Abstract
Stem cell differentiation and lineage specification depend on coordinated programs of gene expression, but our knowledge of the chromatin-modifying factors regulating these events remains incomplete. Ubiquitination of histone H2A (H2A-K119u) is a common chromatin modification associated with gene silencing, and controlled by the ubiquitin-ligase polycomb repressor complex 1 (PRC1) and H2A-deubiquitinating enzymes (H2A-DUBs). The roles of H2A-DUBs in mammalian development, stem cells, and hematopoiesis have not been addressed. Here we characterized an H2A-DUB targeted mouse line Mysm1tm1a/tm1a and demonstrated defects in BM hematopoiesis, resulting in lymphopenia, anemia, and thrombocytosis. Development of lymphocytes was impaired from the earliest stages of their differentiation, and there was also a depletion of erythroid cells and a defect in erythroid progenitor function. These phenotypes resulted from a cell-intrinsic requirement for Mysm1 in the BM. Importantly, Mysm1tm1a/tm1a HSCs were functionally impaired, and this was associated with elevated levels of reactive oxygen species, γH2AX DNA damage marker, and p53 protein in the hematopoietic progenitors. Overall, these data establish a role for Mysm1 in the maintenance of BM stem cell function, in the control of oxidative stress and genetic stability in hematopoietic progenitors, and in the development of lymphoid and erythroid lineages.
Introduction
The process of hematopoiesis generates the cells of the blood and immune system and its disruptions can result in life-threatening disorders, including immunodeficiencies, anemias, BM failures, and malignancies. Hematopoietic cell differentiation starts with a small population of self-renewing and multipotent stem cells and proceeds through a defined series of progenitors, with progressive loss of differentiation potential and progressive acquisition of the specialized characteristics of the different blood cell lineages. Stem cell differentiation and lineage specification are mediated by precisely regulated changes in gene expression and are controlled by the coordinated activities of transcription factors and chromatin remodeling proteins.1-3 For example, the establishment of hematopoiesis requires transcription factors such as Scl/Tal1, Runx1/Aml-1, Lmo2, Gata2, and Meis1,4 while B-cell lineage specification involves progressive activation of E2A, Ebf1, and Pax5, with additional requirement for factors such as Bcl11a and Foxo1.1 The critical roles of chromatin-modifying proteins in the control of hematopoiesis are demonstrated by their frequent translocations in leukemia,5 by studies in transgenic animal models,4,5 and by the recent analyses of the genome-wide histone methylation and acetylation profiles of hematopoietic progenitors.6,7 Nevertheless, the roles of the different histone-modifying factors in cell differentiation and lineage specification in the hematopoietic system and their effects on target gene expression remain poorly understood.
Histone H2A K119-monoubiquitination (H2A-K119u) is an abundant chromatin modification associated with transcriptional silencing and controlled by the activities of ubiquitin-ligating and deubiquitinating enzymes. Polycomb repressor complex 1 (PRC1) is the main H2A ubiquitin ligase8,9 and is essential for normal stem cell differentiation, mammalian development, and hematopoiesis.10 In embryonic stem (ES) cells, PRC1 and H2A-K119u localize to the promoters of key developmental-regulator genes and the loss of PRC1 disrupts normal programs of stem cell differentiation.11-13 Mice harboring mutations in components of PRC1 commonly develop hematopoietic abnormalities, and this is observed in knockout lines for Ring1B,14 Rae28,15 and Mel18.16 Furthermore, PRC1 component Bmi-1 is essential for HSC function,17 lymphocyte differentiation,18,19 and cancer stem cell activity in leukemia.20 Importantly, deregulation in PRC1 is also strongly implicated in the pathogenesis of hematologic malignancies, and mutations in PRC1 and its target genes are common in cancers.21 However, recent reports indicate that some of PRC1 functions as a transcriptional repressor are independent of its H2A ubiquitin ligase catalytic activity.22 Thus, the exact roles of histone H2A ubiquitination in stem cell function and hematopoiesis remain unknown.
The physiologic functions of histone H2A deubiquitinating enzymes (H2A-DUBs) in vivo in a mammalian organism have not been previously investigated. Eight proteins with H2A-DUB activity have been identified, including 6 members of the ubiquitin-specific protease family (Usp3, 12, 16, 21, 22, and 46),23,24 BRCA1-associated protein 1 (BAP1),25 and the Myb-like SWIRM and MPN domains containing protein 1 known as Mysm1.26 Some H2A-DUBs play a role in the regulation of Hox gene expression in Xenopus and Drosophila development,25,27 and responses to injury in hepatocytes.28 However, any role in mammalian stem cells, development, and hematopoiesis remains unidentified.
Here we characterize a novel H2A-DUB–targeted mouse line Mysm1tm1a(KOMP)WTSI/tm1a(KOMP)WTSI and establish a role for Mysm1 in HSC function, in the prevention of oxidative stress and the preservation of genetic stability in hematopoietic progenitors, and in the development of lymphoid and erythroid blood cell lineages.
Methods
Gene targeting and mouse production
Gene targeting was performed as part of the International Knockout Mouse Consortium (www.knockoutmouse.org).29 Targeted ES cells were selected for neomycin resistance and β-galactosidase expression, and the Mysm1tm1a(KOMP)WTSI allele structure was confirmed by long-range PCR and sequencing. A single-integration event was confirmed by neomycin-copy number analysis with quantitative RT-PCR (qRT-PCR). The Mysm1tm1a/tm1a mouse line was derived from the EPD0019_1_A05 ES cell clone and maintained on a C57BL/6 genetic background. The care and use of all mice was in accordance with UK Home Office regulations (United Kingdom Animals Scientific Procedures Act 1986). The mice were maintained in specific pathogen-free conditions, and matched by age and sex within experiments.
Genotyping
DNA was extracted using the DNeasy Blood and Tissue Kit (QIAGEN) and PCR performed using Platinum PCR Supermix (Invitrogen) and the following primers: Mysm1_F, AGCCCTGCATGTTCCAGAAG, Mysm1_R CCTTGCGGTAAAGCCTGTTG, Cassette_R TCGTGGTATCGTTATGCGCC, β-gal_F ATCACGACGCGCTGTATC, β-gal_R ACATCGGG-CAAATAATATCG (Sigma-Aldrich). Three reactions were performed: (1) for Mysm1-wild-type allele with primers Mysm1_F and Mysm1_R, (2) for Mysm1-targeted allele with primers Mysm1_F and Cassette_R, and (3) for the β-galactosidase reporter cassette using the β-gal_F and β-gal_R primer pair.
RNA isolation and qRT-PCR
RNA was isolated using the RNeasy Plus Mini Kit (QIAGEN) and reverse-transcribed with the QuantiTect Reverse Transcription Kit (QIAGEN). qPCRs were performed using the QuantiTect SYBR Green PCR Kit (QIAGEN), the Mysm1 QuantiTect Primer Assay (QIAGEN), and primers Actb_Fw CTAAGGCCAACCGTGAAAAG and Actb_Rv ACCAGAGGCATACAGGGACA (Sigma-Aldrich). The Mysm1 primer assay was designed to span the boundaries of exons 1, 2, and 3 of the Mysm1 coding-transcript ENSMUST00000075872 (QIAGEN). The data were acquired on the StepOnePlus Real-Time PCR system (Applied Biosystems) and analyzed using the ΔΔCt method.
Microarrays
RNA samples were quantified using NanoDrop 1000 (ThermoScientific), and quality-checked by analysis on an Agilent 2100 Bioanalyzer (Agilent Technologies). Five hundred nanograms of RNA were amplified using the Illumina TotalPrep RNA Amplification Kit (Ambion). The biotinylated cRNA in hybridization buffer was then applied to Illumina MouseWG-6 v2 Expression BeadChips, incubated in a humidified atmosphere at 58°C for 16-20 hours, and washed according to standard Illumina protocols. The slides were scanned using an Illumina BeadArray Reader according to the manufacturer's protocols, and the data analyzed using BeadStudio Software (Illumina). Gene ontology term and pathway overrepresentation analyses were performed using InnateDB (www.innatedb.ca).30 All microarray data are available at the Gene Expression Omnibus (GEO) under accession number GSE34285.
Histone extraction and Western blotting
Histones were purified from tissues and cells using either the Q-Proteome Cell Compartment Kit (QIAGEN) or acid-extraction methods previously described.9 Protein concentrations were measured using the QuickStart Bradford Dye Reagent (Bio-Rad). Samples were resolved on the NuPAGE Novex 4%-12% Bis-Tris Gels (Invitrogen), transferred onto nitrocellulose membranes (Bio-Rad), and probed with Abs for ubiquityl-histone H2A (clone E6C5) and histone H2A (both from Upstate-Millipore), followed by rabbit anti–mouse or goat anti–rabbit HRP-conjugated secondary Abs (DakoCytomation). The blots were developed using the ECL Western Blotting Detection Reagent (Amersham/GE Healthcare).
Tissue culture, BM cell differentiation, and in vitro cell stimulation
Detailed protocols are provided in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). ES cells were maintained on mitomycin C (Sigma-Aldrich) inactivated feeder layers, using the media and protocols described by Skarnes et al.29
Hematology
Blood was collected from terminally anesthetized mice at 14 weeks of age into EDTA-coated tubes via the retro-orbital sinus, and analyzed on a ScilVet Animal Blood Counter.
Flow cytometry
Procedures for the tissue collection and cell-surface staining for flow cytometry are provided in supplemental Methods. The data were acquired on FACSAria or LSRII flow cytometers (BD Biosciences). For intracellular staining of hematopoietic cells, BM samples prestained with FITC-conjugated Abs against B220, CD3, CD11b, and TER119, allophycocyanin-conjugated anti-cKit (all from BD Biosciences), and Brilliant Violet 421–conjugated anti-Sca1 (BioLegend), were fixed and permeabilized using PhosFlow Fixation Buffer and Perm Buffer II (BD Biosciences) according to supplier's protocols. The cells were then stained with PE-conjugated Abs against H2AX phospho-Ser139 (clone 20E3; Cell Signaling Technology), or p53 (clone G59-12; BD Biosciences), or p38 MAPK phospho-T180/Y182 (clone 36/p38; BD Biosciences). PE-conjugated mouse IgG1 and rabbit monoclonal isotype controls were used in the control staining protocols. Flow cytometric measurements of β-galactosidase activity were performed using the FluoReporter LacZ Flow Cytometry Kit (Invitrogen/Molecular Probes). For intracellular reactive oxygen species (ROS) measurements, the cells were loaded with carboxy-H2DCFDA (Invitrogen/Molecular Probes) at 5μM in PBS for 15 minutes, and washed twice before analyses. Necrotic and apoptotic cells were detected using 7-AAD and annexin V (BD Pharmingen). The flow cytometric peripheral blood micronucleus assay was performed according to the protocol of Reinholdt et al.31
Tissue staining for β-galactosidase activity
Mice were fixed by cardiac perfusion with 4% (w/v) paraformaldehyde. After dissection, the tissues were fixed again in 4% paraformaldehyde for 30 minutes, rinsed in PBS, and stained in 0.1% (w/v) X-gal solution (bromo-chloro-indolyl-galactopyranoside; Invitrogen) for up to 48 hours. After an additional overnight fixation in 4% paraformaldehyde, the tissues were cleared with 50% (v/v) glycerol and transferred to 70% glycerol for long-term storage. Images were taken using a Leica MZ16A microscope and Imagic software.
Mouse BM transfer experiments
For BM chimeras experiments, irradiated Rag1−/− recipient mice (5 Gy or 2 × 4.5 Gy) were IV injected with 2 × 106 BM cells from CD45.1+ wild-type mice, CD45.2+Mysm1tm1a/tm1a mice, or a 1:1 mix of the two, normalized either by the total cell count or by the numbers of KLS cells (c-kit+lin−Sca1+). The mice were kept on clindamycin (250 mg/L, in drinking water) for 2 weeks and analyzed at 8-20 weeks after reconstitution.
Statistical analyses
Statistical comparisons were performed with Prism 4.0 (GraphPad), using the Student t test or the Mann-Whitney test for comparisons of 2 datasets, and ANOVA for multiple comparisons.
Results
Phenotypic characteristics of the Mysm1-targeted mouse line
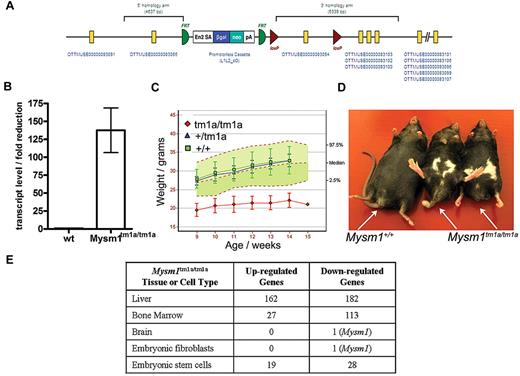
Mysm1 gene targeting was carried out as part of the International Knockout Mouse Consortium, using JM8 ES cells on a C57BL/6N genetic background.29 The targeted Mysm1tm1a(KOMP)WTSI allele incorporated a promoterless gene-trap DNA cassette, encoding the β-galactosidase reporter and neomycin-resistance marker, inserted into the second intron of the gene (Figure 1A, GenBank file available at www.knockoutmouse.org). The targeted allele was predicted to generate a truncated nonfunctional transcript encoding the first 46 of the 819 amino acids of Mysm1 protein, eliminating all structural domains. The allele also contained LoxP and Frt sites to allow the conversion of the allele to a conditional configuration for future studies.29 Analysis of Mysm1tm1a/tm1a mouse tissues (liver, spleen, thymus, skeletal muscle, brain, fibroblasts) demonstrated that the cassette effectively disrupted the splicing of the flanking exons (> 125-fold reduction in mRNA containing the junction of exons 2-3 of Mysm1 ENSMUST00000075872 transcript, Figure 1B). Overall, this indicated that the Mysm1tm1a allele is severely hypomorphic, resulting in a strong but incomplete loss of Mysm1 transcript.
Mysm1tm1a/tm1a mouse line–gene targeting and gross phenotypic characterization. (A) Structure of the Mysm1tm1a(KOMP)WTSI (Mysm1tm1a) allele. (B) Fold reduction in Mysm1-transcript in Mysm1tm1a/tm1a relative to wild-type tissues (liver, spleen, thymus, skeletal muscle, brain, embryonic fibroblasts), analyzed by qRT-PCR using primers spanning the junction of exons 2 and 3 of Mysm1 transcript ENSMUST00000075872. Bars show means ± SEM. (C) Reduced weight of Mysm1tm1a/tm1a mice. Bars show means ± SD; shaded area shows 95% reference range for all wild-type mice of the same strain and sex. Data are from males and are representative of the differences seen for both sexes. (D) Representative images showing reduced body size and dysmorphology in Mysm1tm1a/tm1a mice, compared with a wild-type littermate control. (E) Numbers of significantly up-regulated and down-regulated genes in Mysm1tm1a/tm1a versus wild-type control, by tissue and cell type, based on adjusted P ≤ .05 (tissue and fibroblast comparisons are between 3-4 mice per group; ES cell comparisons are between multiple independently passaged samples of wild-type JM8 line and a Mysm1tm1a/tm1a ES cell line derived from mouse blastocysts).
Mysm1tm1a/tm1a mouse line–gene targeting and gross phenotypic characterization. (A) Structure of the Mysm1tm1a(KOMP)WTSI (Mysm1tm1a) allele. (B) Fold reduction in Mysm1-transcript in Mysm1tm1a/tm1a relative to wild-type tissues (liver, spleen, thymus, skeletal muscle, brain, embryonic fibroblasts), analyzed by qRT-PCR using primers spanning the junction of exons 2 and 3 of Mysm1 transcript ENSMUST00000075872. Bars show means ± SEM. (C) Reduced weight of Mysm1tm1a/tm1a mice. Bars show means ± SD; shaded area shows 95% reference range for all wild-type mice of the same strain and sex. Data are from males and are representative of the differences seen for both sexes. (D) Representative images showing reduced body size and dysmorphology in Mysm1tm1a/tm1a mice, compared with a wild-type littermate control. (E) Numbers of significantly up-regulated and down-regulated genes in Mysm1tm1a/tm1a versus wild-type control, by tissue and cell type, based on adjusted P ≤ .05 (tissue and fibroblast comparisons are between 3-4 mice per group; ES cell comparisons are between multiple independently passaged samples of wild-type JM8 line and a Mysm1tm1a/tm1a ES cell line derived from mouse blastocysts).
The mouse line was characterized using a series of standardized phenotype screens, under the scope of the Sanger Institute Mouse Genetics Project. Mysm1tm1a/tm1a mice displayed significant levels of embryonic mortality, with 10% of offspring from heterozygous matings being of the Mysm1tm1a/tm1a genotype; the Mysm1tm1a/tm1a mice had reduced body size and weight, and abnormalities in the hind limb and tail morphology (Figure 1C-D). Further information on the phenotypic screens performed by the Sanger Mouse Genetics Project and the data are available (http://www.sanger.ac.uk/mouseportal/search?query = mysm1).
Mysm1 expression and tissue-specific roles in transcriptional regulation
The Mysm1tm1a allele can direct the expression of a β-galactosidase reporter from the endogenous Mysm1 promoter, thus facilitating the analysis of Mysm1-promoter activity in different tissues. Staining of tissues from the Mysm1tm1a/+ mice for reporter activity showed widespread Mysm1 expression during early development, for example, in ES cells to 10.5 dpc (days post conception) embryos (data not shown). Mysm1 expression became more restricted at later stages of development from 18.5 dpc embryos to adult mice. A summary of the reporter activity in 37 organs of adult Mysm1tm1a/+ mice is provided in supplemental Table 1 (images available at www.sanger.ac.uk/mouseportal/search?query = mysm1).
To further assess the tissue-specific functions of Mysm1, microarray gene expression analysis was performed on a selection of mouse tissues and significant changes in the transcriptional profiles were observed in the BM and liver of Mysm1tm1a/tm1a compared with wild-type mice (Figure 1E). Pathway overrepresentation analysis of the microarray data indicated significant down-regulation of genes involved in lymphocyte biology and also in translation and ribosome function in Mysm1tm1a/tm1a BM (supplemental Table 2). The genes differentially expressed in Mysm1tm1a/tm1a liver were significantly enriched for complement cascade components and cytochrome P450 enzymes involved in xenobiotic metabolism (supplemental Table 3). The altered transcriptional profiles of Mysm1tm1a/tm1a tissues may include direct changes in the expression of Mysm1-target genes and also possible differences in the cellular composition of Mysm1tm1a/tm1a tissues.
To study the roles of Mysm1 in transcriptional regulation using homogeneous cell populations, Mysm1tm1a/tm1a and control mouse embryonic fibroblasts (MEFs) were derived and their genome-wide transcriptional profiles compared using microarrays. No significant alterations in gene expression were detected in Mysm1tm1a/tm1a MEFs, and the overall levels of ubiquitinated histone H2A (H2A-K119u) were not significantly altered (Figure 1E, supplemental Figure 1), suggesting that Mysm1 does not have global housekeeping roles in transcriptional regulation and its activity may be restricted in a tissue-specific manner.
To assess the role of Mysm1 in ES cells, Mysm1tm1a/tm1a and Mysm1tm1a/+ ES-lines were derived from mouse blastocysts and analyzed in comparison with wild-type and Mysm1tm1a/+ JM8 ES cells. Mysm1tm1a/tm1a ES cells had normal morphology, their growth rate was indistinguishable from wild-type cells, and they expressed normal levels of the pluripotency markers Oct3/4 and SSEA1 (supplemental Figure 2). There were relatively limited differences in the gene expression profile between wild-type and Mysm1tm1a/tm1a ES lines (Figure 1E, list of 52 differentially expressed genes presented in supplemental Table 4). Given the limited changes in gene expression, the overall levels of H2A-K119u were not significantly altered in the Mysm1tm1a/tm1a ES cells (supplemental Figure 1).
Hematologic abnormalities and defect in lymphocyte differentiation in Mysm1tm1a/tm1a mice
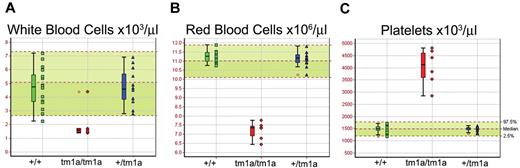
Screening of blood from Mysm1tm1a/tm1a mice demonstrated a reduction in the white and red blood cell counts (∼ 3-fold and ∼ 1.5-fold, respectively, Figure 2A-B). The anemia was further characterized by reduced hematocrit and blood hemoglobin content, increased RBC width distribution, mean corpuscular volume, and mean corpuscular hemoglobin content (supplemental Figure 3A-E), and normal plasma iron levels (data not shown). Mysm1tm1a/tm1a mice also showed elevated platelet counts (∼ 2.5-fold, Figure 2C), with an increase in mean platelet volume (supplemental Figure 3F). This, together with the altered gene expression profile of Mysm1tm1a/tm1a BM, suggested a possible defect in hematopoiesis in Mysm1tm1a/tm1a mice.
Altered blood cell counts in Mysm1tm1a/tm1a mice. (A) White blood cells, (B) red blood cells, and (C) platelets. Bars represent means ± SD; shaded areas represent 95% reference range for all wild-type mice of same strain and sex. Data are from males, at 14 weeks of age, and are representative of the differences seen for both sexes.
Altered blood cell counts in Mysm1tm1a/tm1a mice. (A) White blood cells, (B) red blood cells, and (C) platelets. Bars represent means ± SD; shaded areas represent 95% reference range for all wild-type mice of same strain and sex. Data are from males, at 14 weeks of age, and are representative of the differences seen for both sexes.
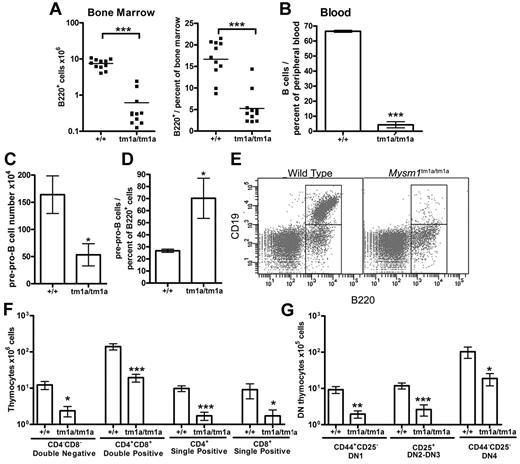
Flow cytometry of the blood and BM of Mysm1tm1a/tm1a mice detected a severe depletion of B lymphocytes (Figure 3A-B), and the absolute number of B220+ B-lineage cells in the BM was reduced > 10-fold. This was associated with a loss of pre-B cell CFU in BM culture assays (supplemental Figure 4A). Evidence of B-cell loss was supported further by the analysis of the microarray data from total BM, showing that the genes down-regulated in Mysm1tm1a/tm1a were significantly enriched for components of lymphocyte receptor signaling pathways and phenotype ontology terms related to arrest in B-cell development (supplemental Table 2E-F, analysis using www.innatedb.ca and www.informatics.jax.org).30
Defects in B and T-cell development in Mysm1tm1a/tm1a mice. (A) Reduced absolute numbers and decreased frequency of B220+ B-lineage cells in the BM of Mysm1tm1a/tm1a mice. (B) Reduced frequency of B cells in the blood of Mysm1tm1a/tm1a mice, gated as CD45+CD19+ cells. (C) Decrease in the absolute number of pre-pro-B cells in Mysm1tm1a/tm1a BM (gated as B220+CD19−CD43+CD11b−DX5−). (D) Relative increase in the proportion of pre-pro-B cells within the B220+ B-cell lineage gate in Mysm1tm1a/tm1a BM. (E) Representative flow cytometric plots of wild-type and Mysm1tm1a/tm1a BM, gated on lymphocytes and demonstrating a preferential loss of B220+CD19+ cells in Mysm1tm1a/tm1a mice. (F) Reduction in the numbers of CD4−CD8− double-negative (DN), CD4+CD8+ double-positive, and CD4+ and CD8+ single-positive thymocytes in the thymus of Mysm1tm1a/tm1a mice. (G) Reduction in the numbers of CD44+CD25− DN1, CD44+/−CD25+ DN2-3, and CD44−CD25− DN4 thymocytes in the thymus of Mysm1tm1a/tm1a mice. Bars represent mean ± SEM. *P < .05, **P < .01, ***P < .001 using the Student t test or the Mann-Whitney test; all BM cell counts are per 1 tibia and femur.
Defects in B and T-cell development in Mysm1tm1a/tm1a mice. (A) Reduced absolute numbers and decreased frequency of B220+ B-lineage cells in the BM of Mysm1tm1a/tm1a mice. (B) Reduced frequency of B cells in the blood of Mysm1tm1a/tm1a mice, gated as CD45+CD19+ cells. (C) Decrease in the absolute number of pre-pro-B cells in Mysm1tm1a/tm1a BM (gated as B220+CD19−CD43+CD11b−DX5−). (D) Relative increase in the proportion of pre-pro-B cells within the B220+ B-cell lineage gate in Mysm1tm1a/tm1a BM. (E) Representative flow cytometric plots of wild-type and Mysm1tm1a/tm1a BM, gated on lymphocytes and demonstrating a preferential loss of B220+CD19+ cells in Mysm1tm1a/tm1a mice. (F) Reduction in the numbers of CD4−CD8− double-negative (DN), CD4+CD8+ double-positive, and CD4+ and CD8+ single-positive thymocytes in the thymus of Mysm1tm1a/tm1a mice. (G) Reduction in the numbers of CD44+CD25− DN1, CD44+/−CD25+ DN2-3, and CD44−CD25− DN4 thymocytes in the thymus of Mysm1tm1a/tm1a mice. Bars represent mean ± SEM. *P < .05, **P < .01, ***P < .001 using the Student t test or the Mann-Whitney test; all BM cell counts are per 1 tibia and femur.
Further flow cytometry demonstrated that B lymphocytes were depleted from the earliest pre-pro-B-cell stage, suggesting a defect in B-cell lineage specification (Figure 3C, gated as B220+CD19−CD43+DX5−CD11b−). Nevertheless, the loss of the pre-pro-B cells was not as severe as that of the later B-cell developmental subsets (Figure 3D-E), suggesting that there may be an additional defect in the progression of pre-pro-B cells to the more mature stages of development. Indeed, the few B220+CD19+ cells present in Mysm1tm1a/tm1a mice expressed higher levels of CD43 (P < .001, data not shown), indicating their less mature state, and there was increased incidence of cell apoptosis and necrosis within this B-cell population (supplemental Figure 4B). B-cell numbers were also severely reduced in the spleen of Mysm1tm1a/tm1a mice (∼ 20-fold), with a more severe effect on the follicular than the transitional and marginal-zone B-cell populations (data not shown).
Mysm1tm1a/tm1a mice also exhibited a severe defect in T-cell development. Thymic cell counts were reduced, with significant losses of the CD4+ and CD8+ single-positive, CD4+CD8+ double-positive, and CD4−CD8− double-negative (DN) thymocytes (> 5-fold, Figure 3F). All developmental subsets of the early DN population were also severely depleted, including DN1 (CD44+CD25−), DN2-3 (CD44+/−CD25+), and DN4 (CD44−CD25−) thymocytes (Figure 3G). Overall, this indicated that the T-cell lineage depletion in Mysm1tm1a/tm1a mice was occurring from the earliest stages of thymocyte differentiation. T cells were similarly severely depleted in the spleen of Mysm1tm1a/tm1a mice (∼ 10-fold, data not shown).
Cell intrinsic requirement for Mysm1 in B- and T-lymphocyte development
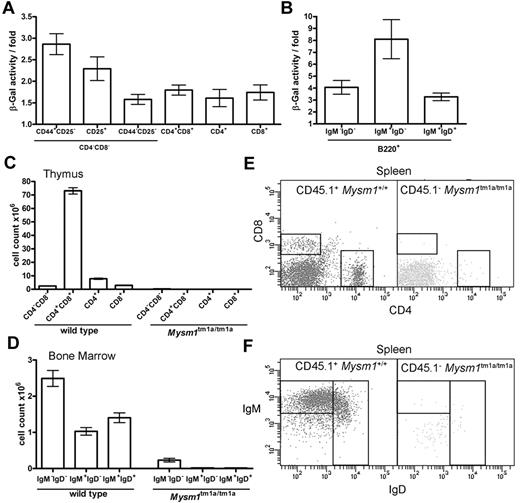
The hematopoietic phenotype of Mysm1tm1a/tm1a mice could be a consequence of a cell-intrinsic requirement for Mysm1 in hematopoietic cells or of complex interactions between multiple cell types. To explore these possibilities, Mysm1 gene expression was analyzed in the developing lymphoid cells, by measuring Mysm1 promoter–driven β-galactosidase reporter activity using a fluorescent substrate and flow cytometry in phenotypically normal Mysm1tm1a/+ cells. The data indicated that the Mysm1 promoter was active at early stages of T-cell differentiation in the thymus and in developing B cells of the BM (Figure 4A-B), suggesting that Mysm1 may have a cell-intrinsic role as a transcriptional regulator in lymphoid cell differentiation. We could also detect Mysm1 transcript in the thymus and in isolated splenic B cells of wild-type mice by qRT-PCR (data not shown).
The defect in lymphocyte development in Mysm1tm1a/tm1a mice is because of a cell-intrinsic function of Mysm1 in hematopoietic cells. (A-B) Mysm1-promoter activity in (A) thymocytes, and (B) B-lineage BM cells. The measurements done on phenotypically normal Mysm1+/tm1a cells that carry a single copy of the Mysm1 promoter–driven β-galactosidase reporter. The data are presented as fold difference in β-galactosidase activity in Mysm1+/tm1a versus background activity in wild-type cells; bars show means ± SEM from ≥ 3 mice. β-galactosidase activity was significantly above background in Mysm1tm1a/+ double-negative DN1-DN4 and double-positive thymocytes, as well as in B-lineage cells including B220+IgM−IgD− pre/pro-B cells, B220+IgM+IgD− immature and B220+IgMlowIgD+ mature B cells (P < .05, t tests), although β-galactosidase activity in the single-positive CD4 and CD8 thymocytes was not significant. (C-F) Irradiated Rag1−/− mice were reconstituted with a 1:1 mix of CD45.1+ wild-type and CD45.2+Mysm1tm1a/tm1a BM for 8 weeks; data were from 4 animals per group and were reproducible in 2 independent experiments. (C) Numbers of thymocytes, and (D) BM B cells, including pro/pre-B220+IgM−IgD−, immature B220+IgM+IgD−, and mature B220+IgMlowIgD+ B cells, in the chimeric mice. Bars represent means ± SEM; ANOVA analyses indicated significantly increased contribution of wild-type versus Mysm1tm1a/tm1a cells to the CD4+CD8+ double-positive (P < .001) and the single-positive CD4 (P < .01) and CD8 (P < .05) populations in the thymus, as well as to the pre/pro-, immature, and mature B cells in the BM (P < .01 for all). (E-F) Representative flow cytometric profiles of the spleen of the chimeric mice, gated on CD45.1+ wild-type cells (left) or CD45.1−Mysm1tm1a/tm1a cells (right), and stained (E) for CD4 and CD8, or (F) for B220, IgM, and IgD.
The defect in lymphocyte development in Mysm1tm1a/tm1a mice is because of a cell-intrinsic function of Mysm1 in hematopoietic cells. (A-B) Mysm1-promoter activity in (A) thymocytes, and (B) B-lineage BM cells. The measurements done on phenotypically normal Mysm1+/tm1a cells that carry a single copy of the Mysm1 promoter–driven β-galactosidase reporter. The data are presented as fold difference in β-galactosidase activity in Mysm1+/tm1a versus background activity in wild-type cells; bars show means ± SEM from ≥ 3 mice. β-galactosidase activity was significantly above background in Mysm1tm1a/+ double-negative DN1-DN4 and double-positive thymocytes, as well as in B-lineage cells including B220+IgM−IgD− pre/pro-B cells, B220+IgM+IgD− immature and B220+IgMlowIgD+ mature B cells (P < .05, t tests), although β-galactosidase activity in the single-positive CD4 and CD8 thymocytes was not significant. (C-F) Irradiated Rag1−/− mice were reconstituted with a 1:1 mix of CD45.1+ wild-type and CD45.2+Mysm1tm1a/tm1a BM for 8 weeks; data were from 4 animals per group and were reproducible in 2 independent experiments. (C) Numbers of thymocytes, and (D) BM B cells, including pro/pre-B220+IgM−IgD−, immature B220+IgM+IgD−, and mature B220+IgMlowIgD+ B cells, in the chimeric mice. Bars represent means ± SEM; ANOVA analyses indicated significantly increased contribution of wild-type versus Mysm1tm1a/tm1a cells to the CD4+CD8+ double-positive (P < .001) and the single-positive CD4 (P < .01) and CD8 (P < .05) populations in the thymus, as well as to the pre/pro-, immature, and mature B cells in the BM (P < .01 for all). (E-F) Representative flow cytometric profiles of the spleen of the chimeric mice, gated on CD45.1+ wild-type cells (left) or CD45.1−Mysm1tm1a/tm1a cells (right), and stained (E) for CD4 and CD8, or (F) for B220, IgM, and IgD.
To confirm the cell-intrinsic requirement for Mysm1 in lymphocyte development, Mysm1tm1a/tm1a and CD45.1-allotype marked wild-type BM were transferred, either separately or mixed at a 1:1 ratio, into irradiated Rag1−/− recipients. The data demonstrated that Mysm1tm1a/tm1a hematopoietic cells were severely impaired in lymphocyte production, even when developing side-by-side with wild-type hematopoietic cells, within wild-type BM and thymic niches (Figure 4C-F). Mysm1tm1a/tm1a lymphocyte development was impaired when the numbers of coinjected wild-type and Mysm1tm1a/tm1a cells were normalized either by the total BM cell count (Figure 4C-F), or by the numbers of HSCs (KLS, gated as Lin−c-kit+Sca1+, data not shown). The defect in lymphocyte development was also observed when Mysm1tm1a/tm1a BM was reconstituted into irradiated Rag1−/− recipients without competing wild-type BM (data not shown). In addition, the mice receiving Mysm1tm1a/tm1a BM showed significantly higher levels of cell apoptosis and necrosis within the developing B- and T-cell populations in the BM and thymus (data not shown). This demonstrated that the defect in lymphocyte development in the Mysm1tm1a/tm1a mice was because of a cell-intrinsic role of Mysm1 in hematopoietic cells.
Role of Mysm1 in the responses of mature B and T lymphocytes to stimulation
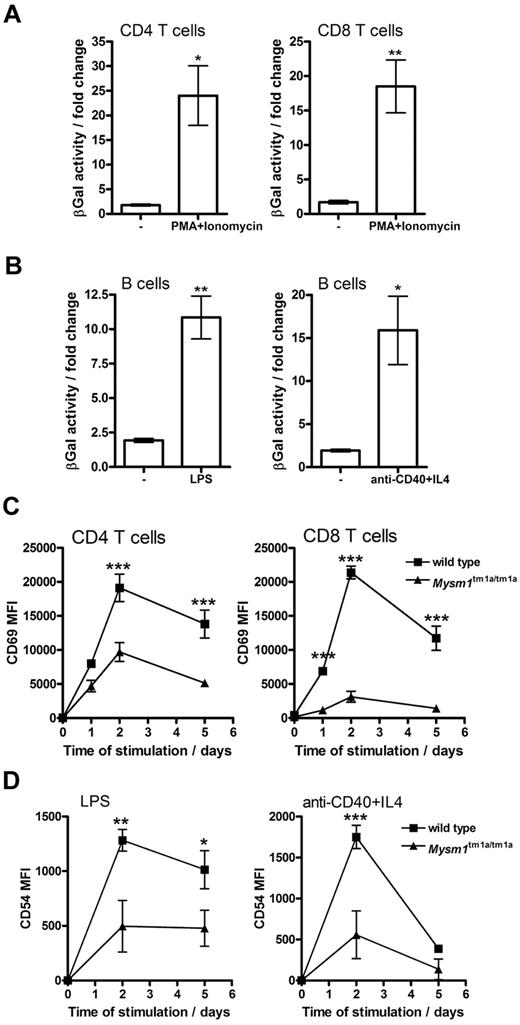
When Mysm1tm1a/+ splenocytes were stimulated in vitro with mitogens or inflammatory stimuli (PMA and ionomycin, LPS, or anti-CD40 and IL-4) an increase in Mysm1 promoter–driven β-galactosidase reporter activity was detected in CD4 and CD8 T cells (∼ 20-fold, Figure 5A), as well as B cells (∼ 10-fold, Figure 5B). Background β-galactosidase activity was unchanged in wild-type cells undergoing identical stimulation. This induction of Mysm1-promoter activity suggested that Mysm1 might play a role, not only in the transcriptional programs of lymphocyte development, but possibly also in lymphocyte activation.
Role of Mysm1 in lymphocyte activation. (A-B) Mysm1 promoter–driven β-galactosidase reporter activity in the splenic (A) T cells and (B) B cells, stimulated in vitro with LPS (1 μg/mL), anti-CD40 agonist Ab (5 μg/mL) and IL-4 (5 ng/mL), or PMA (50 ng/mL) and ionomycin (500 ng/mL), for 48 hours. The data were collected using phenotypically normal Mysm1+/tm1a cells, and presented as fold change versus the background β-galactosidase activity in wild-type cells undergoing identical treatment. Significant induction of Mysm1-driven β-galactosidase reporter activity was observed (*P < .05, **P < .01, using t test). (C) Reduced induction of the CD69 activation marker on Mysm1tm1a/tm1a splenic T cells, stimulated in vitro with PMA and ionomycin, as above, over a 5-day time course. (D) Reduced expression of CD54 on Mysm1tm1a/tm1a splenic B cells, stimulated in vitro with LPS (1 μg/mL), or anti-CD40 (2.5 μg/mL) and IL-4 (5 ng/mL), over 5 days; MFI indicates mean fluorescence intensity. Bars represent means ± SEM from 4 mice; *P < .05, **P < .01, ***P < .001 using 2-way ANOVA.
Role of Mysm1 in lymphocyte activation. (A-B) Mysm1 promoter–driven β-galactosidase reporter activity in the splenic (A) T cells and (B) B cells, stimulated in vitro with LPS (1 μg/mL), anti-CD40 agonist Ab (5 μg/mL) and IL-4 (5 ng/mL), or PMA (50 ng/mL) and ionomycin (500 ng/mL), for 48 hours. The data were collected using phenotypically normal Mysm1+/tm1a cells, and presented as fold change versus the background β-galactosidase activity in wild-type cells undergoing identical treatment. Significant induction of Mysm1-driven β-galactosidase reporter activity was observed (*P < .05, **P < .01, using t test). (C) Reduced induction of the CD69 activation marker on Mysm1tm1a/tm1a splenic T cells, stimulated in vitro with PMA and ionomycin, as above, over a 5-day time course. (D) Reduced expression of CD54 on Mysm1tm1a/tm1a splenic B cells, stimulated in vitro with LPS (1 μg/mL), or anti-CD40 (2.5 μg/mL) and IL-4 (5 ng/mL), over 5 days; MFI indicates mean fluorescence intensity. Bars represent means ± SEM from 4 mice; *P < .05, **P < .01, ***P < .001 using 2-way ANOVA.
To further explore the role of Mysm1 in lymphocyte activation, the responses of Mysm1tm1a/tm1a cells to in vitro stimulation were studied. Splenocytes were magnetically separated into CD19+ and CD19− fractions, stimulated as in the previous paragraph and analyzed for the induction of activation markers, gating on B220+IgD+ B cells or CD4 and CD8 T cells, respectively. Mysm1tm1a/tm1a T cells exhibited reduced induction of the CD69 activation marker in response to PMA and ionomycin stimulation (Figure 5C), while Mysm1tm1a/tm1a B cells expressed lower levels of CD54 in response to LPS or anti-CD40 + IL-4 stimulation (Figure 5D). Overall, this indicated that Mysm1tm1a/tm1a lymphocytes were impaired in their responses to stimulation.
Altered erythroid and megakaryocyte lineage differentiation in Mysm1tm1a/tm1a mice
As Mysm1tm1a/tm1a mice exhibited reduced erythrocyte and increased platelet counts in the blood (Figure 2B-C), their BM was analyzed to establish the role of Mysm1 in the differentiation of these cell lineages. Mysm1tm1a/tm1a BM exhibited a reduction in the numbers of erythroid cells (∼ 4-fold, supplemental Figure 5A) and an increase in megakaryocytes (∼ 2-fold, supplemental Figure 5B). Reductions in cell numbers were observed at all the stages of erythroid lineage differentiation (supplemental Figure 5C) and were associated with a severe decrease in erythroid CFU in Mysm1tm1a/tm1a BM (supplemental Figure 5D), indicating a dysfunction of erythroid progenitors.
The Mysm1 gene was expressed in the cells of erythroid lineage (supplemental Figure 5E), and the anemia phenotype was transferred to irradiated Rag1−/− recipients with Mysm1tm1a/tm1a BM (supplemental Figure 5F), indicating a cell-intrinsic requirement for Mysm1 in erythroid cell production. However, despite Mysm1 gene expression within megakaryocytes (data not shown), thrombocytosis was not transferred to irradiated wild-type mice with Mysm1tm1a/tm1a BM (data not shown), suggesting that abnormalities in the BM niche or signals from other tissues, rather than factors intrinsic to hematopoietic cells, caused Mysm1tm1a/tm1a thrombocytosis.
The numbers of myeloid CD11b+Ly6G+ cells in Mysm1tm1a/tm1a BM were mildly reduced (supplemental Figure 6A), while the frequency of myeloid CFU was comparable with wild type, suggesting unimpaired myeloid progenitor functions (supplemental Figure 6B). Mysm1 promoter–driven reporter activity was detectable in the myeloid cells, although unlike in lymphocytes, its expression was unresponsive to mitogenic or inflammatory stimuli (supplemental Figure 6C, and data not shown).
Impaired stem cell function in Mysm1tm1a/tm1a BM
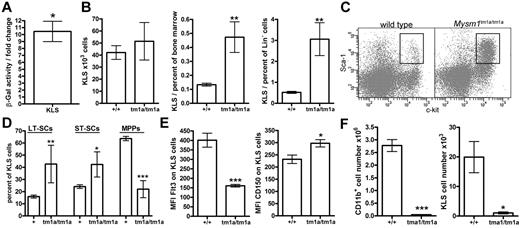
The multi-lineage defect in hematopoiesis in Mysm1tm1a/tm1a mice suggested that Mysm1 may play a critical role at the earliest stages of cell differentiation in the hematopoietic system. Using β-galactosidase reporter activity, we confirmed that the Mysm1 gene was transcriptionally active within the BM KLS cells (lineage−vecKit+Sca1+; Figure 6A), which include HSCs and their earliest progenitors.32 The KLS population was expanded as a proportion of total and lineage-negative BM cells, showing the preferential loss of the more mature cells from the earliest stages of hematopoiesis (Figure 6B-C). Furthermore, within the KLS compartment there was also a preferential preservation of the most-primitive long-term HSCs (LT-HSCs, Flt3−CD34−KLS) and a severe depletion of the more mature multipotent progenitors (MPPs; Flt3+CD34+KLS; Figure 6D-E). This was further supported by increased expression of the CD150 LT-HSC marker on the KLS cells (Figure 6E).32 The loss of Flt3+ KLS cells is also consistent with reduced lymphoid lineage priming within the KLS population.33 Overall, this demonstrated that Mysm1tm1a/tm1a mice had severe defects in the normal programs of cell maintenance and differentiation at the earliest stages of hematopoiesis.
Impaired HSC function in Mysm1tm1a/tm1a mice. (A) Mysm1 gene expression in KLS (cKit+ lineage− Sca1+) cells of mouse BM, as shown by the activity of a Mysm1 promoter–driven β-galactosidase reporter; data are presented as fold increase in reporter activity in Mysm1+/tm1a relative to wild-type cells (*P < .05, one-sample t test). (B) The absolute numbers and proportion of KLS cells in the BM of wild-type and Mysm1tm1a/tm1a mice. (C) Flow cytometric plots of the BM, stained for cKit and Sca1, and gated on the lineage-negative cells, showing a relative expansion of the KLS population in Mysm1tm1a/tm1a mice. (D) Proportion of long-term stem cells (LT-SCs, Flt3−CD34−), short-term stem cells (ST-SCs, Flt3−CD34+), and multipotent progenitors (MPPs, Flt3+CD34+) within the KLS population. (E) Reduced Flt3 and elevated CD150 expression in Mysm1tm1a/tm1a KLS cells. (F) Reduced contribution of Mysm1tm1a/tm1a cells to the myeloid lineage and the primitive KLS population in mixed BM chimeras. CD45.1+ wild-type and CD45.2+Mysm1tm1a/tm1a BM, mixed at 1:1 ratio, were reconstituted into lethally irradiated Rag1−/− recipients. All bars represent means ± SEM. Cell counts per 1 tibia and femur; *P < .05, **P < .01, ***P < .001 using (B,E,F) t test or (D) ANOVA; MFI indicates median fluorescence intensity.
Impaired HSC function in Mysm1tm1a/tm1a mice. (A) Mysm1 gene expression in KLS (cKit+ lineage− Sca1+) cells of mouse BM, as shown by the activity of a Mysm1 promoter–driven β-galactosidase reporter; data are presented as fold increase in reporter activity in Mysm1+/tm1a relative to wild-type cells (*P < .05, one-sample t test). (B) The absolute numbers and proportion of KLS cells in the BM of wild-type and Mysm1tm1a/tm1a mice. (C) Flow cytometric plots of the BM, stained for cKit and Sca1, and gated on the lineage-negative cells, showing a relative expansion of the KLS population in Mysm1tm1a/tm1a mice. (D) Proportion of long-term stem cells (LT-SCs, Flt3−CD34−), short-term stem cells (ST-SCs, Flt3−CD34+), and multipotent progenitors (MPPs, Flt3+CD34+) within the KLS population. (E) Reduced Flt3 and elevated CD150 expression in Mysm1tm1a/tm1a KLS cells. (F) Reduced contribution of Mysm1tm1a/tm1a cells to the myeloid lineage and the primitive KLS population in mixed BM chimeras. CD45.1+ wild-type and CD45.2+Mysm1tm1a/tm1a BM, mixed at 1:1 ratio, were reconstituted into lethally irradiated Rag1−/− recipients. All bars represent means ± SEM. Cell counts per 1 tibia and femur; *P < .05, **P < .01, ***P < .001 using (B,E,F) t test or (D) ANOVA; MFI indicates median fluorescence intensity.
To analyze the functional capacities of Mysm1tm1a/tm1a HSCs, their ability to self-renew and reconstitute hematopoiesis was assessed in direct competition with wild-type cells, by reconstituting lethally irradiated mice with a 1:1 mixture of CD45.1+ wild-type and CD45.2+Mysm1tm1a/tm1a BM. The contribution of Mysm1tm1a/tm1a to all hematopoietic lineages and the KLS compartment itself was severely reduced, so that ∼ 95% of KLS cells in the chimeric mice were wild-type CD45.1+ at both 8 and 20 weeks of reconstitution (Figure 6F, and data not shown). Severely impaired reconstitution of KLS cells was also observed in the recipients reconstituted with Mysm1tm1a/tm1a BM without competing wild-type cells (data not shown); the reduced levels of Flt3 on Mysm1tm1a/tm1a KLS cells in the chimeras further highlighted the impaired production of MPPs from Mysm1-deficient LT-HSCs (P < .01, data not shown).
Genetic instability, oxidative stress, and elevated p53 levels in Mysm1tm1a/tm1a progenitors
As part of the phenotypic screens carried out by the Sanger Mouse Genetics Project, a peripheral blood micronucleus assay31 was performed, demonstrating a significant increase in the frequency of micronucleated normochromatic erythrocytes (NCE) in the blood of Mysm1tm1a/tm1a mice (Figure 7A), suggesting increased genetic instability. As elevated DNA damage and activation of DNA damage response are strongly linked to HSC defects and BM failure,34,35 we further analyzed the levels of γH2AX (H2AX phospho-Ser139, marker of DNA damage foci) in the KLS (lineage−vecKit+Sca1+) stem and progenitor cells of Mysm1tm1a/tm1a BM, using flow cytometry with combination of cell-surface and intracellular staining. This demonstrated elevated levels of γH2AX staining in the KLS cells of Mysm1tm1a/tm1a mice (Figure 7B), indicating increased DNA damage. Activation of DNA damage response was further analyzed, demonstrating increased levels of p53 tumor suppressor in Mysm1tm1a/tm1a KLS cells (Figure 7B). Irradiated wild-type BM was used for positive control staining, and isotype controls for the γH2AX and p53 Abs were used to confirm specificity (data not shown).
Genetic instability, oxidative stress, and elevated p53 protein levels in Mysm1tm1a/tm1a hematopoietic progenitors. (A) Increased frequency of micronucleated cells in the normochromatic erythrocyte (NCE) cell population of Mysm1tm1a/tm1a mouse blood; representative flow cytometric plots and data quantification are presented. (B) Levels of DNA damage γH2AX marker and p53 tumor suppressor protein in the KLS (cKit+ lineage− Sca1+) cells of Mysm1tm1a/tm1a and control mouse BM. MFU indicates mean fluorescence units. (C) Increased levels of reactive oxygen species (ROS) in the progenitor cells of Mysm1tm1a/tm1a BM, measured using carboxy-H2DCFDA ROS-sensitive fluorescent dye. LT-SCs indicates long-term stem cells (KLS, Flt3−CD34−); ST-SCs, short-term stem cells (KLS, Flt3−CD34+); MPPs, multipotent progenitors (KLS, Flt3+CD34+). (D) Increased levels of ROS in Mysm1tm1a/tm1a KLS cells in the BM chimeras. (E) Increase in apoptosis (annexin V+ 7AAD+/−) and necrosis (annexin V− 7AAD+) in Mysm1tm1a/tm1a KLS cells in the BM chimeras. All bars represent means ± SEM; *P < .05, **P < .01, ***P < .001 using (A,B,D) t test or (C,E) ANOVA. (F) Model of the mechanisms of hematopoietic dysfunction in Mysm1tm1a/tm1a mice: (i) ROS induce oxidative damage of DNA as well as other cellular components, and (ii) such damage leads to activation and stabilization of p53, (iii) which is a major inducer of cell apoptosis. (iv) p53 also induces production of ROS, and ROS are implicated in mediating cell-death downstream of p53. (v) Finally, DNA damage and oxidative stress may induce cell death via p53-independent mechanisms.
Genetic instability, oxidative stress, and elevated p53 protein levels in Mysm1tm1a/tm1a hematopoietic progenitors. (A) Increased frequency of micronucleated cells in the normochromatic erythrocyte (NCE) cell population of Mysm1tm1a/tm1a mouse blood; representative flow cytometric plots and data quantification are presented. (B) Levels of DNA damage γH2AX marker and p53 tumor suppressor protein in the KLS (cKit+ lineage− Sca1+) cells of Mysm1tm1a/tm1a and control mouse BM. MFU indicates mean fluorescence units. (C) Increased levels of reactive oxygen species (ROS) in the progenitor cells of Mysm1tm1a/tm1a BM, measured using carboxy-H2DCFDA ROS-sensitive fluorescent dye. LT-SCs indicates long-term stem cells (KLS, Flt3−CD34−); ST-SCs, short-term stem cells (KLS, Flt3−CD34+); MPPs, multipotent progenitors (KLS, Flt3+CD34+). (D) Increased levels of ROS in Mysm1tm1a/tm1a KLS cells in the BM chimeras. (E) Increase in apoptosis (annexin V+ 7AAD+/−) and necrosis (annexin V− 7AAD+) in Mysm1tm1a/tm1a KLS cells in the BM chimeras. All bars represent means ± SEM; *P < .05, **P < .01, ***P < .001 using (A,B,D) t test or (C,E) ANOVA. (F) Model of the mechanisms of hematopoietic dysfunction in Mysm1tm1a/tm1a mice: (i) ROS induce oxidative damage of DNA as well as other cellular components, and (ii) such damage leads to activation and stabilization of p53, (iii) which is a major inducer of cell apoptosis. (iv) p53 also induces production of ROS, and ROS are implicated in mediating cell-death downstream of p53. (v) Finally, DNA damage and oxidative stress may induce cell death via p53-independent mechanisms.
There was also a significant increase in the levels of ROS in the early hematopoietic progenitors of Mysm1tm1a/tm1a BM (Figure 7C), suggesting that the DNA damage and hematopoietic dysfunction in Mysm1tm1a/tm1a mice are linked to oxidative stress. Mysm1tm1a/tm1a KLS cells expressed higher levels of antioxidant peroxiredoxin (qRT-PCR analysis, data not shown); however, there was no increase in the activation of p38 MAPK (p38 phospho-T180/Y182, supplemental Figure 7), which is known to mediate HSC dysfunction downstream of oxidative stress in other mouse models.36 Mysm1tm1a/tm1a KLS and progenitor cells developing in the wild-type niches in chimeric animals also had elevated ROS levels and increased incidences of cell apoptosis and necrosis (Figure 7D-E). Overall, these data linked the hematopoietic phenotype of Mysm1tm1a/tm1a mice to increased DNA damage, oxidative stress, and impaired survival of Mysm1tm1a/tm1a hematopoietic progenitors.
Discussion
In this work, a novel knockout mouse line targeted for an H2A-DUB chromatin-modifying factor Mysm1 was characterized, revealing the critical role of Mysm1 in hematopoiesis. Mysm1tm1a/tm1a mice exhibited impaired HSC function and severe defects in lymphoid and erythroid lineage differentiation. Importantly, Mysm1tm1a/tm1a progenitor cells had elevated levels of ROS, γH2AX DNA damage marker, and p53 tumor suppressor protein, and impaired cell viability. Overall, this established the critical role for Mysm1 in HSC function and the controls of genetic stability and oxidative stress in the hematopoietic system.
The Mysm1 gene expression and the phenotypic defects in the Mysm1tm1a/tm1a mouse line were restricted to specific systems and tissues, with limited global alterations in cell function and changes in gene expression patterns. This indicated that Mysm1 activities are likely restricted to certain cell types, with other H2A-DUBs performing more global housekeeping roles. For example, Usp16 is proposed to perform H2A deubiquitination during mitosis,27 and BAP1 regulates Hox-gene expression in Drosophila development.25 A more specialized role of Mysm1 is also supported by the fact that its orthologs are found only in vertebrates, while other H2A-DUBs are more widely conserved; for example, the Usp22 orthologue Ubp8 is well characterized in yeast. Overall, based on the physiologic and transcriptional changes in the Mysm1tm1a/tm1a mouse line, we identified several tissues and cell types where Mysm1 is functionally active, including particularly the hematopoietic system.
To gain further insight into the mechanisms regulating Mysm1 expression in the hematopoietic system and to explore functional interactions with other hematopoietic transcription factors, ENCODE ChIP datasets were searched for transcription factors binding in the proximity of the Mysm1 gene.37 This provided evidence that the Mysm1 promoter is targeted by several key hematopoietic transcription factors (supplemental Table 5, search limited to hematopoietic cell types). These included the master regulator of B-cell lineage commitment Pax5,1 lymphoid transcription factors Pou2f2 and Tcf12, as well as c-myc, Pu.1, and p65 NFκB. Disruptions in the activities of these factors have been linked to hematopoietic phenotypes with similarities to those exhibited by Mysm1tm1a/tm1a mice (supplemental Table 5, phenotype ontology terms included). Furthermore, recently published datasets of the genome-wide binding sites of 10 key hematopoietic transcriptional regulators indicated that Scl, Pu.1, Erg, Meis1, and Gfi1b all target the Mysm1 gene.38 Overall, this suggests that Mysm1 is induced as part of a network of previously characterized hematopoietic transcriptional regulators, and works together with them to maintain the programs of blood cell differentiation in the BM.
The hematopoietic dysfunction in Mysm1tm1a/tm1a mice was associated with increased ROS, DNA damage, and elevated levels of p53 protein in the hematopoietic progenitors; a proposed model for how these factors interact to cause the loss of Mysm1tm1a/tm1a hematopoietic progenitors is shown in Figure 7F. Importantly, all of these factors, including DNA damage,34,35,39 elevated oxidative stress,40-42 and p53 activation,43 have been demonstrated to cause hematopoietic failure across a range of other mouse models. In particular, elevated ROS were linked to hematopoietic defects in the mice deficient in Bmi-1, a component of histone H2A-ubiquitinase complex PRC1.40 All this strongly suggests that these factors (Figure 7F) are also driving the hematopoietic dysfunction in Mysm1tm1a/tm1a mice.
The exact mechanisms through which Mysm1 limits DNA damage and oxidative stress to maintain BM function are likely complex. First, they may include Mysm1-mediated transcriptional regulation of genes involved in the antioxidant responses, DNA repair, and/or DNA damage response signaling. Second, the possibility of direct interactions between Mysm1 and DNA damage response mediators needs to be highlighted. In particular, histone acetylase pCAF that forms a complex with Mysm126 was previously shown to colocalize to p53-bound promoters, acetylate p53, and stimulate p53 transcriptional activity.44 In addition, Mysm1 was shown to be a target of protein kinase ATM45 that is a key mediator of DNA damage response and a regulator of BM oxidative stress.41 Finally, we can also speculate that Mysm1 H2A-DUB activity may be required for normal execution of DNA repair. Indeed, mono- and poly-ubiquitination of histones H2A and H2AX occur at sites of DNA damage, particularly at DNA breaks, and other H2A-DUBs Usp346 and Usp1647 have been implicated in DNA repair and in transcriptional regulation at the sites of DNA damage. Although inevitably some unanswered questions remain, this study established the role of Mysm1 as a novel regulator of BM stem cell function, oxidative stress, and genetic stability in the hematopoietic system.
The hematopoietic phenotype of the Mysm1tm1a/tm1a mouse line to some extent mimics the symptoms of human aplastic anemia and in particular the 5q− syndrome that presents with anemia, lymphopenia, and thrombocytosis. Although the molecular causes of this condition are incompletely understood, enhanced activation of p53 tumor suppressor is thought to play a role.48 However, despite these phenotypic similarities to human disorders and the reports of genetic variation in MYSM1 in the human population (summarized in supplemental Table 6), no roles for MYSM1 in human disorders of hematopoiesis have been documented to date. MYSM1 genomic region showed association with renal hypodysplasia and diabetic retinopathy,49,50 although similar phenotypes were not seen in the Mysm1tm1a/tm1a mouse line. MYSM1 was also implicated in maintaining abnormal transcriptional programs in prostate cancer cell lines, and this was associated with reduced levels of H2A-K119u in prostate tumors.26 Overall, the roles of MYSM1 in human disorders, in particular of the immune and hematopoietic systems, merit further investigation.
In summary, the current work characterized the first mouse line targeted for an H2A-DUB chromatin-modifying factor Mysm1. The work demonstrated the critical role for Mysm1 in the hematopoietic process, advancing our understanding of the transcriptional and chromatin remodeling machinery of mammalian BM.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors gratefully acknowledge the contribution of the Sanger Institute Research Support Facility and Mouse Informatics Group. Sanger Institute Microarray Facility members who contributed to this work are Peter Ellis, Robert Andrews, Chris McGee, Naomi Hammond, and Ruben Bautista. The authors also thank Prof Steve P. Jackson for helpful discussions.
This work was supported by The Wellcome Trust, European Mouse Disease Clinic (EUMODIC), and Genome Canada. A.N. was a recipient of fellowships from the Canadian Institutes for Health Research and the Michael Smith Foundation. R.E.W.H. is a recipient of a Canada Research Chair in Health and Genomics.
Wellcome Trust
Authorship
Contribution: A.N., S.C., C.H., C.R., R.E.M., A.R.E., D.G., and D.J.A. performed the experiments; A.N., S.C., R.E.W.H., and G.D. supervised the work; A.N., R.E.W.H., and G.D. wrote the manuscript; C.H. and K.Y. derived Mysm1tm1a/tm1a ES cells; L.M., C.P., M.L., and J.E. phenotyped Mysm1tm1a/tm1a mice under the scope of the Sanger Mouse Genetics Project; and N.A., R.R.-S., and J.K.W. managed Mysm1tm1a/tm1a mouse production and phenotyping as part of the Sanger Mouse Genetics Project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anastasia Nijnik, Department of Physiology, McGill University, H3G 0B1 Montreal, QC, Canada; e-mail: anastasiya.nyzhnyk@mcgill.ca.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal