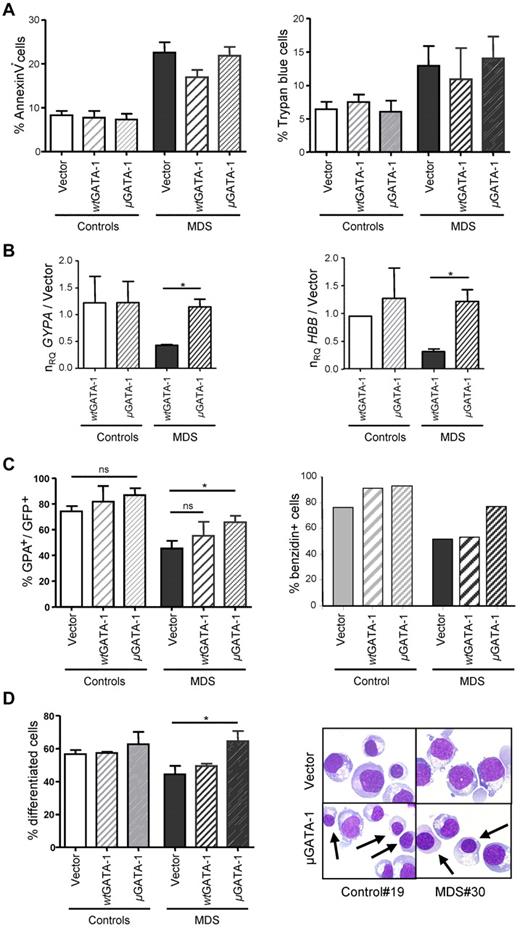
To investigate the implication of inappropriate cleavage of GATA-1 in MDS dyserythropoiesis, we express a mutated, noncleavable μGATA-1, in MDS (n = 5) or control (n = 2) CD34+ progenitors through retroviral transduction. μGATA-1 is able to rescue, more efficiently than wtGATA-1, the level of GATA-1 protein in MDS erythroblasts cultured with Epo for 14 days (supplemental Figure 2). Neither μGATA-1 nor wtGATA-1 affects the level of MDS erythroblast apoptosis or cell viability (Figure 3A). In addition, the expression of BCL2L1 gene is normal in MDS erythroblasts (supplemental Figure 1), suggesting that erythroid genes required for cell survival can escape the control of GATA-1. In contrast, transduction of μGATA-1 significantly increases the expression of GYPA and HBB transcripts in MDS erythroblasts (Figure 3B; P < .05). Accordingly, μGATA-1, but not wtGATA-1 increases the percentage of GPA+ or benzidin+ cells (Figure 3C) and that of differentiated cells represented by polychromatophilic and acidophilic erythroblasts (Figure 3D). Taken together, our findings suggest that caspase-mediated cleavage of GATA-1 that induces a functional inactivation of the protein contributes to MDS dyserythropoiesis by impairing erythroblast differentiation, but not survival.
Retroviral μGATA-1 expression rescues MDS erythroid differentiation. MDS (n = 5) and controls (n = 2) progenitors are transduced with empty, wtGATA-1, or μGATA-1 vectors and analyzed at day 14. (A) Apoptosis and cell mortality. Flow cytometry for apoptosis with annexin V (left panel) and mortality as Trypan blue+ cells (right panel). (B) Quantification of GYPA and HBB transcripts. RT-qPCR in MDS (n = 3) and controls (n = 2) erythroblasts normalized to HPRT. Ratios of mean normalized relative quantity ± SE between wtGATA-1 and μGATA-1–transduced cells to empty vector-transduced cells. (C) GPA expression and hemoglobin synthesis. GPA expression (left panel) expressed as mean percentages ± SE of GFP+ cells. Benzidin labeling (right panel) for hemoglobin synthesis measurement as percentages of positive cells. (D) Erythroblast differentiation. Percentage of differentiated polychromatophilic and acidophilic erythroblasts on MGG-stained cytospins (right panel; *P < .05). Representative microphotographs are shown.
Retroviral μGATA-1 expression rescues MDS erythroid differentiation. MDS (n = 5) and controls (n = 2) progenitors are transduced with empty, wtGATA-1, or μGATA-1 vectors and analyzed at day 14. (A) Apoptosis and cell mortality. Flow cytometry for apoptosis with annexin V (left panel) and mortality as Trypan blue+ cells (right panel). (B) Quantification of GYPA and HBB transcripts. RT-qPCR in MDS (n = 3) and controls (n = 2) erythroblasts normalized to HPRT. Ratios of mean normalized relative quantity ± SE between wtGATA-1 and μGATA-1–transduced cells to empty vector-transduced cells. (C) GPA expression and hemoglobin synthesis. GPA expression (left panel) expressed as mean percentages ± SE of GFP+ cells. Benzidin labeling (right panel) for hemoglobin synthesis measurement as percentages of positive cells. (D) Erythroblast differentiation. Percentage of differentiated polychromatophilic and acidophilic erythroblasts on MGG-stained cytospins (right panel; *P < .05). Representative microphotographs are shown.
Defective nuclear localization of Hsp70 in MDS erythroblasts
Hsp70 has been shown to protect GATA-1 from caspase-mediated cleavage in normal differentiating erythroblasts.14 In the present study, gene expression analysis indicates that several genes encoding large Hsp species of the 70-kDa family (HSP701B, HSP70B′) are up-regulated in MDS erythroblasts (supplemental Table 2b). An ∼ 3-fold up-regulation of HSP701B, which encodes the major Hsp70 protein, is confirmed by RT-qPCRs in day 14 MDS erythroblasts (Figure 4A). Likewise, immunoblot analysis demonstrates that Hsp70 expression was significantly higher along MDS erythroid differentiation compared with normal erythropoiesis (Figure 4B; P < .05). However, increased expression of Hsp70 does not prevent differentiation impairment, apoptosis, and caspase-dependent cleavage of GATA-1.
Defective nuclear localization of Hsp70 in MDS erythroblasts. (A) Quantification of HSP701B transcript. RT-qPCR in 9 MDS ( ) and 7 controls (□) erythroblasts, expressed as mean normalized relative quantity (nRQ) ± SE to GAPDH. (B) Immunoblot for Hsp70 in erythroblasts. (Left panel) Whole-cell lysates. Hsc70 as loading control. Representative of 2 separated experiments. (Right panel) Total Hsp70 expression quantified by densitometry as a ratio to Hsc70 normalized to the ratio of Hsp70 to Hsc70 in UT-7 control cell line for each blot. Student t test for P values; *P < .05. (C) Subcellular localization of Hsp70 by immunofluorescence. MDS with (n = 33) or without dyserythropoiesis (n = 11) or control erythroblasts (n = 19) labeled with Ab to Hsp70. DAPI for nuclei. MGG-stained cytospins are shown (left panel). Nucleo/cytosolic (N/C) ratio of Hsp70 expression at day 14 using the ImageJ software (right panel; **P < .01). (D) Immunoblot analysis of Hsp70. Immunoblot for Hsp70 and Hsc 70 in nuclear and cytosolic extracts (left panel) in MDS with (n = 19) or without (n = 3) dyserythropoiesis and controls (n = 9). Quantification by densitometry of N/C ratio of Hsp70 normalized to N/C ratio of Hsc70 (means ± SE; right panel; *P < .05).
) and 7 controls (□) erythroblasts, expressed as mean normalized relative quantity (nRQ) ± SE to GAPDH. (B) Immunoblot for Hsp70 in erythroblasts. (Left panel) Whole-cell lysates. Hsc70 as loading control. Representative of 2 separated experiments. (Right panel) Total Hsp70 expression quantified by densitometry as a ratio to Hsc70 normalized to the ratio of Hsp70 to Hsc70 in UT-7 control cell line for each blot. Student t test for P values; *P < .05. (C) Subcellular localization of Hsp70 by immunofluorescence. MDS with (n = 33) or without dyserythropoiesis (n = 11) or control erythroblasts (n = 19) labeled with Ab to Hsp70. DAPI for nuclei. MGG-stained cytospins are shown (left panel). Nucleo/cytosolic (N/C) ratio of Hsp70 expression at day 14 using the ImageJ software (right panel; **P < .01). (D) Immunoblot analysis of Hsp70. Immunoblot for Hsp70 and Hsc 70 in nuclear and cytosolic extracts (left panel) in MDS with (n = 19) or without (n = 3) dyserythropoiesis and controls (n = 9). Quantification by densitometry of N/C ratio of Hsp70 normalized to N/C ratio of Hsc70 (means ± SE; right panel; *P < .05).
Defective nuclear localization of Hsp70 in MDS erythroblasts. (A) Quantification of HSP701B transcript. RT-qPCR in 9 MDS ( ) and 7 controls (□) erythroblasts, expressed as mean normalized relative quantity (nRQ) ± SE to GAPDH. (B) Immunoblot for Hsp70 in erythroblasts. (Left panel) Whole-cell lysates. Hsc70 as loading control. Representative of 2 separated experiments. (Right panel) Total Hsp70 expression quantified by densitometry as a ratio to Hsc70 normalized to the ratio of Hsp70 to Hsc70 in UT-7 control cell line for each blot. Student t test for P values; *P < .05. (C) Subcellular localization of Hsp70 by immunofluorescence. MDS with (n = 33) or without dyserythropoiesis (n = 11) or control erythroblasts (n = 19) labeled with Ab to Hsp70. DAPI for nuclei. MGG-stained cytospins are shown (left panel). Nucleo/cytosolic (N/C) ratio of Hsp70 expression at day 14 using the ImageJ software (right panel; **P < .01). (D) Immunoblot analysis of Hsp70. Immunoblot for Hsp70 and Hsc 70 in nuclear and cytosolic extracts (left panel) in MDS with (n = 19) or without (n = 3) dyserythropoiesis and controls (n = 9). Quantification by densitometry of N/C ratio of Hsp70 normalized to N/C ratio of Hsc70 (means ± SE; right panel; *P < .05).
) and 7 controls (□) erythroblasts, expressed as mean normalized relative quantity (nRQ) ± SE to GAPDH. (B) Immunoblot for Hsp70 in erythroblasts. (Left panel) Whole-cell lysates. Hsc70 as loading control. Representative of 2 separated experiments. (Right panel) Total Hsp70 expression quantified by densitometry as a ratio to Hsc70 normalized to the ratio of Hsp70 to Hsc70 in UT-7 control cell line for each blot. Student t test for P values; *P < .05. (C) Subcellular localization of Hsp70 by immunofluorescence. MDS with (n = 33) or without dyserythropoiesis (n = 11) or control erythroblasts (n = 19) labeled with Ab to Hsp70. DAPI for nuclei. MGG-stained cytospins are shown (left panel). Nucleo/cytosolic (N/C) ratio of Hsp70 expression at day 14 using the ImageJ software (right panel; **P < .01). (D) Immunoblot analysis of Hsp70. Immunoblot for Hsp70 and Hsc 70 in nuclear and cytosolic extracts (left panel) in MDS with (n = 19) or without (n = 3) dyserythropoiesis and controls (n = 9). Quantification by densitometry of N/C ratio of Hsp70 normalized to N/C ratio of Hsc70 (means ± SE; right panel; *P < .05).
To investigate this apparent paradox, we have analyzed the subcellular localization of Hsp70 in cultured erythroblasts from 41 MDS with dyserythropoiesis, 10 MDS without dyserythropoiesis, and 19 controls (supplemental Table 1). As expected, Hsp70 is detected in the nucleus of normal cells from basophilic (day 10) to polychromatophilic and acidophilic (day 14) erythroblasts (Figure 4C left panel). In sharp contrast, Hsp70 is absent from the nucleus of polychromatophilic and acidophilic erythroblasts obtained from MDS samples (Figure 4C left panel). Likewise, the ratio of nuclear to cytosolic Hsp70 expression is significantly decreased in MDS compared with control erythroblasts (P < .01; Figure 4C right panel). Immunoblot analysis of Hsp70 protein expression in the nuclear and cytosolic fractions isolated from cultured erythroblast lysates of 19 MDS with dyserythropoiesis (3 MDS without dyserythropoiesis and 9 controls) confirms that while Hsp70 accumulates in the nucleus of normal erythroblasts undergoing differentiation, the protein expression decreases in the nucleus of dysplastic MDS cells (P < .05; Figure 4D). Our results show that a large majority of MDS samples with dyserythropoiesis (30 of 41) exhibits a defective nuclear localization of Hsp70 as evidenced by immunofluorescence in 25 of 33 tested cases including MDS with del(5q) and MDS with ring sideroblasts or by immunoblot on nuclear fractions in 13 of 19 tested cases (Table 1). Independent of the WHO subtype, defective nuclear Hsp70 significantly correlates with dyserythropoiesis (Table 2; χ2 test, P < .0001 and supplemental Table 1).
Clinical and biological characteristics of 42 low-grade MDS with dyserythropoiesis
| Patient no. . | Age, y . | Sex . | WHO diagnosis . | IPSS . | Leukocytes, G/L . | Neutrophils, G/L . | Hb, g/dL . | MCV, fL . | Platelets, G/L . | Karyotype . | % blasts . | % erythroblasts . | % ring sideroblasts . | Dyserythropoiesis . | Hsp70 nuclear localization . | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IF . | WB . | |||||||||||||||
| MDS6 | 65 | M | RA | 0 | 5.6 | 2.8 | 8.5 | 102 | 210 | 45,X,-Y[17]/46,XY[3] | 4 | 31 | 3 | + | Yes | Yes |
| MDS8 | 80 | F | RA | 1 | 7.4 | 6.1 | 10.1 | 100 | 167 | 45,XX,add(1)(p36),del(5)(q13q33)[18] | 4 | 28 | 14 | + | No | |
| MDS15 | 64 | M | RA | 0.5 | 2.8 | 1.6 | 9.2 | 110 | 350 | 46, XY[20] | 3 | 27 | 5 | + | No | |
| MDS17 | 71 | F | RA | 0.5 | 1.4 | 0.7 | 6.5 | 100 | 55 | 46,XX[20] | 2 | 21 | 2 | + | NA | No |
| MDS18 | 68 | M | RA | 0 | 7.5 | 5.7 | 10.6 | 107 | 296 | 46,XY[20] | 3 | 25 | NA | ++ | Yes | |
| MDS21 | 57 | M | RA | 0.5 | 1.6 | 0.6 | 10.4 | 76 | 56 | 46,XY[20] | 3 | 8 | NA | +/− | No | |
| MDS26 | 84 | M | RA | 0 | 8.4 | 7.8 | 7 | 113 | 309 | 45,X,-Y | 4 | 33 | 12 | ++ | NA | Yes |
| MDS28 | 79 | M | RA | 0.5 | 2.2 | 1.0 | 9.8 | 107 | 137 | 46,XY[20] | 3 | 39 | 0 | + | NA | No |
| MDS41 | 67 | M | RA | 4.2 | 2.3 | 9.1 | 102 | 112 | NA | 3 | 35 | 0 | + | No | ||
| MDS43 | 61 | F | RA | 0 | 10.6 | 6.1 | 10.4 | 88 | 327 | 46,XX[20] | 2 | 10 | NA | ++ | No | |
| MDS48 | 84 | M | RA | 0 | 6.1 | 2.0 | 10.8 | 90 | 122 | 46,XY[20] | 3 | 18 | NA | + | No | |
| MDS38 | 80 | F | MDS-U | 0 | 6.4 | 4.2 | 8.7 | 92 | 485 | 46,XX[20] | 2 | 38 | 0 | + | No | |
| MDS45 | 79 | F | MDS-U | NA | 4.6 | 3.0 | 11.2 | 91 | 229 | NA | 2 | 25 | 0 | + | No | No |
| MDS10 | 72 | M | RARS | 0 | 6.9 | 4.7 | 11.4 | 103 | 223 | 45,X,−Y[17]/46,XY[3] | 1 | 35 | 23 | ++ | No | No |
| MDS11 | 91 | F | RARS | 0.5 | 11.4 | 8.3 | 10.4 | 115 | 212 | 47,XX,+8[18]/46,XX[2] | 2 | 44 | 39 | ++ | No | No |
| MDS16 | 71 | M | RARS | 0 | 8.5 | 4.2 | 7.5 | 118 | 186 | 46,XY[20] | 3 | 31 | 45 | ++ | No | |
| MDS24 | 76 | M | RARS | 0 | 8.6 | 4.8 | 8.6 | 104 | 401 | 46,XY[20] | 3 | 41 | 50 | ++ | Yes | |
| MDS36 | 64 | F | RARS | 0 | 3.4 | 2.8 | 7.5 | 110 | 212 | 46,XX[20] | 4 | 51 | 45 | ++ | No | |
| MDS49 | 84 | F | RARS | 0 | 4.2 | 2.5 | 9.1 | 108 | 122 | 46,XX[20] | 3 | 48 | 56 | ++ | Yes | |
| MDS9 | 85 | M | RCMD | 0.5 | 3.8 | 1.62 | 11 | 103 | 78 | 46,XY,del(20)(q12)[18] | 3 | 48 | 4 | + | No | No |
| MDS12 | 70 | F | RCMD | 0 | 4.7 | 2.9 | 11.3 | 98 | 28 | 46,XX[20] | 2 | 49 | 0 | + | No | |
| MDS20 | 79 | M | RCMD | 0.5 | 2.6 | 1.6 | 8.5 | 101 | 175 | 46,XY[20] | 1 | 28 | 6 | + | NA | No |
| MDS23 | 77 | F | RCMD | 0.5 | 2.7 | 1.2 | 9.1 | 84 | 56 | 46,XX[20] | 4 | 42 | NA | ++ | No | |
| MDS33 | 89 | F | RCMD-RS | NA | 7.8 | 5.3 | 9.4 | 79 | 41 | NA | 5 | 48 | 48 | ++ | No | No |
| MDS50 | 90 | M | RCMD | 0.5 | 3.5 | 1.9 | 9.8 | 99 | 85 | 46,XY[20] | 3 | 22 | 0 | + | Yes | |
| MDS1 | 72 | M | RAEB1 | 1 | 3.2 | 1.4 | 8.6 | 105 | 73 | 46,XY[20] | 8 | 15 | NA | + | Yes | Yes |
| MDS3 | 75 | F | RAEB1 | 0.5 | 6.0 | 4.0 | 10.3 | 101 | 49 | 46,XX[20] | 6 | 20 | NA | +/− | Yes | Yes |
| MDS4 | 81 | M | RAEB1 | NA | 8.2 | 4.9 | 9.2 | 96 | 150 | NA | 7 | 33 | 0 | + | No | No |
| MDS5 | 59 | F | RAEB1 | 0.5 | 3.5 | 1.6 | 9.7 | 95 | 168 | 46,XX,del(5)(q14q34)[15]/46,XX[3] | 7 | 10 | NA | + | Yes | |
| MDS7 | 57 | F | RAEB1 | 1 | 1.2 | 0.9 | 10.5 | 94 | 14 | 46,XX[21] | 6 | 24 | 0 | +/− | NA | Yes |
| MDS13 | 80 | F | RAEB1 | 1 | 2.4 | 1.1 | 9.8 | 125 | 296 | 46,XX,del(5)(q13;q34),del(11)¿Üq14)[12]/46,XX[8 | 6 | 30 | 0 | + | No | |
| MDS14 | 61 | F | RAEB1 | 1.5 | 4.6 | 2.7 | 8 | 107 | 301 | 46,XX,del(5)(q13q34)[5]/47,idem, + 21[8]/46,XX,del(11)(q13;q24)[3] | 5 | 43 | 0 | +/− | No | No |
| MDS19 | 79 | M | RAEB1 | NA | 3.6 | 1.8 | 10.8 | 100 | 152 | NA | 6 | 28 | 0 | +/− | No | |
| MDS22 | 84 | F | RAEB1 | 1 | 1.8 | 0.4 | 8.1 | 107 | 242 | 46,XX[20] | 7 | 48 | 55 | ++ | NA | No |
| MDS25 | 36 | F | RAEB1 | 0.5 | 3.4 | 1.5 | 11.3 | 108 | 227 | 46,XX,del(5)(q13;q33)[20] | 7 | 14 | NA | + | No | |
| MDS27 | 78 | F | RAEB1 | 1 | 2.8 | 0.8 | 10.3 | 104 | 181 | 46,XX[20] | 9 | 49 | 30 | ++ | No | No |
| MDS29 | 86 | M | RAEB1 | 2.4 | 0.9 | 10.4 | 98 | 111 | 45,X,−Y[20] | 5 | 4 | NA | + | NA | Yes | |
| MDS30 | 87 | M | RAEB1 | NA | 5.6 | 3.2 | 9.2 | 99 | 187 | NA | 8 | 23 | 0 | + | NA | No |
| MDS37 | 88 | F | RAEB1 | 0.5 | 7.6 | 5.1 | 10.4 | 102 | 62 | 46,XX[20] | 6 | 23 | 70 | ++ | No | |
| MDS39 | 83 | M | RAEB1 | 1 | 1.1 | 0.47 | 9.3 | 104 | 16 | 46,XY,del(5q)(q13q33)[2]/45,X,−Y[4]/46,XY[14] | 5 | 29 | 3 | + | No | |
| MDS46 | 69 | M | RAEB1 | 0.5 | 6 | 3.3 | 9.7 | 98 | 290 | 46,XY[20] | 5 | 23 | 33 | + | No | |
| Patient no. . | Age, y . | Sex . | WHO diagnosis . | IPSS . | Leukocytes, G/L . | Neutrophils, G/L . | Hb, g/dL . | MCV, fL . | Platelets, G/L . | Karyotype . | % blasts . | % erythroblasts . | % ring sideroblasts . | Dyserythropoiesis . | Hsp70 nuclear localization . | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IF . | WB . | |||||||||||||||
| MDS6 | 65 | M | RA | 0 | 5.6 | 2.8 | 8.5 | 102 | 210 | 45,X,-Y[17]/46,XY[3] | 4 | 31 | 3 | + | Yes | Yes |
| MDS8 | 80 | F | RA | 1 | 7.4 | 6.1 | 10.1 | 100 | 167 | 45,XX,add(1)(p36),del(5)(q13q33)[18] | 4 | 28 | 14 | + | No | |
| MDS15 | 64 | M | RA | 0.5 | 2.8 | 1.6 | 9.2 | 110 | 350 | 46, XY[20] | 3 | 27 | 5 | + | No | |
| MDS17 | 71 | F | RA | 0.5 | 1.4 | 0.7 | 6.5 | 100 | 55 | 46,XX[20] | 2 | 21 | 2 | + | NA | No |
| MDS18 | 68 | M | RA | 0 | 7.5 | 5.7 | 10.6 | 107 | 296 | 46,XY[20] | 3 | 25 | NA | ++ | Yes | |
| MDS21 | 57 | M | RA | 0.5 | 1.6 | 0.6 | 10.4 | 76 | 56 | 46,XY[20] | 3 | 8 | NA | +/− | No | |
| MDS26 | 84 | M | RA | 0 | 8.4 | 7.8 | 7 | 113 | 309 | 45,X,-Y | 4 | 33 | 12 | ++ | NA | Yes |
| MDS28 | 79 | M | RA | 0.5 | 2.2 | 1.0 | 9.8 | 107 | 137 | 46,XY[20] | 3 | 39 | 0 | + | NA | No |
| MDS41 | 67 | M | RA | 4.2 | 2.3 | 9.1 | 102 | 112 | NA | 3 | 35 | 0 | + | No | ||
| MDS43 | 61 | F | RA | 0 | 10.6 | 6.1 | 10.4 | 88 | 327 | 46,XX[20] | 2 | 10 | NA | ++ | No | |
| MDS48 | 84 | M | RA | 0 | 6.1 | 2.0 | 10.8 | 90 | 122 | 46,XY[20] | 3 | 18 | NA | + | No | |
| MDS38 | 80 | F | MDS-U | 0 | 6.4 | 4.2 | 8.7 | 92 | 485 | 46,XX[20] | 2 | 38 | 0 | + | No | |
| MDS45 | 79 | F | MDS-U | NA | 4.6 | 3.0 | 11.2 | 91 | 229 | NA | 2 | 25 | 0 | + | No | No |
| MDS10 | 72 | M | RARS | 0 | 6.9 | 4.7 | 11.4 | 103 | 223 | 45,X,−Y[17]/46,XY[3] | 1 | 35 | 23 | ++ | No | No |
| MDS11 | 91 | F | RARS | 0.5 | 11.4 | 8.3 | 10.4 | 115 | 212 | 47,XX,+8[18]/46,XX[2] | 2 | 44 | 39 | ++ | No | No |
| MDS16 | 71 | M | RARS | 0 | 8.5 | 4.2 | 7.5 | 118 | 186 | 46,XY[20] | 3 | 31 | 45 | ++ | No | |
| MDS24 | 76 | M | RARS | 0 | 8.6 | 4.8 | 8.6 | 104 | 401 | 46,XY[20] | 3 | 41 | 50 | ++ | Yes | |
| MDS36 | 64 | F | RARS | 0 | 3.4 | 2.8 | 7.5 | 110 | 212 | 46,XX[20] | 4 | 51 | 45 | ++ | No | |
| MDS49 | 84 | F | RARS | 0 | 4.2 | 2.5 | 9.1 | 108 | 122 | 46,XX[20] | 3 | 48 | 56 | ++ | Yes | |
| MDS9 | 85 | M | RCMD | 0.5 | 3.8 | 1.62 | 11 | 103 | 78 | 46,XY,del(20)(q12)[18] | 3 | 48 | 4 | + | No | No |
| MDS12 | 70 | F | RCMD | 0 | 4.7 | 2.9 | 11.3 | 98 | 28 | 46,XX[20] | 2 | 49 | 0 | + | No | |
| MDS20 | 79 | M | RCMD | 0.5 | 2.6 | 1.6 | 8.5 | 101 | 175 | 46,XY[20] | 1 | 28 | 6 | + | NA | No |
| MDS23 | 77 | F | RCMD | 0.5 | 2.7 | 1.2 | 9.1 | 84 | 56 | 46,XX[20] | 4 | 42 | NA | ++ | No | |
| MDS33 | 89 | F | RCMD-RS | NA | 7.8 | 5.3 | 9.4 | 79 | 41 | NA | 5 | 48 | 48 | ++ | No | No |
| MDS50 | 90 | M | RCMD | 0.5 | 3.5 | 1.9 | 9.8 | 99 | 85 | 46,XY[20] | 3 | 22 | 0 | + | Yes | |
| MDS1 | 72 | M | RAEB1 | 1 | 3.2 | 1.4 | 8.6 | 105 | 73 | 46,XY[20] | 8 | 15 | NA | + | Yes | Yes |
| MDS3 | 75 | F | RAEB1 | 0.5 | 6.0 | 4.0 | 10.3 | 101 | 49 | 46,XX[20] | 6 | 20 | NA | +/− | Yes | Yes |
| MDS4 | 81 | M | RAEB1 | NA | 8.2 | 4.9 | 9.2 | 96 | 150 | NA | 7 | 33 | 0 | + | No | No |
| MDS5 | 59 | F | RAEB1 | 0.5 | 3.5 | 1.6 | 9.7 | 95 | 168 | 46,XX,del(5)(q14q34)[15]/46,XX[3] | 7 | 10 | NA | + | Yes | |
| MDS7 | 57 | F | RAEB1 | 1 | 1.2 | 0.9 | 10.5 | 94 | 14 | 46,XX[21] | 6 | 24 | 0 | +/− | NA | Yes |
| MDS13 | 80 | F | RAEB1 | 1 | 2.4 | 1.1 | 9.8 | 125 | 296 | 46,XX,del(5)(q13;q34),del(11)¿Üq14)[12]/46,XX[8 | 6 | 30 | 0 | + | No | |
| MDS14 | 61 | F | RAEB1 | 1.5 | 4.6 | 2.7 | 8 | 107 | 301 | 46,XX,del(5)(q13q34)[5]/47,idem, + 21[8]/46,XX,del(11)(q13;q24)[3] | 5 | 43 | 0 | +/− | No | No |
| MDS19 | 79 | M | RAEB1 | NA | 3.6 | 1.8 | 10.8 | 100 | 152 | NA | 6 | 28 | 0 | +/− | No | |
| MDS22 | 84 | F | RAEB1 | 1 | 1.8 | 0.4 | 8.1 | 107 | 242 | 46,XX[20] | 7 | 48 | 55 | ++ | NA | No |
| MDS25 | 36 | F | RAEB1 | 0.5 | 3.4 | 1.5 | 11.3 | 108 | 227 | 46,XX,del(5)(q13;q33)[20] | 7 | 14 | NA | + | No | |
| MDS27 | 78 | F | RAEB1 | 1 | 2.8 | 0.8 | 10.3 | 104 | 181 | 46,XX[20] | 9 | 49 | 30 | ++ | No | No |
| MDS29 | 86 | M | RAEB1 | 2.4 | 0.9 | 10.4 | 98 | 111 | 45,X,−Y[20] | 5 | 4 | NA | + | NA | Yes | |
| MDS30 | 87 | M | RAEB1 | NA | 5.6 | 3.2 | 9.2 | 99 | 187 | NA | 8 | 23 | 0 | + | NA | No |
| MDS37 | 88 | F | RAEB1 | 0.5 | 7.6 | 5.1 | 10.4 | 102 | 62 | 46,XX[20] | 6 | 23 | 70 | ++ | No | |
| MDS39 | 83 | M | RAEB1 | 1 | 1.1 | 0.47 | 9.3 | 104 | 16 | 46,XY,del(5q)(q13q33)[2]/45,X,−Y[4]/46,XY[14] | 5 | 29 | 3 | + | No | |
| MDS46 | 69 | M | RAEB1 | 0.5 | 6 | 3.3 | 9.7 | 98 | 290 | 46,XY[20] | 5 | 23 | 33 | + | No | |
Immunofluorescence and immunoblot analysis of Hsp70 localization. Dyserythropoiesis: +/− indicates mild; +, moderate; and ++, strong.
MDS indicates myelodysplastic syndrome; MCV, mean corpuscular volume; IF, immunofluoresence; WB, Western blot; M, male; F, female; RCMD, refractory cytopenia with multilineage dysplasia; RAEB, RA with excess of blasts; CTL, control; cyta, cytapheresis; RA, refractory anemia; RARS, RA with sideroblasts; IPSS, International Prognosis Scoring System; and NA, not available.
Defective nuclear localization of Hsp70 correlates with MDS dyserythropoiesis
| . | MDS with dyserythropoiesis . | MDS without dyserythropoiesis . | Controls . | χ2 test . |
|---|---|---|---|---|
| Nuclear Hsp70 | 11 | 9 | 18 | P < .0001 |
| Cytosolic Hsp70 | 30 | 1 | 2 |
| . | MDS with dyserythropoiesis . | MDS without dyserythropoiesis . | Controls . | χ2 test . |
|---|---|---|---|---|
| Nuclear Hsp70 | 11 | 9 | 18 | P < .0001 |
| Cytosolic Hsp70 | 30 | 1 | 2 |
MDS erythroblast samples with (n = 41) or without (n = 10) dyserythropoiesis and 20 were tested for Hsp70 subcellular localization by immunofluorescence or immunoblot. χ2 test for statistical analysis.
MDS indicates myelodysplastic syndrome.
Hsp70 does not colocalize with GATA-1 and caspase-3 in the nucleus of MDS erythroblasts
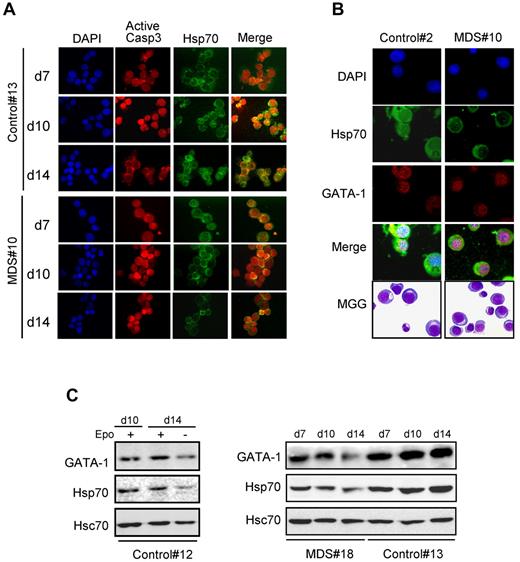
We have then investigated the subcellular localization of Hsp70 and active caspase-3 by coimmunofluorescence experiments. In the culture conditions used, caspase-3 activation is detected at day 10 of the differentiation of normal CD34+ cells, and Hsp70 colocalizes with active caspase-3 in erythroblast nuclei, as previously described.14 In MDS erythroblasts, activated caspase-3 is detected in some cells at day 10 of culture and in most cells at day 14. Hsp70 does not colocalize with the active caspase (Figure 5A) or with GATA-1 (Figure 5B) in the nucleus of day 14 MDS erythroblasts in which the stress protein fails to accumulate. By immunoblot analysis, we confirm that both GATA-1 and Hsp70 expression decreases in the nuclear fraction of MDS erythroblasts mimicking the fate of normal erythroblasts on Epo starvation (Figure 5C). Thus, MDS erythroblasts demonstrate, despite high levels of Hsp70, a decrease in nuclear Hsp70 which may account for an increased cleavage of GATA-1.
Hsp70 does not colocalize with active caspase-3 or GATA-1 in the nucleus of MDS erythroblasts. Erythroblasts from MDS (n = 7) and controls (n = 4) harvested at day 14. (A) Immunolocalization of Hsp70 and active caspase-3. DAPI for nuclei. Merge represents both active caspase-3 (red) and Hsp70 (green). (B) Immunolocalization of GATA-1 and Hsp70 in polychromatophilic erythroblasts. GATA-1 (red), Hsp70 (green), and DAPI (blue). Merge represents the superimposition of all 3 colors. (C) Expression of GATA-1 and Hsp70 in MDS erythroblast nuclei. MDS or control cells are harvested at day 7, 10, and 14. Immunoblot for GATA-1 or Hsp70 in the nuclear fraction. Hsc70 as loading control. Representative experiments are shown.
Hsp70 does not colocalize with active caspase-3 or GATA-1 in the nucleus of MDS erythroblasts. Erythroblasts from MDS (n = 7) and controls (n = 4) harvested at day 14. (A) Immunolocalization of Hsp70 and active caspase-3. DAPI for nuclei. Merge represents both active caspase-3 (red) and Hsp70 (green). (B) Immunolocalization of GATA-1 and Hsp70 in polychromatophilic erythroblasts. GATA-1 (red), Hsp70 (green), and DAPI (blue). Merge represents the superimposition of all 3 colors. (C) Expression of GATA-1 and Hsp70 in MDS erythroblast nuclei. MDS or control cells are harvested at day 7, 10, and 14. Immunoblot for GATA-1 or Hsp70 in the nuclear fraction. Hsc70 as loading control. Representative experiments are shown.
Inhibition of apoptosis by Bcl-2 does not influence Hsp70 subcellular localization
Dyserythropoietic MDS cells exhibit high level of apoptosis together with low level of Hsp70 in the nucleus. To explore the link between the excess of apoptosis and the defect of nuclear Hsp70, we have used lentiviral transduction to ectopically express wtBcl-2 in 5 MDS with dyserythropoiesis, 4 MDS without dyserythropoiesis, and 2 control samples. We have previously shown that wtBcl-2 inhibits Fas-dependent activation of caspases in MDS cells.19 Here, we show that wtBcl-2 expression inhibits mitochondrial membrane depolarization, as indicated by a 30% to 50% reduction of HIDClow cells, and subsequent apoptosis, as shown by a 50% to 70% reduction of phosphatidylserine exposure in GFP+ MDS erythroblasts, but not in normal erythroblasts (Figure 6A left panel). Overexpression of wtBcl-2 also decreases GPA expression, both in GFP+-MDS and control erythroblasts, confirming the slowing effect of Bcl2 on erythroid cell maturation (Figure 6A right panel). Immunofluorescence experiments show that Hsp70, which is absent from the nucleus in dyserythropoietic MDS samples, remains in the cytoplasm of wtBcl-2–transduced erythroblasts. wtBcl-2 has no impact on Hsp70 nuclear localization in control cells, as confirmed by the quantification of signals (Figure 6B). Whatever, the stage of erythroid maturation, Bcl2 does not influence Hsp70 localization (supplemental Figure 3). Altogether, the defect in Hsp70 migration from the cytoplasm to the nucleus of MDS erythroblasts is independent of the apoptotic process.
Inhibition of apoptosis does not influence Hsp70 subcellular localization.wtBcl2 was expressed by lentiviral transduction in 9 MDS and 2 control samples. (A) wtBcl2 inhibits apoptosis and delays erythroid cell differentiation. Flow cytometry for mitochondrial membrane depolarization as % HIDClow cells and phosphatidylserine exposure as the percentage of annexin V+ cells in the GFP+ cell population (left panel) and glycophorin A expression as the percentage of positive cells (right panel). (B) Immunofluorescence for Hsp70 in 9 MDS and 2 control erythroblast samples at day 14 of the culture. Representative experiments (1 control and 3 MDS) are shown. Nucleus countercolored with DAPI (left panel). Quantification of Hsp70 nucleo/cytosolic ratio by using the ImageJ software.
Inhibition of apoptosis does not influence Hsp70 subcellular localization.wtBcl2 was expressed by lentiviral transduction in 9 MDS and 2 control samples. (A) wtBcl2 inhibits apoptosis and delays erythroid cell differentiation. Flow cytometry for mitochondrial membrane depolarization as % HIDClow cells and phosphatidylserine exposure as the percentage of annexin V+ cells in the GFP+ cell population (left panel) and glycophorin A expression as the percentage of positive cells (right panel). (B) Immunofluorescence for Hsp70 in 9 MDS and 2 control erythroblast samples at day 14 of the culture. Representative experiments (1 control and 3 MDS) are shown. Nucleus countercolored with DAPI (left panel). Quantification of Hsp70 nucleo/cytosolic ratio by using the ImageJ software.
Nucleus-targeted Hsp70 rescues MDS erythroblast differentiation
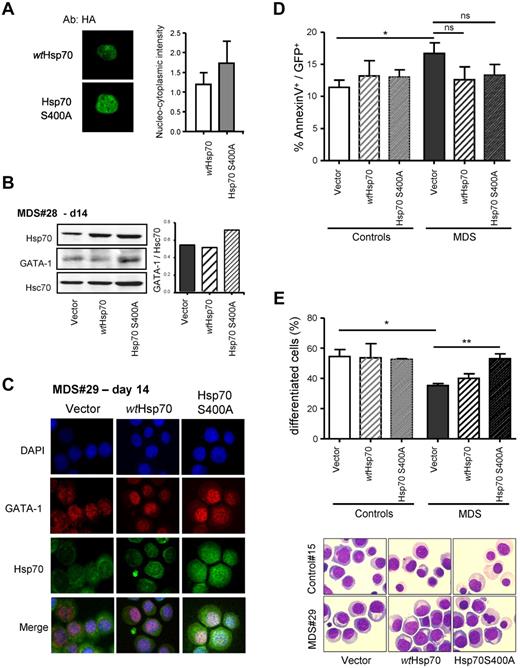
The functional nuclear export sequence (NES), L394DVTPLSL,401 identified in Hsc54/Hsc70 protein is highly conserved in the Hsp70 family proteins. Mutation of one amino acid of this sequence abrogates the export of Hsp70 which, thus, accumulates in the nucleus.20 To establish the contribution of a lack of nuclear Hsp70 to MDS impairment of differentiation, we have disrupted the NES by converting serine 400 into alanine. Plasmids encoding HA-tagged wtHsp70 or HA-tagged Hsp70 S400A are transfected into primary erythroblasts. Immunofluorescence experiments using anti-HA Abs show that wtHsp70 is detected both in the cytoplasm and the nucleus, while Hsp70 S400A accumulates in the nucleus (Figure 7A). wtHsp70 and Hsp70 S400A constructs have been then inserted into lentiviral vectors. After transduction with empty, wtHsp70, or Hsp70 S400A vectors, MDS CD34+ progenitors are allowed to grow and differentiate along the erythroid lineage. By Western blot, we observe that expression of Hsp70 is increased in MDS cells infected with either wtHsp70 or Hsp70 S400A compared with the empty vector, while GATA-1 expression increases only in MDS cells infected with the Hsp70 S400A vector (Figure 7B). Immunofluorescence microscopy analysis showed that, at day 14 of MDS cell culture, Hsp70 S400A is mainly present in the nucleus and colocalizes with GATA-1. Erythroblasts ectopically expressing Hsp70 S400A demonstrate an increased level of GATA-1 compared with erythroblasts infected with empty or wtHsp70 vectors (Figure 7C). These results suggest that GATA-1 expression can be rescued by the nuclear accumulation of Hsp70. In agreement with our hypothesis, we demonstrate that rescuing GATA-1 expression efficiently improves terminal erythroid differentiation in MDS as shown by the significant increment of the percentage of differentiated cells in Hsp70 S400A-transfected cultures (58%) compared with empty (37%) or wtHsp70 (40%) vector on MGG-colored cytospins. However, expression of Hsp70 S400A has no impact on MDS erythroblast apoptosis rate (Figure 7D). Altogether, these results show that protecting GATA-1 from cleavage by expression of a nuclear targeted Hsp70 rescues MDS erythroid differentiation, and does not modify the apoptotic phenotype.
Mutant S400A of Hsp70 rescues MDS erythroblast differentiation. (A) Hsp70 S400A preferentially localizes to the nucleus. Normal erythroblasts transfected with HA-wtHsp70 or HA-Hsp70 S400A plasmids. Immunofluorescence anti-HA analyzed by confocal microscopy. (B) Immunoblot analysis of Hsp70 and GATA-1 expression. MDS erythroblasts infected with wtHsp70 or Hsp70 S400A or empty lentiviral vector at day 14. (Top panel) Representative immunoblot. (Bottom panel) Quantification of GATA-1 normalized to Hsc70 by densitometry. (C) Immunofluorescence for Hsp70 and GATA-1 in day 14 erythroblasts. MDS (n = 9) or control (n = 4) erythroblasts were infected with wtHsp70 or Hsp70 S400A or empty lentiviral vector. Nuclei countercolored with DAPI. (D) Apoptosis. Annexin V labeling in the GFP+ cells. (E) Erythroid differentiation. Differentiated polychromatophilic and acidophilic erythroblasts numbered on MGG-stained cytospins. Representative microphotographs are shown. Student t test for P values; *P < .05; **P < .01; ns indicates not significant.
Mutant S400A of Hsp70 rescues MDS erythroblast differentiation. (A) Hsp70 S400A preferentially localizes to the nucleus. Normal erythroblasts transfected with HA-wtHsp70 or HA-Hsp70 S400A plasmids. Immunofluorescence anti-HA analyzed by confocal microscopy. (B) Immunoblot analysis of Hsp70 and GATA-1 expression. MDS erythroblasts infected with wtHsp70 or Hsp70 S400A or empty lentiviral vector at day 14. (Top panel) Representative immunoblot. (Bottom panel) Quantification of GATA-1 normalized to Hsc70 by densitometry. (C) Immunofluorescence for Hsp70 and GATA-1 in day 14 erythroblasts. MDS (n = 9) or control (n = 4) erythroblasts were infected with wtHsp70 or Hsp70 S400A or empty lentiviral vector. Nuclei countercolored with DAPI. (D) Apoptosis. Annexin V labeling in the GFP+ cells. (E) Erythroid differentiation. Differentiated polychromatophilic and acidophilic erythroblasts numbered on MGG-stained cytospins. Representative microphotographs are shown. Student t test for P values; *P < .05; **P < .01; ns indicates not significant.
Discussion
GATA-1 plays a central role in erythrocyte and megakaryocyte differentiation.9,21 Synthesis of a functionally inactive GATA-1 has been previously involved in transient myeloproliferative disorder associated with Down syndrome,22 X-linked familial macrocytic anemia, or dyserythropoietic anemia in X-linked thrombocytopenia.23,24 We show here that functional inactivation of GATA-1 is involved in the impairment of MDS erythroblast maturation through the decrease of target gene transcription. However, inactivation of GATA-1 appears not to alter the transcription of all its target genes considering that BCL2L1 which encodes the antiapoptotic protein BclxL, is normally expressed in MDS erythroblasts (supplemental Figure 1). As shown previously,25,26 the BCL2L1 gene is also under the control of the transcription factor STAT5 whose DNA-binding activity is normal in MDS erythroblasts.4 This can explain the maintenance of a normal BclxL expression. Furthermore, μGATA-1 mutant and a nuclear-targeted mutant of Hsp70 both rescue MDS erythroblasts from defective differentiation, but not from apoptosis. Conversely, inhibition of apoptosis by wtBcl2 has no impact on Hsp70 subcellular localization. Taken together, caspase-mediated cleavage of GATA-1 is not sufficient for the onset of MDS apoptotic phenotype, while it supports the defect of differentiation, thus uncoupling apoptosis and differentiation in dysplastic MDS erythroblasts.
Proteins of the Hsp70 family are ATP-dependent chaperones assisting the folding of newly synthesized polypeptides, the assembly of multiprotein complexes and the transport of proteins across cellular membranes.27 Consistently with the up-regulation of HSP701B gene and the overexpression of Hsp70 in AML and high-risk MDS samples,28,29 we found a high level of expression of HSP701B RNA and Hsp70 protein in MDS erythroblasts. However, Hsp70 does not localize into the nucleus of MDS late erythroblasts. Hsp70 shuttling is tightly regulated, and clearly implicates a conserved leucine-rich NES in the C-terminal part of the protein.19 This NES is homologous to the characterized NES in several proteins including other members of Hsp70 families of widely divergent species from Escherichia coli to mammals. Mutation of one of leucine or valine comprised between amino acids 394 and 401 abolishes the nuclear export of Hsp70.20 We show here that mutating serine 400 to alanine also allows Hsp70 targeting to the nucleus. This suggests that substitution of the serine is as effective as that of leucine or valine to abrogate the functionality of NES. Furthermore, when serine is mutated to aspartate mimicking a phosphorylated residue, Hsp70 is restricted to the cytoplasm (J.V., unpublished data, 2010). This suggests that phosphorylation of this residue could be crucial for determining the subcellular localization of the protein. Previous studies have shown that the inhibition of protein dephosphorylation by serine/threonine or tyrosine phosphatase inhibitors does prevent Hsc70 accumulation in the nucleus.30 This finding suggests that Hsp70 could be retained in the cytoplasm by a phosphorylation event and that the subcellular localization of Hsps may be controlled by a phosphorylation/dephosphorylation process. Evidence for Hsp phosporylation and its upstream regulators are still lacking. One possibility, considering the timing of caspase activation in normal erythroblasts, is that Hsp70 relocalization is driven by Epo stimulation, thus involving protein-protein interaction or posttranslational modifications or both. If Epo signaling is implicated in the control of Hsp70 localization, then the alteration identified in MDS samples with dyserythropoiesis may indicate a defect in Epo signaling. Investigating the complexity of this regulation in normal and MDS erythroblasts is an important challenge that will certainly shed light on the regulation of normal and pathologic erythropoiesis.
We show here that the defect of Hsp70 nuclear accumulation on Epo stimulation contributes to the inhibition of MDS erythroblast maturation. In normal erythroblasts, differentiation is blocked at the basophilic stage by inhibition of caspase activation. A limited cleavage of caspase substrates, sparing GATA-1, is required for acquisition of maturation features on Epo stimulation. In contrast, when erythroblasts are starved of Epo, caspases are fully activated leading to the cleavage of all substrates including GATA-1, and Hsp70 is mainly retained in the cytoplasm impeding the protection of GATA-1 from caspases. In MDS, caspases are strongly activated at least partly downstream of Fas3,31 and, despite the presence of Epo, Hsp70 is maintained in the cytoplasm. Furthermore, wtBcl2 transduction does not modify Hsp70 subcellular localization, suggesting that the contribution of inappropriate activation of death pathway to defective expression of Hsp70 in the nucleus is unlikely. Thus, we cannot rule out that Epo signaling either is partially defective or might not be the sole pathway that controls Hsp70 localization. These 2 hypotheses are currently under investigation.
Defining the defect of Epo response in MDS is a major goal. Indeed, 30% to 40% of anemic patients with low-grade MDS are primary resistant to erythropoiesis-stimulating agent (ESA). We have prospectively shown that patients presenting at diagnosis with a profound diminution of the number of BFU-E/CFU-E and/or a decreased expression of activated MAPK ERK1/2 in the BM are those who fail to response to ESA treatment.32 Whether the nonresponse to ESA treatment correlates with the lack of nuclear Hsp70 localization is still a matter of investigation and should contribute to elucidate the pathway that triggers Hsp70 redistribution in the nucleus of erythroblasts undergoing Epo-driven differentiation. Finally, our preliminary results suggest that restoring the nucleocytosolic trafficking of Hsp70 could be a biomarker of lenalidomide as well as demethylating agent activity. Thus, targeting this trafficking may be a new therapeutic approach in the management of early MDS anemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr R. De Maria (Department of Hematology, Oncology and Molecular Medicine, Istituto Superiore di Sanita, Rome, Italy) who provided us the wt and μGATA-1 vectors; Prof Didier Bouscary, Dr Sophie Park, Dr Jérome Tamburini (Cochin, Paris, France) ; Prof Odile Beyne-Rauzy (Toulouse, France) ; Prof Bruno Quesnel (Lille, France); Prof Mathilde Hunault-Berger (Angers, France); Dr Borhane Slama (Avignon, France); Dr Ingrid Lafon (Dijon, France) for recording patients; Céline Marnier (Cochin, Paris, France) for technical assistance; Rosa Sapena (Cochin, Paris, France), Edith Mortera (Toulouse, France), Céline Marie (Angers, France), Monique Granjouan (Dijon, France), and Lamya Slama (Cochin, Paris, France) for recording clinical data and banking; Maryline Delattre (Unité de Recherche Clinique Paris Center); and Dr Nicolas Cagnard (Bioinformatics Platform, Paris Descartes University).
This work was supported by grants from la Direction Régionale de la Recherche Clinique de l'Assistance Publique-Hôpitaux de Paris (PHRC Cancer 2005), la Ligue contre le Cancer, l'ANR Blanc program, la Fondation pour la Recherche Médicale, le Cancéropole d'Ile de France.
E.F. was a recipient of the Ministère de la Recherche and the Société Française d'Hématologie. J.V. is a recipient of the Ministère de la Recherche and the Ligue nationale contre le Cancer. A.-S.G. is a recipient of Agence Nationale de la Recherche.
Authorship
Contribution: E.F., J.V., A.d.T., A.-S.G., O.K., P.V., J.-A.R., P.M., E.S., C.G., O.H., and M.F. designed the study and experiments; E.F., J.V., A.d.T., C.P.-E., A.S., J.-B.A., C.F., E.G., A.-S.G., and G.C. performed experiments; E.G., F.D., and M.F. provided BM samples; E.F., J.V., A.d.T., C.P.-E., A.S., E.G., G.C., O.K., P.V., J.-A.R., Y.Z., C.L., P.M., E.S., C.G., O.H., and M.F. analyzed experiments; E.F., J.V., C.L., P.M., E.S., C.G., O.H., and M.F. wrote the manuscript; and E.S., O.H., and M.F. validated the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michaela Fontenay, Service d'Hématologie Biologique, Hôpital Cochin, 27, rue du Faubourg Saint-Jacques, F75679 Paris Cedex 14, France; e-mail: michaela.fontenay@inserm.fr; or Olivier Hermine, Service d'hématologie clinique, Hôpital Necker, 149 rue de Sèvres, 75743 Paris cedex 15, France; e-mail: ohermine@gmail.com.
References
Author notes
E.F. and J.V. contributed equally.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal