Abstract
Myeloid cell leukemia-1 (Mcl-1) protein is an anti-apoptotic Bcl-2 family protein that plays essential roles in multiple myeloma (MM) survival and drug resistance. In MM, it has been demonstrated that proteasome inhibition can trigger the accumulation of Mcl-1, which has been shown to confer MM cell resistance to bortezomib-induced lethality. However, the mechanisms involved in this unwanted Mcl-1 accumulation are still unclear. The aim of the present study was to determine whether the unwanted Mcl-1 accumulation could be induced by the unfolded protein response (UPR) and to elucidate the role of the endoplasmic reticulum stress response in regulating Mcl-1 expression. Using quantitative RT-PCR and Western blot, we found that the translation of activating transcription factor-4 (ATF4), an important effector of the UPR, was also greatly enhanced by proteasome inhibition. ChIP analysis further revealed that bortezomib stimulated binding of ATF4 to a regulatory site (at position −332 to −324) at the promoter of the Mcl-1 gene. Knocking down ATF4 was paralleled by down-regulation of Mcl-1 induction by bortezomib and significantly increased bortezomib-induced apoptosis. These data identify the UPR and, more specifically, its ATF4 branch as an important mechanism mediating up-regulation of Mcl-1 by proteasome inhibition.
Introduction
Multiple myeloma (MM), an incurable hematologic malignancy, is characterized by clonal proliferation of malignant plasma cells in the BM, leading to one or more clinical manifestations of bone destruction, anemia, hypercalcemia, and renal insufficiency.1 MM comprises approximately 1% of all cancers, but it is the second most common hematologic cancer after lymphoma and accounts for 13.4% of all hematologic malignancies diagnosed, 19% of all deaths resulting from hematologic malignancies, and 2% of all cancer deaths. In the past decade, the outcome of patients with MM has dramatically improved due to the introduction of new and more effective treatments, the wider use of high-dose therapy, and better appreciation of potential complications and their management. The new drugs thalidomide, lenalidomide, and bortezomib have improved overall responses and response duration, progression-free survival, and overall survival for patients with newly diagnosed, relapsed, and refractory MM.2,3 Bortezomib (PS-341, Velcade) is a first-in-class boronic acid dipeptide small molecule that reversibly inhibits the proteasome, which has a critical role in such diverse processes as cell-cycle regulation and Ag processing. The efficacy of bortezomib in the treatment of MM has clearly demonstrated the role of proteasome inhibitors in this disease; however, some patients do not respond and others may relapse or become refractory. Clearly, the mechanisms mediating resistance to bortezomib are still not fully understood.4,5
Myeloid cell leukemia-1 (Mcl-1) is an anti-apoptotic protein of the Bcl-2 family. Among Bcl-2 family members, Mcl-1 distinguishes itself by its ability to oppose several apoptotic stimuli. Mcl-1 protects tumor cells against spontaneous and chemotherapy-induced apoptosis. In MM, it was demonstrated that Mcl-1, rather than Bcl-2 or Bcl-xl, plays essential roles in promoting MM cell survival.6-8 Specific down-regulation or repression of Mcl-1 has been proposed to initiate apoptosis in MM.9,10 The decrease of Mcl-1 caused by cleavage is tightly associated with the induction of apoptosis by bortezomib in MM.11,12 In contrast, it was speculated that proteasome inhibition could prevent Mcl-1 turnover and accordingly decrease the rate of apoptosis.13 Accumulated and cleaved Mcl-1 products by proteasome inhibition have either a pro- or an anti-apoptotic function, highlighting the complexity and pivotal role of Mcl-1 regulation. Until now, the mechanism of Mcl-1 accumulation by proteasome inhibition was still not clear.
MM is a cancer of Ab-producing plasma cells, which continue to secrete immunoglobulins in a normal fashion but exhibit uncontrolled growth in the BM. Proteins are folded properly in the endoplasmic reticulum (ER). In the secretory pathway, protein synthesis at the ER occurs concomitantly with translocation, modification, and folding with the assistance of molecular chaperones. When proteins are misfolded, there are 2 branches of ER quality control that address the situation.14 One is the ER-associated degradation pathway to dispose of misfolded proteins, which involves retro-translocation, polyubiquitination, and degradation in the cytosol through the 26S proteasome. A second major response is called the unfolded protein response (UPR), which refers to the transcriptional up-regulation of genes that are thought to enable the cell to cope with and fold misfolded proteins.15 Various stress stimuli such as hypoxia, ischemia, and starvation interfere with ER function, causing ER stress, which is defined by the accumulation of unfolded proteins in the ER. As a selective and potent inhibitor of the 26S proteasome, the inhibition of the 26S proteasome by bortezomib leads to the accumulation of misfolded proteins, resulting in ER stress followed by a coordinated cellular UPR. Inherently, extensive immunoglobulin production sensitizes myeloma cells for proteasome inhibition.16,17
In the present study, we hypothesized that bortezomib-induced unwanted Mcl-1 accumulation was mediated by the UPR. To elucidate the role of ER stress in regulating Mcl-1 expression in MM, we systematically compared the expression of Mcl-1 and associated molecules of the key UPR arms at both the protein and mRNA levels after treatment with bortezomib. We also investigated the mechanisms of regulation of Mcl-1 expression and explored the role of transcription activating factor-4 (ATF4) in this process. Our data demonstrate that bortezomib-induced Mcl-1 up-regulation is mediated through ATF4 and clarify one of the molecular mechanisms for bortezomib resistance in MM.
Methods
Cells and cell culture
The human MM cell lines RPMI-8226, U266, OPM2, LP1, and MMS1 and the murine MM cell line 5T33vt were used in this study. These cell lines were maintained in RPMI 1640 medium (Lonza) supplemented with 10% heat-inactivated FBS (HyClone), l-glutamine (2mM) and antibiotics (penicillin 100 U/mL and streptomycin 50 μg/mL; Lonza).
Reagents and Abs
The polyclonal Abs used were: rabbit anti–beta-actin, rabbit anti–Mcl-1, rabbit anti–Bcl-2, rabbit anti–Bcl-xl, anti–caspase-3, and HRP-conjugated goat anti–rabbit (all from Cell Signaling Technology); rabbit anti-CCAAT/enhancer-binding protein homologous protein (anti-CHOP), rabbit anti-ATF4, rabbit anti–GRP-78, rabbit anti–p-eIF2α, XBP1, and ATF6α (all from Santa Cruz Biotechnology); and rabbit anti-ATF3 (Novus Biologicals). Bortezomib and tunicamycin were bought from Millennium Pharmaceuticals and Enzo Life Sciences, respectively; the pan-caspase inhibitor Z-VAD-FMK was from R&D Systems. All drugs were dissolved in DMSO and aliquots of stock solutions were stored at −20°C. For the experiments, drugs were diluted to the appropriate concentrations and the final concentration of DMSO was < 0.01% in the medium.
Western blot analysis
Western blot analysis was performed as described previously.18 Briefly, cells were lysed in 10mM Tris-HCl (pH 7.05), 50mM NaCl, 50mM NaF, 30mM sodium pyrophosphate, 1% Triton X-100, 5μM ZnCl2, 100μM Na3VO4, 1mM DTT, 20mM β-glycerophosphate, 20mM p-nitrophenol phosphate, 20 μg/mL of aprotinin, 2.5 μg/mL of leupeptin, 0.5mM PMSF, 0.5mM benzamidine, 5 μg/mL of pepstatin, and 50nM okadaic acid. Lysates were resolved on 10% or 12.5% SDS-PAGE and transferred to a nitrocellulose membrane. Membranes were blocked for 1 hour at room temperature with 5% nonfat milk, and then incubated with primary Ab followed by a HRP-conjugated secondary Ab. The protein band was detected using the Western Lightning Plus-ECL detection system (PerkinElmer). Densitometric analyses were performed using National Institutes of Health ImageJ Version 1.45 software.
Quantitative real-time PCR (qRT-PCR)
RNA extraction was performed using the RNeasy kit (QIAGEN). RNA was analyzed using an Agilent 2100 bioanalyzer. Total RNA was reverse transcribed using the Verso cDNA synthesis kit (Thermo Scientific) according to the manufacturer's instructions. qRT-PCR was performed with Maxima SYBR Green/ROX qPCR Master Mix (Fermantas) on an ABI Prism 7900 Fast instrument using gene-specific primers (primer sequences can be found in Table 1). The thermal cycling conditions included 2 minutes at 50°C and 10 minutes at 95°C, followed by 40 cycles of 95°C for 0.15 minutes and 60°C for 1 minute. Ct values were collected for β-actin and the genes of interest during the log phase of the cycle. Quantification of given genes expressed as the mRNA level was normalized to β-actin RNA using the ΔΔCt method.
Primer sequences used for qRT-PCR
| Gene symbol . | GenBank accession no. . | Primer set sequence (5′-> 3′) . | Amplicon size, bp . |
|---|---|---|---|
| Beta-actin (h) | NM_001101 | Forward: ATCGTGCGTGACATTAAGGAGAAG Reverse: AGGAAGGAAGGCTGGAAGAGTG | 179 |
| BCL-2 (h) | NM_000633 | Forward: TTCAACACAGACCCACCCAGAG Reverse: GCAGGATAGCAGCACAGGATTG | 276 |
| BCL-XL (h) | NM_138578 | Forward: GCAGCCGAGAGCCGAAAGG Reverse: GGATGTGGTGGAGCAGAGAAGG | 161 |
| MCL-1 (h) | NM_021960 | Forward: AAGAGGCTGGGATGGGTTTGTG Reverse: TTGGTGGTGGTGGTGGTTGG | 178 |
| ATF3 (h) | NM_001674 | Forward: AGCAGCAGAGAACCATCAAGGC Reverse: GAAAGGCGGGCAGGACACC | 184 |
| ATF4 (h) | NM_001675 | Forward: TCCGAATGGCTGGCTGTGG Reverse: AGTGTAGTCTGGCTTCCTATCTCC | 420 |
| ATF6 (h) | NM_007348 | Forward: AGTTCGTCAGTTCCTCCTTACCTC Reverse: AGTTCTCTGCCTGCCACCAAG | 226 |
| CHOP (h) | NM_004083 | Forward: TGCTTCTCTGGCTTGGCTGAC Reverse: CCGTTTCCTGGTTCTCCCTTGG | 145 |
| GRP-78 (h) | NM_005347 | Forward: AGGAGGAGGACAAGAAGGAGGAC Reverse: CAGGAGTGAAGGCGACATAGGAC | 156 |
| Beta-actin (m) | NM_007393 | Forward: GCGACAGCAGTTGGTTGGAG Reverse: TTTGGGAGGGTGAGGGACTTC | 165 |
| Bcl-2 (m) | NM_177410 | Forward: CTGGTTGAATGAGTCTGGGCTTTG Reverse: AGTGTTGGAGGTCTGGTGCTTAC | 337 |
| Bcl-xl (m) | NM_009743 | Forward: ACTGTGGCTGGTGTGGTTCTG Reverse: AGTGGTTCTCCTGGTAGCAATGG | 139 |
| Mcl-1 (m) | NM_008562 | Forward: GCGTGTTATGCTCCCAGTTCC Reverse: TGCCAATCCAAGAATGCCAATCC | 240 |
| Atf3 (m) | NM_007498 | Forward: GTGGAGCAGGCAGGAGCATC Reverse: CTTGGAAGGGCGTTGTCAGC | 204 |
| Atf4 (m) | NM_009716 | Forward: GCCTGACTCTGCTGCTTACATTAC Reverse: CCCTTGCCTTACGGACCTCTTC | 452 |
| Atf6 (m) | NM_001081304 | Forward: CAGAGGCAGCACACGCATTC Reverse: GGTCAGCAGGAGCAGAGAAGG | 279 |
| Chop (m) | NM_007837 | Forward: CATCTTCATACACCACCACACCTG Reverse: CTCGTTCTCCTGCTCCTTCTCC | 432 |
| Grp-78 (m) | NM_022310 | Forward: GTCTGCTTCGTGTCTCCTCCTG Reverse: TCCTCCTTCTTGTCCTCCTCCTC | 220 |
| Gene symbol . | GenBank accession no. . | Primer set sequence (5′-> 3′) . | Amplicon size, bp . |
|---|---|---|---|
| Beta-actin (h) | NM_001101 | Forward: ATCGTGCGTGACATTAAGGAGAAG Reverse: AGGAAGGAAGGCTGGAAGAGTG | 179 |
| BCL-2 (h) | NM_000633 | Forward: TTCAACACAGACCCACCCAGAG Reverse: GCAGGATAGCAGCACAGGATTG | 276 |
| BCL-XL (h) | NM_138578 | Forward: GCAGCCGAGAGCCGAAAGG Reverse: GGATGTGGTGGAGCAGAGAAGG | 161 |
| MCL-1 (h) | NM_021960 | Forward: AAGAGGCTGGGATGGGTTTGTG Reverse: TTGGTGGTGGTGGTGGTTGG | 178 |
| ATF3 (h) | NM_001674 | Forward: AGCAGCAGAGAACCATCAAGGC Reverse: GAAAGGCGGGCAGGACACC | 184 |
| ATF4 (h) | NM_001675 | Forward: TCCGAATGGCTGGCTGTGG Reverse: AGTGTAGTCTGGCTTCCTATCTCC | 420 |
| ATF6 (h) | NM_007348 | Forward: AGTTCGTCAGTTCCTCCTTACCTC Reverse: AGTTCTCTGCCTGCCACCAAG | 226 |
| CHOP (h) | NM_004083 | Forward: TGCTTCTCTGGCTTGGCTGAC Reverse: CCGTTTCCTGGTTCTCCCTTGG | 145 |
| GRP-78 (h) | NM_005347 | Forward: AGGAGGAGGACAAGAAGGAGGAC Reverse: CAGGAGTGAAGGCGACATAGGAC | 156 |
| Beta-actin (m) | NM_007393 | Forward: GCGACAGCAGTTGGTTGGAG Reverse: TTTGGGAGGGTGAGGGACTTC | 165 |
| Bcl-2 (m) | NM_177410 | Forward: CTGGTTGAATGAGTCTGGGCTTTG Reverse: AGTGTTGGAGGTCTGGTGCTTAC | 337 |
| Bcl-xl (m) | NM_009743 | Forward: ACTGTGGCTGGTGTGGTTCTG Reverse: AGTGGTTCTCCTGGTAGCAATGG | 139 |
| Mcl-1 (m) | NM_008562 | Forward: GCGTGTTATGCTCCCAGTTCC Reverse: TGCCAATCCAAGAATGCCAATCC | 240 |
| Atf3 (m) | NM_007498 | Forward: GTGGAGCAGGCAGGAGCATC Reverse: CTTGGAAGGGCGTTGTCAGC | 204 |
| Atf4 (m) | NM_009716 | Forward: GCCTGACTCTGCTGCTTACATTAC Reverse: CCCTTGCCTTACGGACCTCTTC | 452 |
| Atf6 (m) | NM_001081304 | Forward: CAGAGGCAGCACACGCATTC Reverse: GGTCAGCAGGAGCAGAGAAGG | 279 |
| Chop (m) | NM_007837 | Forward: CATCTTCATACACCACCACACCTG Reverse: CTCGTTCTCCTGCTCCTTCTCC | 432 |
| Grp-78 (m) | NM_022310 | Forward: GTCTGCTTCGTGTCTCCTCCTG Reverse: TCCTCCTTCTTGTCCTCCTCCTC | 220 |
h indicates human; and m, mouse.
XBP1 splicing assay
The region of X-box binding protein 1 (XBP1) containing the splice junction was amplified by PCR using the XBP1 primers (Table 2) with the cycling conditions of 94°C for 5 minutes, followed by 40 cycles of 94°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds. The PCR products were analyzed on 1.2% agarose gel.
Primer sequences used for RT-PCR analysis of XBP1 splicing
| Gene symbol . | Primer sequence (5′-> 3′) . | Amplicon size, bp . |
|---|---|---|
| Xbp1 (h) | Forward: TTGCTGAAGAGGAGGCGGAAG Reverse: GGTCCAAGTTGTCCAGAATGC | Unspliced (210)/spliced (184) |
| Xbp1 (m) | Forward: GAACACGCTTGGGAATGGACAC Reverse: AGAAAGGGAGGCTGGTAAGGAAC | Unspliced (343)/spliced (327) |
| Gene symbol . | Primer sequence (5′-> 3′) . | Amplicon size, bp . |
|---|---|---|
| Xbp1 (h) | Forward: TTGCTGAAGAGGAGGCGGAAG Reverse: GGTCCAAGTTGTCCAGAATGC | Unspliced (210)/spliced (184) |
| Xbp1 (m) | Forward: GAACACGCTTGGGAATGGACAC Reverse: AGAAAGGGAGGCTGGTAAGGAAC | Unspliced (343)/spliced (327) |
h indicates human; and m, mouse.
Transient transfection of siRNA
U266, RPMI-8226, and MMS1 cells were transiently transfected with ATF4 (sc-35112) or ATF6 (sc-37699), XBP1 siRNA (sc-38627; Santa Cruz Biotechnology) using the Amaxa Nucleofector II (Lonza) according to the manufacturer's instructions. Scrambled siRNA (sc-37007; Santa Cruz Biotechnology) was used as a mock control. Briefly, 2 × 106 MM cells were resuspended in 100 μL of R Nucleofector solution containing 10μM siRNA and electroporated with the X-005 (U266) and T-005 (RPMI-8226, MMS1) Nucleofector program, respectively. Cells were transferred to culture plates and cultured at 1 × 106 cells/mL for 3 hours before incubation with bortezomib. Sixteen to 20 hours after electroporation, cells were subjected to RNA isolation and Western blotting in the presence or absence of bortezomib.
Detection of apoptotic cells
After exposure to bortezomib and/or knockdown, apoptotic cells were evaluated using annexin V–FITC apoptosis detection kit (BD Biosciences). Briefly, 1 × 106 cells were washed twice with PBS and stained with 5 μL of 7-amino-actinomycin D and 5 μL of annexin V–FITC in 100 μL of binding buffer, and incubated at 4°C for 15 minutes. Cells were then resuspended in 400 μL of binding buffer and immediately analyzed using a FACScan flow cytometer (BD Biosciences); 10 000 events were recorded and analyzed.
Bioinformatic analysis
Detection of transcription factor binding consensus sequences was carried out using the web-based MATCH program (http://www.gene-regulation.com/cgi-bin/pub/programs/match/bin/match.cgi) that uses the publicly available TRANSFAC 6.0 database (http://www.gene-regulation.com/cgi-bin/pub/databases/transfac/search.cgi).19
ChIP assays
ChIP assays were performed using the protocol for the SimpleChIP Enzymatic Chromatin IP Kit (Magnetic Beads; Cell Signaling Technology). Briefly, 4 × 107 LP1 cells for each experiment (treated or untreated with 50nM bortezomib) were cross-linked by the addition of 37% formaldehyde into the medium at a final concentration of 1% and incubation for 10 minutes at room temperature. Cross-linking was stopped by the addition of glycine to a final concentration 0.125M and incubation for 5 minutes at room temperature. Cells were then washed with ice-cold PBS + PMSF, the cell pellet was resuspended in 10 mL of ChIP lysis buffer (buffer A) with protease inhibitors, and incubated on ice for 10 minutes. Pelleted nuclei were resuspended in 1 mL of buffer B supplemented with DTT and digested with micrococcal nuclease for 20 minutes at 37°C to produce 150- to 900-bp DNA fragments. The digestion was stopped by adding 100 μL of 0.5M EDTA solution. After washing, the digested nuclei were then diluted in 1 mL of ChIP dilution buffer, followed by immunoprecipitation with Ab against ATF4 (5 μg) or control IgG (5 μg) at 4°C overnight with rotation. Each immunoprecipitation contained 500 μL of digested nuclei, and 10 μL of digested nuclei were removed to be used as 2% input. Immune complexes were collected with 30 μL of ChIP grade protein G magnetic beads and washed with low-salt and high-salt washing buffers. Next, immune complexes were eluted using freshly prepared elution buffer. Cross-links were reversed by heating at 65°C in the presence of NaCl followed by proteinase K treatment. The DNA was purified with ChIP DNA Clean and Concentrator kit (Zymo Research). ChIP DNA (1 μL) was next used as a template for quantitative real-time PCR using primer pairs, as shown in Table 3.
Primer sequences used for ChIP analysis
| Primer set no. . | Primer sequence (5′-> 3′) . | Location . | Amplicon size, bp . |
|---|---|---|---|
| MCL-1 #1 | Forward: ACAGAGGTAGCCACGAGAAGG Reverse: TGGAAGGAAGCGGAAGTGAGAAG | promoter −423 promoter −56 | 367 |
| MCL-1 #2 | Forward: CCAACATCGTGAAAACCCCATCTC Reverse: GCCTCCCTGCTTGCTTCTCC | promoter −1033 promoter −731 | 302 |
| MCL-1 #3 | Forward: CCAAGCCAAGCCATCGCATC Reverse: TGGGATAGGTGGTGAAGTTGAGAG | promoter −1568 promoter −1296 | 272 |
| Primer set no. . | Primer sequence (5′-> 3′) . | Location . | Amplicon size, bp . |
|---|---|---|---|
| MCL-1 #1 | Forward: ACAGAGGTAGCCACGAGAAGG Reverse: TGGAAGGAAGCGGAAGTGAGAAG | promoter −423 promoter −56 | 367 |
| MCL-1 #2 | Forward: CCAACATCGTGAAAACCCCATCTC Reverse: GCCTCCCTGCTTGCTTCTCC | promoter −1033 promoter −731 | 302 |
| MCL-1 #3 | Forward: CCAAGCCAAGCCATCGCATC Reverse: TGGGATAGGTGGTGAAGTTGAGAG | promoter −1568 promoter −1296 | 272 |
Statistical analysis
All data presented are derived from at least 3 independent determinations unless otherwise noted. A 2-tailed paired t test was used for paired samples with Prism Version 5.0 software (GraphPad). P < .05 was considered statistically significant. For Western blotting, data are representative of 3 independent experiments.
Results
Bortezomib induces up-regulation of Mcl-1 in MM
For these studies, we used bortezomib at concentrations lower than their respective IC50 for each cell line. The IC50 for each cell line were as follows: OPM2, 8.1nM; U266, 5.7nM; 5T33vt, 6.8nM; MMS1, 33.5nM; RPMI-8226, 58.0nM; LP1, 201.8nM (as measured using Cell-Titer Glo). As shown in Figure 1A, bortezomib significantly increased the protein level of full-length of Mcl-1L(s) in human myeloma cell lines and in Mcl-1L/v murine 5T33vt cells; however, changes in the levels of Bcl-2 and Bcl-xl were negligible. In contrast, bortezomib also time-dependently induced the cleaved Mcl-1 fragments in bortezomib-sensitive RPMI-8226 and 5T33vt cells other than bortezomib-resistant LP1 cells, in which pro-apoptotic BH3 protein Bim is absent (data not shown). To address the contribution of caspases to the mechanism of Mcl-1 down-regulation, we used the pan-caspase inhibitor Z-VAD-FMK. When OPM2 cells were pretreated with 50μM Z-VAD-FMK, along with the decrease of active caspase-3, the formation of cleaved Mcl-1 fragments was partially inhibited, and accordingly the full-length Mcl-1L and Mcl-1s were increased (Figure 1B). To further clarify the mechanism by which Mcl-1 is accumulated on treatment with bortezomib, we compared the MCL-1, BCL-2, and BCL-XL mRNA expression levels in different MM cell lines treated with bortezomib (Figure 1C-E). Bortezomib treatment led to a significant increase of MCL-1 in all tested human and murine MM cell lines as measured by quantitative RT-PCR. The expression levels of BCL-2 and BCL-XL were not significantly disturbed (P > .05).
Western blot and qRT-PCR analysis of Mcl-1, Bcl-2, and Bcl-xl expression in MM cells during bortezomib treatment. (A) Top panel: Western blot analysis of bortezomib-treated (for 0, 6, 12, and 24 hours) human LP1 and RPMI-8226 cells and murine 5T33vt cells. The blots were probed with Mcl-1, Bcl-2, Bcl-xl, and β-actin Abs. The concentrations of bortezomib used in human LP1 and RPMI-8226 and murine 5T33vt cells were 50, 10, and 5nM, respectively. The data are representative of 3 independent experiments. Bottom panel: amounts of Mcl-1, Bcl-2, and Bcl-xl were quantified by densitometric analysis of Western blot using ImageJ Version 1.45 software. (B) The pan caspase inhibitor Z-VAD-FMK abrogates the cleavage of Mcl-1. Left panel: Western blot analysis of the Mcl-1 and caspase-3 expression in OPM2 cells. OPM2 cells were untreated (control) or were treated with 5nM bortezomib for 16 hours with or without 2 hours of pretreatment with 50μM Z-VAD-FMK as indicated. Right panel: densitometric analysis of Western blots. (C-E) qRT-PCR analysis of MCL-1, BCL-2, and BCL-XL expression at the mRNA level. The MM cell lines RPMI-8226 (10nM), MMS1 (10nM), U266 (5nM), LP1 (100nM), OPM2 (5nM), and 5T33vt (2.5nM) were treated with bortezomib for 24 hours. RNA was isolated and subjected to qRT-PCR using primers for MCL-1, BCL-2, BCL-XL, and β-actin. Beta-actin was used as an internal control. n = 3 in all experiments. *P < .05 and **P < .01 versus vehicle-treated samples. The columns show the mean results representative of 3 similar experiments; the bars indicate SD.
Western blot and qRT-PCR analysis of Mcl-1, Bcl-2, and Bcl-xl expression in MM cells during bortezomib treatment. (A) Top panel: Western blot analysis of bortezomib-treated (for 0, 6, 12, and 24 hours) human LP1 and RPMI-8226 cells and murine 5T33vt cells. The blots were probed with Mcl-1, Bcl-2, Bcl-xl, and β-actin Abs. The concentrations of bortezomib used in human LP1 and RPMI-8226 and murine 5T33vt cells were 50, 10, and 5nM, respectively. The data are representative of 3 independent experiments. Bottom panel: amounts of Mcl-1, Bcl-2, and Bcl-xl were quantified by densitometric analysis of Western blot using ImageJ Version 1.45 software. (B) The pan caspase inhibitor Z-VAD-FMK abrogates the cleavage of Mcl-1. Left panel: Western blot analysis of the Mcl-1 and caspase-3 expression in OPM2 cells. OPM2 cells were untreated (control) or were treated with 5nM bortezomib for 16 hours with or without 2 hours of pretreatment with 50μM Z-VAD-FMK as indicated. Right panel: densitometric analysis of Western blots. (C-E) qRT-PCR analysis of MCL-1, BCL-2, and BCL-XL expression at the mRNA level. The MM cell lines RPMI-8226 (10nM), MMS1 (10nM), U266 (5nM), LP1 (100nM), OPM2 (5nM), and 5T33vt (2.5nM) were treated with bortezomib for 24 hours. RNA was isolated and subjected to qRT-PCR using primers for MCL-1, BCL-2, BCL-XL, and β-actin. Beta-actin was used as an internal control. n = 3 in all experiments. *P < .05 and **P < .01 versus vehicle-treated samples. The columns show the mean results representative of 3 similar experiments; the bars indicate SD.
Induction of GRP-78 and CHOP, 2 key markers of ER stress, by bortezomib in MM
To further investigate the mechanism of regulating Mcl-1 expression, we next compared the effect of bortezomib on the induction of the UPR. mRNA levels of the ER stress markers GRP-78 and CHOP were significantly increased in bortezomib-treated cells (Figure 2A-B). Furthermore, we confirmed by Western blot that bortezomib elevated the GRP-78 and CHOP expression at the protein level in a time-dependent manner (Figure 2C).
Bortezomib activates the UPR in MM cells. (A-B) qRT-PCR analysis of the ER stress markers GRP-78 and CHOP expression at the mRNA level. The MM cell lines RPMI-8226 (10nM), MMS1 (10nM), LP1 (100nM), OPM2 (5nM), and 5T33vt (2.5nM) were treated with bortezomib for 24 hours. n = 3 for all experiments. *P < .05 and **P < .01 versus vehicle-treated samples. (C) Left panel: Western blot analysis of GRP-78 and CHOP expression. Whole-cell extracts were prepared from bortezomib-treated (for 0, 6, 12, and 24 hours) human LP1 and MMS1 cells and murine 5T33vt cells; the concentrations of bortezomib used were 100, 10, and 5nM, respectively. The data are representative of 3 independent experiments. Right panel: amounts of GRP-78 and CHOP were quantified by densitometric analysis of Western blot using ImageJ Version 1.45 software. (D) Western blot analysis of the UPR arm of ATF4 activation in bortezomib-treated MM cells. The doses of bortezomib were used at 10nM in both RPMI-8226 and MMS1 cells and at 2.5nM in 5T33vt cells. Left panels show the representative blots of 3 independent experiments; right panels show densitometric analysis. (E-F) qRT-PCR analysis of ATF4 and its target gene ATF3 expression in bortezomib-treated MM cells. The MM cell lines RPMI-8226 (10nM), MMS1 (10nM), LP1 (100nM), OPM2 (5nM), and 5T33vt (2.5nM) were treated with bortezomib for 24 hours. Data represent the means ± SD for 3 separate experiments. *P < .05 and ** P < .01 versus vehicle-treated samples.
Bortezomib activates the UPR in MM cells. (A-B) qRT-PCR analysis of the ER stress markers GRP-78 and CHOP expression at the mRNA level. The MM cell lines RPMI-8226 (10nM), MMS1 (10nM), LP1 (100nM), OPM2 (5nM), and 5T33vt (2.5nM) were treated with bortezomib for 24 hours. n = 3 for all experiments. *P < .05 and **P < .01 versus vehicle-treated samples. (C) Left panel: Western blot analysis of GRP-78 and CHOP expression. Whole-cell extracts were prepared from bortezomib-treated (for 0, 6, 12, and 24 hours) human LP1 and MMS1 cells and murine 5T33vt cells; the concentrations of bortezomib used were 100, 10, and 5nM, respectively. The data are representative of 3 independent experiments. Right panel: amounts of GRP-78 and CHOP were quantified by densitometric analysis of Western blot using ImageJ Version 1.45 software. (D) Western blot analysis of the UPR arm of ATF4 activation in bortezomib-treated MM cells. The doses of bortezomib were used at 10nM in both RPMI-8226 and MMS1 cells and at 2.5nM in 5T33vt cells. Left panels show the representative blots of 3 independent experiments; right panels show densitometric analysis. (E-F) qRT-PCR analysis of ATF4 and its target gene ATF3 expression in bortezomib-treated MM cells. The MM cell lines RPMI-8226 (10nM), MMS1 (10nM), LP1 (100nM), OPM2 (5nM), and 5T33vt (2.5nM) were treated with bortezomib for 24 hours. Data represent the means ± SD for 3 separate experiments. *P < .05 and ** P < .01 versus vehicle-treated samples.
The e-IF2α–ATF4 arm of the UPR was activated in bortezomib-treated MM cells
To assess the role of ATF4 in bortezomib-induced UPR, we investigated the kinetics of activation of the e-IF2α–ATF4 signaling of UPR during bortezomib treatment. Western blot analysis revealed the phosphorylation kinetics of e-IF2α in 3 MM cell lines (Figure 2D). Bortezomib markedly increased the e-IF2α phosphorylation level. Consistent with this change, expression of ATF4 protein and its downstream ATF3 translation were also increased. Moreover, we found that ATF4 mRNA was also elevated during bortezomib treatment, with the magnitude of induction greatest in response to bortezomib in all tested MM cell lines, consistent with the observed protein levels (Figure 2E). As a target gene of ATF4, ATF3 was induced in response to bortezomib, but the induction was more marked compared with ATF4 (Figure 2F).
Transcriptional up-regulation of Mcl-1 is mediated by ATF4
To predict the interaction of ATF4 with Mcl-1, we subjected Mcl-1 sequence to analysis using MATCH, a publicly available web tool that allows the detection of known transcription factor consensus-binding sequences (matrices) based on TRANSFAC datasets.20 We examined the presence of ATF4-binding sites using stringent conditions in which the core binding site similarity equaled 100% (core similarity = 1), and revealed 3 potential ATF4 consensus sites within the human MCL-1 promoter and 1 potential ATF4 consensus site within the murine Mcl-1 promoter (Figure 3A). The use of ChIP analysis to gain an understanding of in vitro transcription factor-DNA interactions in several cell systems has revolutionized the study of DNA-binding proteins and their role in regulating gene expression. To gain an understanding of the role of ATF4 in mediating high levels of Mcl-1 during bortezomib treatment, we performed ChIP analysis to validate the binding of ATF4 at the Mcl-1 promoter. As is evident in Figure 3B, ChIP analysis using anti-ATF4 was carried out in LP1 cells treated and untreated with bortezomib for 16 hours. Our data show the persistent ATF4 binding to the “−332 to −335” site of the MCL-1 promoter in both treated and untreated LP1 cells, indicating that ATF4 is involved in transcriptional up-regulation of MCL-1.
ChIP analysis showing the interaction of ATF4 with the Mcl-1 promoter. (A) Human Mcl-1 upstream region showing 3 consensus sites for ATF4. There is only 1 consensus site for ATF4 at the promoter of murine Mcl-1. The core consensus motifs are boxed. (B) ChIP analysis of ATF4 binding in bortezomib-treated LP1 cells. LP1 cells were treated with 100nM bortezomib for 16 hours. Immunoprecipitation was performed using control IgG or anti-ATF4 Ab. Samples were analyzed by qRT-PCR using primers for region −1521 to −1297, −1033 to −733, and −423 to −80 nucleotides. *P < .05 versus control IgG-immunoprecipitated samples (n = 3).
ChIP analysis showing the interaction of ATF4 with the Mcl-1 promoter. (A) Human Mcl-1 upstream region showing 3 consensus sites for ATF4. There is only 1 consensus site for ATF4 at the promoter of murine Mcl-1. The core consensus motifs are boxed. (B) ChIP analysis of ATF4 binding in bortezomib-treated LP1 cells. LP1 cells were treated with 100nM bortezomib for 16 hours. Immunoprecipitation was performed using control IgG or anti-ATF4 Ab. Samples were analyzed by qRT-PCR using primers for region −1521 to −1297, −1033 to −733, and −423 to −80 nucleotides. *P < .05 versus control IgG-immunoprecipitated samples (n = 3).
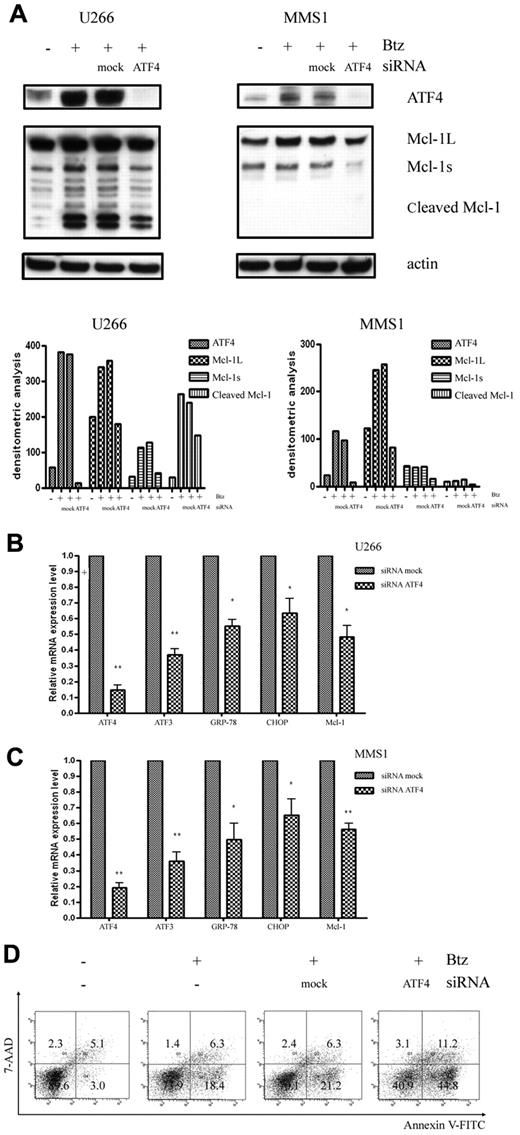
Knockdown of ATF4 renders MM cells sensitive to bortezomib-induced apoptosis
To further address the involvement of the ATF4 branch of the UPR in Mcl-1 up-regulation, we analyzed the correlation between the expression levels of ATF4 and Mcl-1. We found that knockdown of ATF4 with siRNA specifically and markedly inhibited bortezomib-induced Mcl-1 protein accumulation in U266 cells (Figure 4A). A similar down-regulation effect of ATF4 siRNA on bortezomib-induced up-regulation of Mcl-1 was also reproducibly observed in MMS1 cells (Figure 4A). Knockdown of ATF4 was paralleled by down-regulation of Mcl-1 induction, further suggesting that ATF4 could play a role in regulating cellular Mcl-1 level. In support of this notion, we further analyzed the correlation between the expression levels of ATF4 and Mcl-1 and other ATF4 downstream genes such as ATF3, GRP-78, and CHOP after knocking down ATF4 in U266 and MMS1 cells (Figure 4B-C). Similarly, knockdown of ATF4 also prevented the expression of ATF4 target gene such as ATF3, GRP-78, and CHOP, suggesting a possibility of common mechanisms for the regulation of these genes. Accumulated Mcl-1 was believed to confer MM cell protection and drug resistance in response to bortezomib treatment; however, the simultaneously induced CHOP by ATF4 favors apoptosis in response to ER stress. To clarify the effects of the knockdown of ATF4 on MM cell survival, we measured apoptosis using flow cytometry by annexin V and 7-amino-actinomycin D staining. As shown in Figure 4D, knocking down ATF4 markedly increased the number of apoptotic cells in the presence of bortezomib in U266 cells. These findings further support a prosurvival function of ATF4 in MM cells.
Knockdown of ATF4 increases sensitivity to bortezomib. (A) Western blot analysis of ATF4 and Mcl-1 expression levels in bortezomib-treated U266 cells (2.5nM for 16 hours) and MMS1 cells (5nM for 16 hours). Top panels show the representative blots of 3 independent experiments; bottom panels show densitometric analysis. (B-C) qRT-PCR measurement of ATF4 downstream gene-expression levels after knockdown ATF4 in the presence of bortezomib in U266 cells (2.5nM for 16 hours; B) and MMS1 cells (5nM for 16 hours). n = 3 for all experiments. *P < .05 and **P < .01 versus si-mock samples. Data represent the means ± SD. (D) Knockdown of ATF4 induces apoptosis in U266 cells. ATF4 knockdown or control U266 cells were treated with 2.5nM bortezomib for 16 hours. Bortezomib was added in the medium 4 hours after electroporation-mediated RNAi. Apoptosis of each sample was examined by flow cytometry after annexin V–FITC/7-amino-actinomycin D staining. FACS data shown are representative of 3 independent experiments.
Knockdown of ATF4 increases sensitivity to bortezomib. (A) Western blot analysis of ATF4 and Mcl-1 expression levels in bortezomib-treated U266 cells (2.5nM for 16 hours) and MMS1 cells (5nM for 16 hours). Top panels show the representative blots of 3 independent experiments; bottom panels show densitometric analysis. (B-C) qRT-PCR measurement of ATF4 downstream gene-expression levels after knockdown ATF4 in the presence of bortezomib in U266 cells (2.5nM for 16 hours; B) and MMS1 cells (5nM for 16 hours). n = 3 for all experiments. *P < .05 and **P < .01 versus si-mock samples. Data represent the means ± SD. (D) Knockdown of ATF4 induces apoptosis in U266 cells. ATF4 knockdown or control U266 cells were treated with 2.5nM bortezomib for 16 hours. Bortezomib was added in the medium 4 hours after electroporation-mediated RNAi. Apoptosis of each sample was examined by flow cytometry after annexin V–FITC/7-amino-actinomycin D staining. FACS data shown are representative of 3 independent experiments.
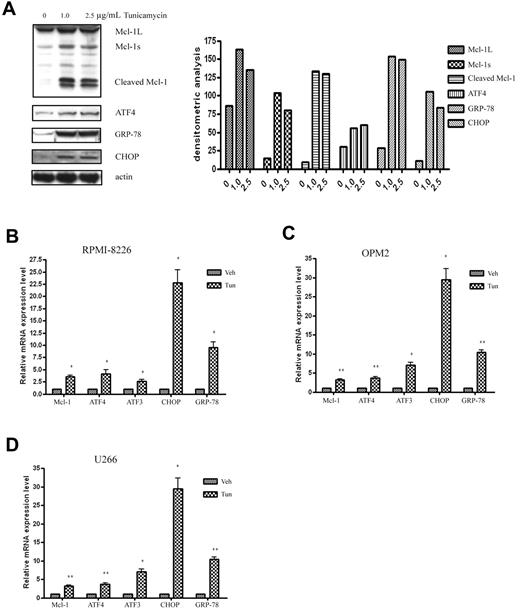
Mcl-1 gene expression is up-regulated in response to the ER stress inducer tunicamycin
To further confirm that the up-regulation of Mcl-1 depends on UPR activation, we used tunicamycin, a nonselective UPR inducer, to treat MM cells and investigated the association between the expression of Mcl-1 and the UPR components. As shown in Figure 5A, consistent with the results of bortezomib-treated MM cells, tunicamycin induced Mcl-1 accumulation and cleavage at the protein level. Moreover, tunicamycin also up-regulated MCL-1 transcription at the mRNA level in all tested RPMI-8226 (Figure 5B), OPM2 (Figure 5C), and U266 (Figure 5D) cells. The induced MCL-1 paralleled the high expression levels of the UPR activation components CHOP, GRP-78, and ATF4 and its target gene ATF3. These results suggest that Mcl-1 up-regulation is tightly associated with UPR activation.
The ER inducer tunicamycin increases Mcl-1 expression and triggers UPR activation in MM cells. (A) Tunicamycin treatment induces Mcl-1 accumulation and cleavage in OPM2 cells. OPM2 cells were treated with different doses of tunicamycin for 24 hours. Cell extracts were probed with Mcl-1, ATF4, GRP-78, CHOP, and β-actin pAbs. Left panel shows the representative blots of 3 independent experiments; right panel shows densitometric analysis. (B-D) qRT-PCR analysis of the expression of Mcl-1 and the UPR-related components in response to tunicamycin treatment in RPMI-8226 (B), OPM2 (C), and U266 (D) cells. RPMI-8226, OPM2, and U266 cells were treated with 2.5 μg/mL of tunicamycin for 24 hours. RNA isolation, cDNA synthesis, and qRT-PCR were performed as described in “Methods.” *P < .05 and **P < .01 versus vehicle-treated samples. Data represent the means ± SD of 3 separate experiments, each performed in triplicate.
The ER inducer tunicamycin increases Mcl-1 expression and triggers UPR activation in MM cells. (A) Tunicamycin treatment induces Mcl-1 accumulation and cleavage in OPM2 cells. OPM2 cells were treated with different doses of tunicamycin for 24 hours. Cell extracts were probed with Mcl-1, ATF4, GRP-78, CHOP, and β-actin pAbs. Left panel shows the representative blots of 3 independent experiments; right panel shows densitometric analysis. (B-D) qRT-PCR analysis of the expression of Mcl-1 and the UPR-related components in response to tunicamycin treatment in RPMI-8226 (B), OPM2 (C), and U266 (D) cells. RPMI-8226, OPM2, and U266 cells were treated with 2.5 μg/mL of tunicamycin for 24 hours. RNA isolation, cDNA synthesis, and qRT-PCR were performed as described in “Methods.” *P < .05 and **P < .01 versus vehicle-treated samples. Data represent the means ± SD of 3 separate experiments, each performed in triplicate.
XBP1 splicing in response to bortezomib in MM cells
As another major regulator of the UPR, XBP1 is a basic leucine zipper type transcription factor. XBP1 is activated by spliceosome-independent mRNA splicing initiated by inositol-requiring enzyme 1-α (IRE1α) on the cytosolic surface of the ER membrane. The endoribonuclease IRE1α cleaves a 26-nt fragment from an unspliced form of XBP1 mRNA, inducing a frameshift of the open reading frame of the message. XBP1 protein translated from the unspliced mRNA (XBP1U) has no transcriptional activity, whereas XBP1 protein from the spliced mRNA (XBP1S) is a potent transcription factor that induces the expression of UPR-related genes.21-23 To directly address the regulation of XBP1 splicing in bortezomib-treated MM cells, we carried out RT-PCR analysis to amplify the region of XBP1 that encompasses the splice junction. As shown in Figure 6A through F, all tested MM cells showed low levels of constitutive splicing of XBP1, whereas bortezomib slightly decreased the splicing of XBP1 in human OPM2, MMS1, U266, and murine 5T33vt cells. However, in RPMI-8226 and LP1 cells, XBP1 splicing was markedly increased by bortezomib. Tunicamycin can trigger XBP1 splicing in most MM cell lines except U266. Moreover, using Western blot, we confirmed that bortezomib can increase and decrease XBP1 splicing in RPMI-8226 and MMS1 cells, respectively (Figure 6G). In addition, knocking down XBP1 in U266 cells showed that the expression levels of the UPR activation markers CHOP and GRP-78 were significantly down-regulated; however, the expression of MCL-1 and ATF4 and its target gene ATF3 was not disturbed (Figure 6H). These results suggest that XBP1 and XBP1 splicing are not directly involved in bortezomib induced Mcl-1 expression.
The role of XBP1 in UPR-mediated Mcl-1 expression. (A-F) RT-PCR analysis of XBP1 splicing in different MM cell lines. The MM cell lines OPM2, MMS1, U266, 5T33vt, RPMI-8226, and LP1 were treated with bortezomib and tunicamycin for 24 hours at varying doses. RNA isolation and RT-PCR were performed as described in “Methods.” In the human MM cell lines OPM2, MMS1, U266, RPMI-8226, and LP1, the 210- and 184-bp DNA fragments correspond to unspliced and spliced human XBP1 mRNAs, respectively. In 5T33vt cells, the 343- and 327-bp DNA fragments correspond to unspliced and spliced mouse Xbp1 mRNAs, respectively. (G) Western blot analysis of XBP1 splicing in bortezomib-treated RPMI-8226 and MMS1 cells. Total protein extracts from RPMI-8226 and MMS1 cells treated with 10nM of bortezomib for 0, 6, 12, and 24 hours were electrophoresed and subjected to Western blot analysis using XBP1 Abs. Left panels show the representative blots of 3 independent experiments; right panels show densitometric analysis. (H) qRT-PCR measurement of MCL-1 and UPR-related downstream genes in U266 cells with XBP1 knockdown. n = 3 for all experiments. *P < .05 and **P < .01 versus siRNA mock samples. Data represent the means ± SD.
The role of XBP1 in UPR-mediated Mcl-1 expression. (A-F) RT-PCR analysis of XBP1 splicing in different MM cell lines. The MM cell lines OPM2, MMS1, U266, 5T33vt, RPMI-8226, and LP1 were treated with bortezomib and tunicamycin for 24 hours at varying doses. RNA isolation and RT-PCR were performed as described in “Methods.” In the human MM cell lines OPM2, MMS1, U266, RPMI-8226, and LP1, the 210- and 184-bp DNA fragments correspond to unspliced and spliced human XBP1 mRNAs, respectively. In 5T33vt cells, the 343- and 327-bp DNA fragments correspond to unspliced and spliced mouse Xbp1 mRNAs, respectively. (G) Western blot analysis of XBP1 splicing in bortezomib-treated RPMI-8226 and MMS1 cells. Total protein extracts from RPMI-8226 and MMS1 cells treated with 10nM of bortezomib for 0, 6, 12, and 24 hours were electrophoresed and subjected to Western blot analysis using XBP1 Abs. Left panels show the representative blots of 3 independent experiments; right panels show densitometric analysis. (H) qRT-PCR measurement of MCL-1 and UPR-related downstream genes in U266 cells with XBP1 knockdown. n = 3 for all experiments. *P < .05 and **P < .01 versus siRNA mock samples. Data represent the means ± SD.
Discussion
The main finding of this study is that bortezomib transcriptionally up-regulates the expression of the major anti-apoptotic protein Mcl-1 via activation of the UPR and, more specifically, its ATF4 branch, to promote MM cell viability under stress. First, we demonstrated that bortezomib-induced Mcl-1 accumulation in MM cells depends on up-regulation at the transcription level. Second, we proved by ChIP analysis that ATF4 indeed binds to the MCL-1 promoter. Third, after knockdown of ATF4 in MM cells, the expression of Mcl-1 in response to bortezomib was significantly impaired and, in agreement with this scenario, the bortezomib-induced apoptosis was promoted. The survival of MM cells depends on the UPR-mediated homeostatic-apoptotic switch. Our findings suggest that the bortezomib-induced Mcl-1 level determines the likelihood of resistance and survival of MM cells to a certain degree.
Bortezomib is particularly toxic to MM cells. Although it has been assumed from the start that bortezomib would have diverse effects on cancer cell biology, the most common mechanism attributed to its antitumor actions is related to the inhibition of NF-κB.24 However, a recent work proved that bortezomib actually activates NF-κB, providing compelling evidence that NF-κB inhibition is probably not involved in the effects of bortezomib in MM cells.25 Growing evidence suggests that the selectivity of bortezomib for MM cells may be explained by an increased susceptibility of MM cells to ER stress-induced apoptosis.16 One of the defining features of plasma cells is an expansive and highly developed ER that is specialized for the production and secretion of large numbers of Abs.26 The efficient functioning of the ER is essential for proper cellular activity and survival. In most cases, conditions that interfere with ER function cause the accumulation and aggregation of unfolded proteins. Indeed, the development and survival of plasma cells tightly depend on the UPR, which is also an intriguing target for novel treatments for MM. For example, bortezomib treatment of MM cells can increase ER stress and activate the UPR.27 Downstream consequences of these events included induction of CHOP, inhibition of cellular proliferation, and promotion of cellular death through JUN N-terminal kinase (JNK) activation and caspase cleavage.27-29 Cells experiencing irremediable ER stress commit to apoptosis through the mitochondria-mediated intrinsic apoptosis pathway, which is typically regulated by the Bcl-2 protein family, including multidomain pro-apoptotic proteins (Bax and Bak), anti-apoptotic proteins (Bcl-2, Bcl-xl, Mcl-1, A1, Bcl-B, and Bcl-w), and pro-apoptotic BH3-only proteins (Bid, Bad, Bim, Noxa, Puma, and Hrk).30,31 In response to ER stress, the pro-apoptotic BH3-only proteins are transcriptionally or posttranslationally activated to stimulate pro-apoptotic Bax and Bak either directly or indirectly through antagonizing anti-apoptotic members. Among these BH3-only family members, Puma, Noxa, Bid, and Bim have been found to mediate apoptosis triggered by ER stress.32-34 However, it is still unclear whether and how UPR signaling components communicate with the Bcl-2 family members or other pro- and anti-apoptotic signaling molecules to initiate apoptosis.
Recent evidence indicates that the down-regulation or reduction of Mcl-1 plays very important roles in response to drug-induced apoptotic stimuli. Mcl-1 differs from its other family members in its regulation at both the transcriptional and posttranslational levels.35 It is known that the human MCL-1 gene undergoes differential splicing and yields 2 mRNAs encoding anti-apoptotic Mcl-1L and pro-apoptotic Mcl-1s.35,36 Recently, Mcl-1ES, a new variant of Mcl-1, was also identified in human cells. Mcl-1ES can interact with Mcl-1L and induce mitochondrial cell death.37 In mice, Mcl-1 and a new splicing variant, Mcl-1v, are expressed in a variety of murine normal and tumor cell lines and tissues. Mcl-1v has the same localization and anti-apoptotic activity as Mcl-1, but has higher stability than Mcl-1 in the cells undergoing apoptosis. In addition to the transcriptional regulation mechanisms, Mcl-1 activity still can be regulated by the complex posttranslational modifications such as phosphorylation and cleavage.35 For example, phosphorylation at Thr163, the conserved MAP kinase/ERK site located within the PEST region, slows Mcl-1 protein turnover.38 Alternatively, some agents can significantly increase the cleavage of Mcl-1 by caspases, enhance its turnover, and impair its ability to counteract Bim and Noxa-induced apoptosis.11,39 Of particular interest is the recent observation that proteasome inhibition may lead to the accumulation of the anti-apoptotic Mcl-1.13 Mcl-1 is a short-lived protein, with a half-life in cells and tissues of approximately 30 minutes to 3 hours.35,40 The accumulation of Mcl-1 by proteasome inhibition was speculated to be because of prolongation of its half-life, but this is still disputed.
In the present study, we have demonstrated that Mcl-1 can be up-regulated via ATF4 by bortezomib, providing a novel mechanism for Mcl-1 regulation under stress. As shown in Figure 1, we confirmed that bortezomib can induce Mcl-1 accumulation, and the level of accumulated Mcl-1 can be down-regulated by caspase-mediated cleavage. More importantly, we revealed that transcriptional up-regulation is related to the Mcl-1 accumulation in MM cells. Further, we demonstrated that bortezomib activates the UPR in MM cells, as evidenced by the significantly increased expression of the ER stress markers GRP-78 and CHOP at both the transcriptional and translational levels (Figure 2A-C). We also found that the ATF4 branch of the UPR is involved in the bortezomib-induced UPR response. The upstream regulator of ATF4, the phosphorylation level of e-IF2α kinase, was enhanced and, accordingly, the translation of ATF4 was also up-regulated by bortezomib (Figure 2D-E). Although the translational up-regulation of ATF4 has received the most attention, its transcriptional up-regulation should also play a very important role because it provides higher amounts of mRNA for translation and thereby constantly activates the UPR. To address the relationship between ATF4 and Mcl-1, we analyzed the MCL-1 promoter by bioinformatics analysis, and the results showed that there are 3 potential binding sites of ATF4 in humans. Further studies using ChIP analysis demonstrated that ATF4 specifically binds to the regulatory site at the position −332 to −324 in the promoter of the MCL-1 gene (Figure 3). Moreover, as shown in Figure 4, knockdown of ATF4 in both U266 and MMS1 cells decreased the expression of Mcl-1 at both the mRNA and protein levels. Simultaneously, down-regulation of ATF4 renders MM cells sensitive to bortezomib-induced apoptosis, suggesting the protective roles of ATF4 and Mcl-1 in bortezomib-induced apoptosis. To further clarify the role of UPR-activated ATF4 in regulating Mcl-1 expression, we evaluated the expression of Mcl-1 in another in vitro UPR model triggered by tunicamycin in MM.28 As confirmed in Figure 5, the induced Mcl-1 expression is in accordance with the highly activated UPR components GRP-78, CHOP, and ATF4 and its downstream gene ATF3. Similar results were obtained in MM cells treated with another proteasome inhibitor, MG132 (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). More importantly, in U266-Btz, a stable, bortezomib-resistant sub-cell line established by long-term exposure to bortezomib, we further confirmed the regulatory relationship between ATF4 and Mcl-1 (supplemental Figure 2). Our results support the notion that ATF4 up-regulates Mcl-1 expression through a transcriptional mechanism.
Although our study clearly demonstrates the involvement of ATF4 signaling in bortezomib-induced Mcl-1, it is evident that inhibition of this pathway does not completely abrogate Mcl-1 induction or cell death. This suggests that additional factors contribute to Mcl-1 induction during ER stress. In addition to ATF4, we also investigated whether bortezomib can activate the other 2 key UPR branches, XBP1 and ATF6, and whether these 2 pathways may also contribute to Mcl-1 induction. As shown in Figure 6, we investigated the role of XBP1 splicing in regulating Mcl-1 expression. In all tested MM cell lines, bortezomib showed widely divergent effects on XBP1 splicing: in human OPM2, MMS1, and U266 cells and in murine 5T33vt cells, bortezomib decreased XBP1 splicing, which agrees with previous studies showing that proteasome inhibition prevents XBP1 splicing in some MM cell lines.28,41,42 However, in LP1 and RPMI-8226 cells, we found that XBP1 splicing was trigged by bortezomib, which is also in agreement with previous reports,43,44 suggesting that the effects of bortezomib on XBP1 splicing is cell-context dependent. Another UPR inducer, tunicamycin, can trigger efficient XBP1 splicing in most MM cell lines except U266. Moreover, our present results also show that low levels of constitutive XBP1 splicing exists in all tested MM cells. However, no matter whether bortezomib increases or decreases XBP1 splicing, all of the tested MM cell lines showed Mcl-1 up-regulation, suggesting that XBP1 is not directly involved in the regulation of Mcl-1 expression. To validate this notion, XBP1 was knocked down with siRNA in U266 cells in the absence of bortezomib, and XBP1 silencing was shown to decrease the expressions of its target genes, the UPR marker GRP-78 and CHOP, but could not disturb the expression of MCL-1 and ATF4. These data show that XBP1 is not involved in Mcl-1 up-regulation.
After activation of the UPR, ATF6 is transported to the Golgi apparatus, where it is cleaved to generate a 50-kDa, cytosolic b-ZIP–containing fragment that migrates to the nucleus to activate transcription of UPR target genes.45 In the present study, we confirmed that bortezomib can trigger ATF6α activation by cleavage (supplemental Figure 3A). Moreover, by knockdown of ATF6 with siRNA, we found that the Mcl-1 transcription level could be down-regulated, and this was accompanied by a decreased expression of the UPR marker GRP-78 and of CHOP, suggesting that ATF6 also plays a role in regulating Mcl-1 expression in response to bortezomib treatment (supplemental Figure 3B). As previously demonstrated in human melanoma cells, activation of the ATF6, IRE1α, and MEK/ERK signaling pathways are all involved in the transcriptional up-regulation of Bcl-2 and Mcl-1 expression on the tunicamycin- and thapsigargin-induced UPR.46 Interestingly, we only found Mcl-1 to be up-regulated in MM cells; the expressions of Bcl-2 and Bcl-xl were slightly down-regulated or were not disturbed (Figure 1A; tunicamycin data not shown). Nevertheless, repression of Mcl-1 expression was also found to be coupled to tunicamycin, UV light exposure, elevated osmotic pressure, and arsenite-induced ER stress in several cancer cell lines.47 In addition, it has been demonstrated that ATF5, another member of the CREB family of transcription factors, also has a key role in promoting Mcl-1 expression in human malignant glioma.48,49 In response to the RAS-mitogen-activated protein kinase or PI3K signaling cascade–mediated induction of cAMP response element-binding protein-3-like-2 (CREB3L2), ATF5 is activated to promote survival by stimulating the transcription of Mcl-1 in human malignant glioma.50 However, we found that the expression level of ATF5 is very low in MM cells, and bortezomib treatment cannot markedly affect its expression (data not shown). Our results suggest that the mechanisms of regulating Mcl-1 expression and UPR-induced Mcl-1 expression are related to the cell context and the diversity of stimuli. More details on the mechanism how ATF6 is involved in Mcl-1 regulation during ER stress should be studied further in the future.
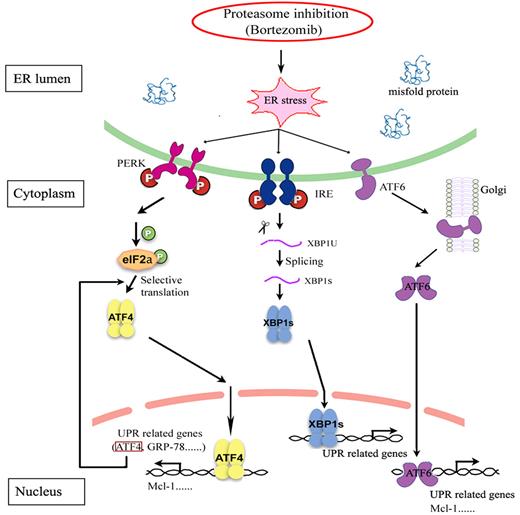
In summary, our results demonstrate that ATF4 is involved in ER stress–induced Mcl-1 up-regulation in response to bortezomib in MM (Figure 7). These results shed light on our understanding of the complex regulation of the mechanism of MM cell resistance to proteasome inhibition, allowing us to reveal novel therapeutic targets and to develop improved treatment strategies to overcome chemoresistance of MM to bortezomib.
Schematic diagram depicting the joint roles of the UPR in regulating Mcl-1 expression by proteasome inhibition. In response to bortezomib, e-IF2α is phosphorylated and the ATF4 branch is activated to trigger protective effects for cell survival by up-regulating the expression of the anti-apoptotic protein Mcl-1. Bortezomib shows completely contrary effects, either promoting or inhibiting XBP1 splicing in different MM cell lines, suggesting that XBP1 splicing is not directly involved in bortezomib-induced Mcl-1 expression. Bortezomib can increase the cleavage of ATF6 to form active ATF6α, which also plays a role in regulating Mcl-1.
Schematic diagram depicting the joint roles of the UPR in regulating Mcl-1 expression by proteasome inhibition. In response to bortezomib, e-IF2α is phosphorylated and the ATF4 branch is activated to trigger protective effects for cell survival by up-regulating the expression of the anti-apoptotic protein Mcl-1. Bortezomib shows completely contrary effects, either promoting or inhibiting XBP1 splicing in different MM cell lines, suggesting that XBP1 splicing is not directly involved in bortezomib-induced Mcl-1 expression. Bortezomib can increase the cleavage of ATF6 to form active ATF6α, which also plays a role in regulating Mcl-1.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank A. Willems and C. Seynaeve for expert technical assistance.
This work was supported by Vlaamse Kankerliga, Fonds voor Wetenschappelijk Onderzoek-Vlaanderen, Stichting Tegen Kanker, and Vrije Universiteit Brussel-Onderzoeksraad. J.H. was a recipient of a scholarship from the China Scholarship Council. E.V.V., E.M., and E.D.B. are postdoctoral fellows of Fonds voor Wetenschappelijk Onderzoek-Vlaanderen.
Authorship
Contribution: J.H., N.D., and K.V. designed the research; J.H. and N.D. performed the research, analyzed the data, and wrote the manuscript; E.V.V., E.M., and D.X. provided essential reagents, analyzed the data, and revised the manuscript; E.D.B. revised the manuscript; and K.V. and B.V.C. critically revised the manuscript for important intellectual content.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Karin Vanderkerken, Department of Hematology and Immunology, Myeloma Center Brussels, Vrije Universiteit Brussel, Laarbeeklaan 103, 1090 Brussels, Belgium; e-mail: karin.vanderkerken@vub.ac.be.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal