Abstract
Self-renewal activity is essential for the maintenance and regeneration of the hematopoietic system. The search for molecules capable of promoting self-renewal and expanding hematopoietic stem cells (HSCs) has met with limited success. Here, we show that a short isoform (AML1a) of RUNX1/AML1 has such activities. Enforced AML1a expression expanded functionally defined HSCs, with an efficiency that was at least 20 times greater than that of the control in vivo and by 18-fold within 7 days ex vivo. The ex vivo–expanded HSCs could repopulate hosts after secondary transplantations. Moreover, AML1a expression resulted in vigorous and long-term (> 106-fold at 4 weeks) ex vivo expansion of progenitor cell populations capable of differentiating into multilineages. Gene expression analysis revealed that AML1a expression was associated with up-regulation of genes, including Hoxa9, Meis1, Stat1, and Ski. shRNA-mediated silencing of these genes attenuated AML1a-mediated activities. Overall, these findings establish AML1a as an isoform-specific molecule that can influence several transcriptional regulators associated with HSCs, leading to enhanced self-renewal activity and hematopoietic stem/progenitor cell expansion ex vivo and in vivo. Therefore, the abilities of AML1a may have implications for HSC transplantation and transfusion medicine, given that the effects also can be obtained by cell-penetrating AML1a protein.
Introduction
Although HSC transplantation is increasingly being used to treat various malignant and genetic diseases, the relative inability to expand hematopoietic stem cells ex vivo limits the widespread application of HSC transplantation. This is especially the case where the number of available stem cells is limited, as in the case of the transplantation of cord blood-derived stem cells into adult patients. The modest expansion of stem cells is expected to circumvent these drawbacks. One approach to expand HSCs focuses on externally acting growth factors and related molecules,1,2 whereas a complementary approach involves identification of intrinsic regulatory molecules that promote self-renewal divisions.3,4 However, only a few intrinsic regulators have thus far been documented as promoters of HSC expansion. Among these, Hoxb4 is the best documented regulator capable of expanding HSCs in vivo and ex vivo. Subsequently, other members of the Hox family of transcription factors and their chimeric proteins have been shown to have an activity similar to that of Hoxb4, but no intrinsic regulators other than Hox-related molecules have been well explored. Recently, Deneault et al conducted a functional screening to identify intrinsic regulators with an activity similar to that of Hox4,5 but assessment of their capacity to expand HSCs awaits further study.
AML1/RUNX1 was originally identified as a component of the fusion gene generated as a consequence of the t(8;21) chromosomal translocation characteristic of a subset of acute myelogenous leukemia, and encodes a transcription factor.6,7 Aml1 expression marks long-term repopulating HSCs in the mid-gestation mouse embryo, and gene targeting experiments have unequivocally shown that Aml1 is essential for the development of hematopoiesis.8 In sharp contrast, conditional gene-targeting experiments have revealed that Aml1 is not essential for the maintenance of adult HSCs. Aml1 deficiency in adult stages is accompanied by increases in myeloid progenitors, impaired development of lymphoid and megakaryocytic cells, and compromised competitive repopulation activity after transplantation,9,10 although interestingly it is associated with increased numbers of HSCs.11
The human AML1 gene generates 3 alternatively spliced variants: AML1a, AML1b, and AML1c. AML1b and AML1c possess the DNA-binding region at the amino terminus, as well as the carboxyl-terminal transcriptional regulatory domains, and they are therefore considered broadly similar in function. In contrast, AML1a retains the DNA-binding domain but lacks the transcriptional regulatory domains. AML1a is thus considered a potential functional antagonist of AML1b and AML1c.6 AML1a is preferentially expressed in immature hematopoietic cell compartments in cord blood, and experimental manipulation of murine and human hematopoietic cells to express AML1a achieved enhanced engraftment of the cells after transplantation into mice,12 suggesting a role for AML1a in stem/progenitor cells.
Here, we first addressed questions of how AML1a expression affects HSC activities and explored potential use of the activity of AML1a in expanding functionally defined HSCs ex vivo. We then explored the activity of AML1a in expanding progenitor cells ex vivo. Finally, we provide evidence that AML1a influences several transcription factors associated with HSCs to achieve the expansion.
Methods
Mice
Mice were purchased from Charles River and SLC. Ly5.1(CD45.1)–C57BL6 mice were provided by Dr Toshitada Takahashi (Aichi Cancer Center Research Institute, Nagoya, Japan). All experiments on mice in this study were performed with the approval of and in accordance with Aichi Cancer Center's Administrative Panel on Laboratory Animal Care.
Plasmids
Plasmids were constructed as described in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The MSCV-IRES-GFP/HOXB4 plasmid was generously provided by Dr R. K. Humphries (Terry Fox Laboratory, Vancouver, BC).3
Cell culture
Cells were cultured in IMDM (Invitrogen) supplemented with 10% fetal calf serum in the presence of murine SCF (100 ng/mL), IL-3 (10 ng/mL), and IL6 (10 ng/mL) for retroviral infection, as described previously.12 Differentiation induction was performed as described in supplemental Methods.
Transplantation and CRU assay
Evaluation of the frequency of cells with long-term repopulating potential was performed using the competitive repopulation unit (CRU) assay, as described in supplemental Methods.
Flow cytometry
Flow cytometry and flow sorting were conducted with an FACSCalibur system (BD Biosciences) and JSAN (Bay Bioscience). Data were analyzed using FlowJo software Version 7.5.5 (TreeStar).
Southern blot analysis of genomic DNA
Southern blot analysis was conducted as described previously.13
Purification of PTD-HA-AML1a fusion protein and determination of uptake, subcellular localization, and DNA-binding ability of the protein
PTD-HA-AML1a fusion protein was purified as described in supplemental Methods. Cells were treated with PTD-HA-AML1a protein, fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, stained with anti-hemagglutinin (HA) antibody (HA-7; Sigma-Aldrich), and visualized by anti-mouse IgG (H+L) conjugated with fluorescein (Invitrogen) under a BX60 fluorescence microscope (Olympus). The protein also was analyzed by immunoblotting with anti-HA antibody. The DNA-binding assay was conducted as described in supplemental Methods.
Micrographs
Cytospine preparation of cells and colonies were examined using BH-2 and CKX31 microscopes (Olympus), respectively. Immunofluorescence staining was examined using a BX60 microscope (Olympus). Images were obtained with a C-4040 digital camera (Olympus). The images were then processed by Photoshop Elements Version 2.0 software (Adobe Systems).
Microarray and bioinformatics analyses
These analyses were performed as described in supplemental Methods. Gene set enrichment analyses14 were performed with gene set enrichment analysis (GSEA) Version 2.0 software (Broad Institute). Microarray data are available in the ArrayExpress database under accessions E-MEXP-3179.
Quantitative PCR and shRNA
Oligonucleotides used for shRNA and quantitative PCR are described in supplemental Methods.
Results
AML1a expression expands HSCs in vivo
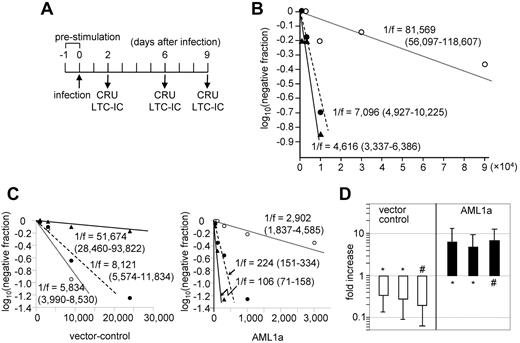
We demonstrated previously that retrovirally expressed AML1a confers hematopoietic cells with enhanced engraftment activity after transplantation,12 but the cells responsible for the enhanced engraftment activity were not specified. Therefore, we first sought to determine whether AML1a expression affects the number of functionally defined HSCs in vivo. Although the engraftment activity was enhanced, we observed an underrepresentation of B- and T-lymphoid cell compartments in the periphery of mice that received a transplant of AML1a-infected bone marrow (BM) cells. We hypothesized that AML1a expression could increase the number of HSCs but may simultaneously inhibit the development of B and T cells. Given that functionally defined HSCs rely on the detection of multilineages of peripheral cells as read-outs in addition to the self-renewal activity, the underrepresentation in B- and T-cell compartments could result in an underestimation of the number of HSCs. We therefore devised a new retrovirus vector that allowed AML1a expression under the control of doxycycline (Dox; hereafter designated as TetOFF-GFP-tetO-AML1a; Figure 1A) to determine whether silencing AML1a expression reverses the underrepresentation. BM cells collected from 5-fluorouracil–treated mice were infected with the TetOFF-GFP-tetO-AML1a virus, sorted for green fluorescent protein (GFP) labeling, and cultured with SCF and IL-3. The cells expressed AML1a in the absence of Dox, but the expression ceased in 24 hours after Dox treatment (Figure 1B). Similarly, mice that received a transplant with TetOFF-GFP-tetO-AML1a–infected cells expressed AML1a in BM (supplemental Figure 1A) and spleen in the absence of Dox, but this expression was silenced by Dox administration, thus establishing the authenticity of the vector. Mice that received a transplant with TetOFF-GFP-tetO-AML1a–infected cells exhibited no apparent abnormalities in the granulocyte/macrophage differentiation program but exhibited an underrepresentation of B- and T-cell lineages in the absence of Dox; therefore, the cells expressed AML1a (supplemental Figure 1B). The underrepresentation of B cells was, at least in part, because of a partial blockade in the differentiation of B cells from the B220+CD43+ pro-B to B220+CD43− pre-B or immature B-cell stages (supplemental Figure 1B). In contrast, T cells were not apparently altered during differentiation in the thymus, but the contribution of AML1a-expressing cells to the T-cell compartment was considerably lesser in the periphery than in the thymus, suggesting that AML1a expression was associated with inhibition of the egress of T cells from the thymus. After Dox treatment for silencing AML1a expression, the underrepresentation of B cells in GFP-positive compared with GFP-negative compartments of BM was substantially, although incompletely, restored; this effect also was observed for T cells (supplemental Figure 1B). To examine the origin of the B and T cells that emerged because of Dox treatment, Southern blot analyses of proviral integration sites were performed using DNA isolated from BM (> 90% of cells in BM were neither T nor B cells) and B220-positive B cells isolated from spleen and thymocytes. Results showed identical inserts in these different cell populations, suggesting that B, T and non-T and non-B cells were derived from the common ancestors representing HSCs (supplemental Figure 1C). Overall, these findings suggest that AML1a expression inhibits HSCs from contributing to the B- and T-cell compartment, which is released after the silencing of AML1a. The TetOFF-GFP-tetO-AML1a system now allows the enumeration of functionally defined HSCs.
In vivo expansion of HSCs by AML1a expression. (A) Retrovirus vector (TetOFF-GFP-tetO-AML1a) allowing Dox-controllable expression of AML1a (not to scale). (B) Expression of AML1a in BM-derived hematopoietic cells infected with TetOFF-GFP-tetO-AML1a retrovirus, in the absence (0 hours) and after the addition of Dox at the indicated time. (C) Transplanted CRUs, and the fold-increase of CRUs after 4 or 3 (*) months in vivo after transplantation, are presented at the bottom. Corresponding CRUs after 4 or 3 (*) months in vivo are presented for vector-only virus (□) and TetOFF-GFP-tetO-AML1a–infected (■) cells. Bars represent 95% CIs. Results of 3 independent experiments each for the control and TetOFF-GFP-tetO-AML1a are presented.
In vivo expansion of HSCs by AML1a expression. (A) Retrovirus vector (TetOFF-GFP-tetO-AML1a) allowing Dox-controllable expression of AML1a (not to scale). (B) Expression of AML1a in BM-derived hematopoietic cells infected with TetOFF-GFP-tetO-AML1a retrovirus, in the absence (0 hours) and after the addition of Dox at the indicated time. (C) Transplanted CRUs, and the fold-increase of CRUs after 4 or 3 (*) months in vivo after transplantation, are presented at the bottom. Corresponding CRUs after 4 or 3 (*) months in vivo are presented for vector-only virus (□) and TetOFF-GFP-tetO-AML1a–infected (■) cells. Bars represent 95% CIs. Results of 3 independent experiments each for the control and TetOFF-GFP-tetO-AML1a are presented.
We therefore enumerated functional HSCs in BM of mice with CRU assays. TetOFF-GFP-tetO-AML1a–infected or control cells (both were GFP-positive) were transplanted into mice. The number of functionally defined HSCs (CRU) that the recipient mice received was estimated with CRU assays: mice were administered Dox before assessing the contribution of the infected cells to the lineages. After 4 months, GFP-positive cells were sorted from BM of the recipient mice and used for the CRU assay to enumerate CRUs contained in the GFP-positive fraction of BM of the recipient mice (supplemental Figure 2). Figure 1C summarizes the estimated number of CRUs in the recipient mice and in the GFP-positive fraction of their BM after 4 months. Based on these estimations, the extent of HSC (in terms of CRU) expansion in the recipient mice during the 4 months was calculated. Control cells with 2.7 to 4.3 CRUs expanded up to 110-fold, whereas TetOFF-GFP-tetO-AML1a–infected cells with 1.3 or 7.7 CRUs expanded 2300- to 2800-fold at 4 months. The number of CRUs in control and TetOFF-GFP-tetO-AML1a–infected cells in the recipient mice was not identical; therefore, a direct comparison was difficult. Nevertheless, these findings suggest that AML1a expression expanded HSCs with an efficiency at least 20 times that of the control in vivo for 4 months. In an additional experiment, 3.1 CRU of TetOFF-GFP-tetO-AML1a–infected cells expanded 500-fold at 3 months. These results indicate that AML1a expression expands functionally defined HSCs in vivo with an efficiency considerably greater than that of the control, thus accounting, at least in part, for the enhancement in engraftment activity of hematopoietic cells.
AML1a expression expands HSCs ex vivo
Given the ability of AML1a to expand HSCs in vivo, we next tested whether AML1a expression could expand functionally defined HSCs ex vivo. To test this possibility, we infected mouse hematopoietic cells with GFP-only control or TetOFF-GFP-tetO-AML1a retrovirus, cultured them ex vivo, and then we assayed the cells for CRUs (Figure 2A,B,D). In addition, the long-term culture-initiating cell (LTC-IC) assay, an ex vivo surrogate for estimating HSC activity, was performed (Figure 2C). Our initial experiment made use of fetal liver cells as a stem cell source. Ter119-depleted e14 fetal liver cells were infected with TetOFF-GFP-tetO-AML1a (day 0), sorted for GFP positivity after 2 days (day 2), and cultured for up to an additional 7 days. CRU and LTC-IC assays were conducted on days 2 (the day of sorting), 6, and 9 (cultured for 4 and 7 days, respectively, after the sorting; Figure 2A) using 1/3 aliquots of the culture. Results of the CRU assays (Figure 2B) show that although the CRU frequency on day 2 was 1/81 569, it was 1/7 096 on day 6 and 1/4616 on day 9 (cell numbers equivalent to those on day 2). Thus, CRU increased 11.5- and 17.7-fold during 4 and 7 days of culture, respectively. Consistent with these increments, the LTC-ICs of TetOFF-GFP-tetO-AML1a–infected cells in culture showed a similar trend, that is, increasing 13- and 27.4-fold during 4 and 7 days of culture, respectively (Figure 2C). In contrast, the LTC-ICs of GFP-only control cells decreased 8.9-fold during 7 days of culture (Figure 2C). The results were confirmed by additional assays comparing CRUs in culture on days 2 and 6 using fetal liver and BM as stem cell sources. TetOFF-GFP-tetO-AML1a–infected cells showed significantly increased CRUs, whereas control cells showed reduced CRUs during 4 days of culture (Figure 2D).
Ex vivo expansion of HSCs by AML1a expression. (A) Schedule of retrovirus infection and CRU and LTC-IC assays. Cells were sorted for GFP+ on day 2. (B) Result of the CRU assay using fetal liver cells conducted on days 2 (○), 6 (●), and 9 (▴) of TetOFF-GFP-tetO-AML1a virus-infected cells. Cell numbers are presented as day 2 equivalents. (C) Results of LTC-IC assays conducted on days 2 (○), 6 (●), and 9 (▴) of vector-control (left) and TetOFF-GFP-tetO-AML1a (right) virus-infected cells. Cell numbers are presented as day 2 equivalents. Three experiments for each group yielded a similar result. (D) Summary of CRU assays conducted on days 2 and 6 of vector-control (□) and TetOFF-GFP-tetO-AML1a (■) virus-infected cells. Results are presented as the fold-increase of CRUs on day 6 relative to day 2. Fetal liver (*) and adult mouse BM (#) cells were used as cell sources. Bars represent 95% CIs. Results of 3 independent experiments each for the control and AML1a are presented.
Ex vivo expansion of HSCs by AML1a expression. (A) Schedule of retrovirus infection and CRU and LTC-IC assays. Cells were sorted for GFP+ on day 2. (B) Result of the CRU assay using fetal liver cells conducted on days 2 (○), 6 (●), and 9 (▴) of TetOFF-GFP-tetO-AML1a virus-infected cells. Cell numbers are presented as day 2 equivalents. (C) Results of LTC-IC assays conducted on days 2 (○), 6 (●), and 9 (▴) of vector-control (left) and TetOFF-GFP-tetO-AML1a (right) virus-infected cells. Cell numbers are presented as day 2 equivalents. Three experiments for each group yielded a similar result. (D) Summary of CRU assays conducted on days 2 and 6 of vector-control (□) and TetOFF-GFP-tetO-AML1a (■) virus-infected cells. Results are presented as the fold-increase of CRUs on day 6 relative to day 2. Fetal liver (*) and adult mouse BM (#) cells were used as cell sources. Bars represent 95% CIs. Results of 3 independent experiments each for the control and AML1a are presented.
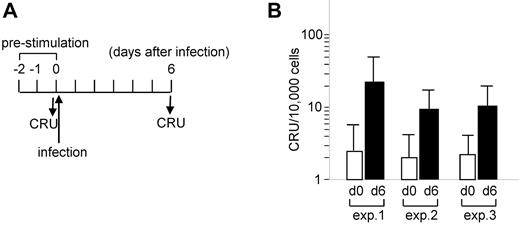
Furthermore, the increment in CRUs in culture was evident in experiments designed to compare cultures before infection and 6 days after infection (Figure 3A). Ly5.2-donor BM cells were infected and transplanted into Ly5.1 mice without sorting (%GFP-positive, ∼ 25% in transplanted cells). The number of CRUs, estimated based on the contribution of Ly5.2 cells to Ly5.1-recipient mice, increased 5- to 7-fold at day 6 (Figure 3B). Southern blot analysis of the provirus integration sites using DNA isolated from BM cells of transplanted mice revealed inserts that were shared in some mice as well as complex multiple inserts that seemed to be specific to a given mouse (supplemental Figure 3). These findings suggest that TetOFF-GFP-tetO-AML1a–infected cells self-renewed and expanded in short-term culture ex vivo. The expansion was not because of a small number of selected clones dominating the culture, but because of the HSC progeny of many originally transduced cells.
Comparison of CRUs of BM cells before and after infection with TetOFFeGFP-AML1a virus. (A) Schedule of retrovirus infection and CRU assays. BM cells were infected with TetOFF-GFP-tetO-AML1a virus and left unsorted. (B) Summary of 3 independent CRU assays on days 0 (before infection) and 6. Cell numbers are presented as day 0 equivalents. Bars represent 95% CIs.
Comparison of CRUs of BM cells before and after infection with TetOFFeGFP-AML1a virus. (A) Schedule of retrovirus infection and CRU assays. BM cells were infected with TetOFF-GFP-tetO-AML1a virus and left unsorted. (B) Summary of 3 independent CRU assays on days 0 (before infection) and 6. Cell numbers are presented as day 0 equivalents. Bars represent 95% CIs.
Finally, we compared TetOFF-GFP-tetO-AML1a–infected cells with fresh BM cells in terms of the number of CRUs (Table 1). The CRU frequency of the unmanipulated, fresh Ly5.2 BM cells was enumerated as 1/39 922. After infection with the TetOFF-GFP-tetO-AML1a virus and 4 days of ex vivo culture, the frequency of CRU increased to 1/7959, even though the %GFP (therefore, the percentage of TetOFF-GFP-tetO-AML1a–infected cells) in Ly5.2 donor cells used for the CRU assays was only 30%. In addition, HSCs expanded ex vivo by AML1a had the ability to repopulate hosts after secondary transplantation. The Ly5.2 donor cells were isolated using MACS columns from BM (femora and tibiae) of each Ly5.1 primary recipient mouse that received a transplant with TetOFF-GFP-tetO-AML1a–infected Ly5.2 BM cells. The isolated Ly5.2 cells were then transplanted into each new Ly5.1 secondary recipient mouse. The Ly5.1 secondary recipient mice were then assayed to investigate the contribution of the Ly5.2 cells toward hematopoiesis after 5 months (supplemental Figure 4, experimental procedure). In total, 46 of 47 Ly5.1 secondary recipient mice exhibited engraftment of Ly5.2 cells; 1 secondary recipient mouse that did not exhibit engraftment of Ly5.2 cells was among the secondary recipient mice that received a transplant with Ly5.2 cells taken from the primary recipient mice that received a transplant with the lowest number of Ly5.2 donor test cells (Table 1). A CRU of 1/8941 was estimated based on the engrafting ability and contribution to hematopoiesis in the secondary recipient mice; this value was very close to that enumerated using the primary recipient mice. These findings demonstrate the capacity of ex vivo–expanded functional HSCs by AML1a to sustain stem cell activity in vivo.
AML1a expression increases secondarily transplantable HSCs
| Cell no.* . | Unmanipulated BM (1°) . | AML1a-transduced BM (1°) . | AML1a-transduced BM (2°) . |
|---|---|---|---|
| 500 000 | 12/12† | 12/12 | 12/12 |
| 200 000 | 11/11 | 12/12 | 12/12 |
| 50 000 | 9/12 | 12/12 | 12/12 |
| 12 500 | 3/12 | 8/10 | 8/10 |
| 3125 | 0/11 | 3/10 | 2/10 |
| HSC | 1/39 922‡ | 1/7 959 | 1/8,941 |
| 95% CI | 1/68 165 ∼ 374;1/23 381 | 1/14 545 ∼ 374;1/4 355 | 1/16 261 ∼ 374;1/4 916 |
| Cell no.* . | Unmanipulated BM (1°) . | AML1a-transduced BM (1°) . | AML1a-transduced BM (2°) . |
|---|---|---|---|
| 500 000 | 12/12† | 12/12 | 12/12 |
| 200 000 | 11/11 | 12/12 | 12/12 |
| 50 000 | 9/12 | 12/12 | 12/12 |
| 12 500 | 3/12 | 8/10 | 8/10 |
| 3125 | 0/11 | 3/10 | 2/10 |
| HSC | 1/39 922‡ | 1/7 959 | 1/8,941 |
| 95% CI | 1/68 165 ∼ 374;1/23 381 | 1/14 545 ∼ 374;1/4 355 | 1/16 261 ∼ 374;1/4 916 |
Number of “test” cells equivalent to unmanipulated BM cells.
Number of mice positive for engraftment/number of total mice used for the primary transplantation.
The HSC frequency and its 95% CI were calculated using Poisson statistics.
Cell-penetrating AML1a protein expands HSCs ex vivo
Although all mice that received a transplant with functional HSCs expanded by AML1a did not develop leukemia during the extended observation period (18 months), the use of a retrovirus is accompanied by potential risks, such as insertional mutagenesis. We therefore investigated the use of a cell-penetrating protein to express AML1a in cells. To this end, we created a plasmid that drives the expression of AML1a fused to a protein transduction domain (PTD)15 in bacteria. A 6× histidine tag and HA tag were included in the fusion protein to facilitate the purification and detection of the protein (Figure 4A). Bacterially expressed PTD-AML1a protein was purified and tested for uptake using 32D mouse myeloid cells. Immunoblotting (Figure 4B) and immunofluorescence staining (Figure 4C) analysis of cells incubated with the fusion protein revealed that the fusion protein penetrated cells and was predominantly located within the nuclei. The PTD-AML1a protein in nuclear extracts of the cells exhibited DNA-binding activity toward the AML1-binding motif, but not to its mutant (Figure 4D; supplemental Figure 5A). Moreover, when lineage-depleted mouse BM cells were cultured with SCF and IL-3, the PTD-AML1a fusion protein retarded the differentiation of cells to granulocytes-macrophages (supplemental Figure 5B-C), which is consistent with the effects of retrovirally expressed AML1a (supplemental Figure 6B). These results indicate that PTD-AML1a readily penetrates cells and functions as expected in a manner similar to virally expressed AML1a. We therefore finally tested the PTD-AML1a protein for ex vivo expansion of HSCs. Ly5.2 mouse BM cells were cultured with SCF, IL-3, and IL-6. PTD-AML1a or bovine serum albumin (BSA; control) was added to the culture 4 times a day for 4 days (Figure 4E), and the culture was assayed for CRU using Ly5.1 mice as recipients. The results showed that CRUs increased 2.6-fold in PTD-AML1a–treated cells, whereas CRUs decreased 2- to 3-fold in BSA-treated control cells (Figure 4F). Thus, these results indicated the feasibility of using AML1a to expand functionally defined HSCs ex vivo without using retroviruses.
Ex vivo expansion of HSCs by cell-penetrating AML1a protein. (A) Schematic drawing of cell-penetrating AML1a protein (not to scale). (B) Immunoblotting of 32D cells treated with the cell-penetrating PTD-HA-AML1a protein with anti-HA antibody. The anti-tubulin blot allows comparison of the protein amount loaded. Asterisk (*) depicts degradation products. (C) Immunofluorescence staining of 32D cells treated with BSA (control) and PTD-HA-AML1a protein with anti-HA antibody. Nuclei are counterstained with propidium iodide (PI). Original magnification, ×200 (UPlan Apo 20×/0.70 objective). (D) DNA binding assays of nuclear extracts of cells treated with PTD-HA-AML1a protein. Nuclear extracts were incubated with biotinylated double-stranded oligo-DNA for the AML1-binding motif, or its mutant. The DNA-bound protein was visualized by immunoblotting with anti-HA antibody. WT indicates wild-type, and MT, mutont. (E) Schedule of PTD-HA-AML1a protein treatment. Bone marrow cells from Ly5.2 mice administered with 5-fluorouracil 4 days earlier were cultured with SCF, IL-3, and IL-6. The protein (10nM) was added 4 times a day (8°, 13°, 18°, 23°) for 4 days after prestimulation and transplanted into Ly5.1 recipient mice for CRU assay. (F) Results of 2 independent CRU assays of pretreatment (○) and after control (BSA; ▵) and PTD-HA-AML1a (●) treatments. Contribution of Ly5.2 donor cells to hematopoiesis in Ly5.1 recipient mice were analyzed 4 (experiment shown left) and 6 (experiment shown right) months after the transplantation.
Ex vivo expansion of HSCs by cell-penetrating AML1a protein. (A) Schematic drawing of cell-penetrating AML1a protein (not to scale). (B) Immunoblotting of 32D cells treated with the cell-penetrating PTD-HA-AML1a protein with anti-HA antibody. The anti-tubulin blot allows comparison of the protein amount loaded. Asterisk (*) depicts degradation products. (C) Immunofluorescence staining of 32D cells treated with BSA (control) and PTD-HA-AML1a protein with anti-HA antibody. Nuclei are counterstained with propidium iodide (PI). Original magnification, ×200 (UPlan Apo 20×/0.70 objective). (D) DNA binding assays of nuclear extracts of cells treated with PTD-HA-AML1a protein. Nuclear extracts were incubated with biotinylated double-stranded oligo-DNA for the AML1-binding motif, or its mutant. The DNA-bound protein was visualized by immunoblotting with anti-HA antibody. WT indicates wild-type, and MT, mutont. (E) Schedule of PTD-HA-AML1a protein treatment. Bone marrow cells from Ly5.2 mice administered with 5-fluorouracil 4 days earlier were cultured with SCF, IL-3, and IL-6. The protein (10nM) was added 4 times a day (8°, 13°, 18°, 23°) for 4 days after prestimulation and transplanted into Ly5.1 recipient mice for CRU assay. (F) Results of 2 independent CRU assays of pretreatment (○) and after control (BSA; ▵) and PTD-HA-AML1a (●) treatments. Contribution of Ly5.2 donor cells to hematopoiesis in Ly5.1 recipient mice were analyzed 4 (experiment shown left) and 6 (experiment shown right) months after the transplantation.
Enforced expression of AML1a expands the immature hematopoietic cell population with potential to differentiate into multilineages ex vivo
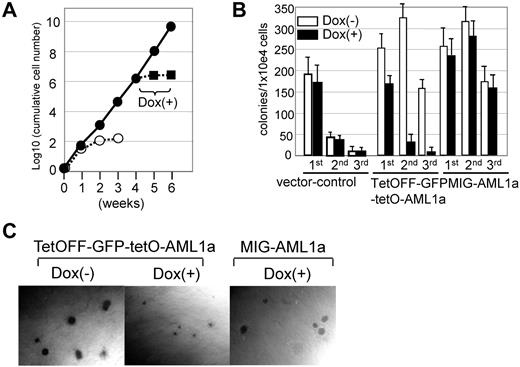
Finally, we investigated whether AML1a expression can be exploited to efficiently produce blood cell components ex vivo. In a long-term cytokine-driven culture with SCF and IL-3 (Figure 5A), TetOFF-GFP-tetO-AML1a–infected cells vigorously expanded 106-fold in 4 weeks, whereas control cells ceased to grow in 2 weeks. The growth of TetOFF-GFP-tetO-AML1a–infected cells was nevertheless evidently inhibited by Dox treatment (ie, by silencing AML1a expression; Figure 5A). Similarly, in replating colony formation assays (assays for self-renewal), AML1a expression augmented the replating ability of BM cells compared with the control. Silencing of AML1a expression in TetOFF-GFP-tetO-AML1a–infected cells by Dox treatment, however, notably diminished the replating ability (Figure 5B). Dox did not affect colony formation and the replating ability of cells infected with retrovirus for vector-only control or constitutively expressing AML1a (MIG-AML1a virus; Figure 5B). Furthermore, this silencing evidently inhibited the growth of individual TetOFF-GFP-tetO-AML1a–infected colonies while sparing MIG-AML1a–infected colonies (Figure 5C). Thus, AML1a expression can promote self-renewal and expand BM cells ex vivo.
Effects of AML1a expression on growth of hematopoietic cells ex vivo. (A) Fold-increases of cumulative cell number are presented on a logarithmic scale versus time. BM cells infected with TetOFF-GFP-tetO-AML1a (●) or control (○) retrovirus were sorted for GFP+ and cultured in the presence of SCF and IL-3. Cells were counted and replated at 105 cells/mL every 7 days. After 4 weeks, Dox was added to an aliquot of culture of TetOFF-GFP-tetO-AML1a–infected cells (■), which showed inhibited cell growth. Three experiments yielded similar results. (B) Frequency of colony-forming cells in serial replating experiments. BM cells infected with the indicated retrovirus, sorted for GFP+ and plated in the multimyeloid condition in the presence or absence of Dox. Colonies were counted and cells were harvested on day 7; 1 × 104 cells were replated. Representative data from 3 independent experiments conducted in triplicate are presented. (C) Photomicrographs show relative sizes of colonies generated in the multimyeloid condition. BM cells infected with the indicated retrovirus, sorted for GFP+, and assayed for colony formation in the presence or absence of Dox. Original magnification ×40 (UPlan FLN4×/0.13 PhP objective).
Effects of AML1a expression on growth of hematopoietic cells ex vivo. (A) Fold-increases of cumulative cell number are presented on a logarithmic scale versus time. BM cells infected with TetOFF-GFP-tetO-AML1a (●) or control (○) retrovirus were sorted for GFP+ and cultured in the presence of SCF and IL-3. Cells were counted and replated at 105 cells/mL every 7 days. After 4 weeks, Dox was added to an aliquot of culture of TetOFF-GFP-tetO-AML1a–infected cells (■), which showed inhibited cell growth. Three experiments yielded similar results. (B) Frequency of colony-forming cells in serial replating experiments. BM cells infected with the indicated retrovirus, sorted for GFP+ and plated in the multimyeloid condition in the presence or absence of Dox. Colonies were counted and cells were harvested on day 7; 1 × 104 cells were replated. Representative data from 3 independent experiments conducted in triplicate are presented. (C) Photomicrographs show relative sizes of colonies generated in the multimyeloid condition. BM cells infected with the indicated retrovirus, sorted for GFP+, and assayed for colony formation in the presence or absence of Dox. Original magnification ×40 (UPlan FLN4×/0.13 PhP objective).
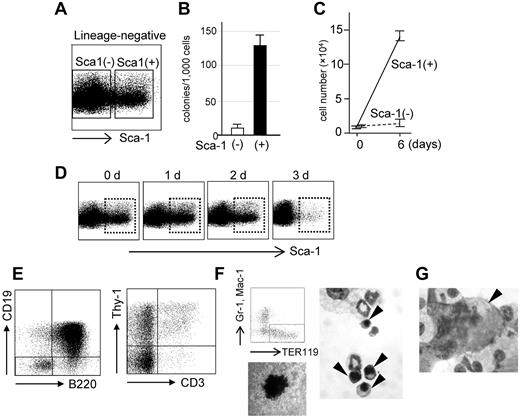
To identify cells capable of self-renewing ex vivo, we performed flow cytometric analysis. The cytokine-driven culture (with SCF and IL-3) of TetOFF-GFP-tetO-AML1a–infected cells contained blastlike cells along with granulocytes or monocytes or macrophages marked by Gr-1 expression, Mac-1 expression, or both (supplemental Figure 6A-B) but lacked cells expressing any erythroid- or megakaryocytic- or lymphoid-affiliated markers such as Ter119, CD41, B220, CD19, Thy-1, and CD3. The blastlike cells represented a lineage marker (Lin)-negative (Lin−: in this case Gr-1- and Mac-1–negative) and c-kit-positive (c-kit+) fraction (supplemental Figure 6A-C) and comprised Sca-1-positive (Sca-1+) and -negative (Sca-1−) fractions (Figure 6A); the Sca-1+ cells were Flt3− and CD34+ and clearly exhibited visible nucleoli (supplemental Figure 6C). Colony-forming ability was highly enhanced in the Sca-1+ fraction (Figure 6B) and also exclusively showed growth in liquid culture, whereas Sca-1− (Figure 6C) and Lin+ fractions essentially showed no growth. When Sca-1+ cells were sorted and cultured, Lin−Sca-1− and Lin+ cells emerged, yet the Sca-1+ fraction remained in the culture. The percentage of Sca-1+ cells in the culture remained largely unchanged in the long term; therefore, the Sca-1+ cells were deemed to have expanded 106-fold in 4 weeks. The maintenance of this Sca-1+ fraction was strictly dependent on AML1a expression as the silencing of AML1a by the addition of Dox extinguished the fraction (Figure 6D; supplemental Figure 6D). Thus, Sca-1+ cells are self-renewing because of AML1a expression and are necessary as well as sufficient for the maintenance and expansion of the whole culture.
Proliferation and differentiation potentials of AML1a-expressing hematopoietic cells ex vivo. (A) FACS profile of the lineage-negative fraction of bone marrow cells infected with the TetOFF-GFP-tetO-AML1a retrovirus and cultured ex vivo. (B-C) Colony formation abilities and cell proliferation in liquid culture of Sca-1+ and Sca-1− cells in the Lin− fraction. BM cells infected with TetOFF-GFP-tetO-AML1a retrovirus were sorted for GFP+. Lin− cells were further fractionated into Sca-1+ and Sca-1− and assayed for colony formation (B) and growth in liquid culture (C). (D) FACS profiles of the lineage-negative fraction at the indicated time after Dox treatment. (E) Lymphoid differentiation potential of Sca-1+ cells. B-cell (left) and T-cell (right) differentiation abilities of Sca-1+ cells in the presence of Dox and cultured with IL-7 + Flt-3 ligand on OP9 (left) and OP9-DLL1 (right) stroma cell layers. Erythroid (F) and (G) megakaryocytic differentiation of Sca-1+ cells in the presence of Dox and induced by erythropoietin and thrombopoietin, respectively. FACS analysis for erythroid-specific TER119 expression, hemoglobin staining of a colony, and morphology of erythroid cells (arrow heads) are presented in panel F. A photomicrograph showing a megakaryocyte (arrowhead) is presented in panel G. (F-G) May-Grunwald Giemsa staining. Original magnifications, ×1000 (SPlan100 100×/1.25 oil 160/0.17 objective). Typical results from at least 3 independent experiments are presented in panels A through G.
Proliferation and differentiation potentials of AML1a-expressing hematopoietic cells ex vivo. (A) FACS profile of the lineage-negative fraction of bone marrow cells infected with the TetOFF-GFP-tetO-AML1a retrovirus and cultured ex vivo. (B-C) Colony formation abilities and cell proliferation in liquid culture of Sca-1+ and Sca-1− cells in the Lin− fraction. BM cells infected with TetOFF-GFP-tetO-AML1a retrovirus were sorted for GFP+. Lin− cells were further fractionated into Sca-1+ and Sca-1− and assayed for colony formation (B) and growth in liquid culture (C). (D) FACS profiles of the lineage-negative fraction at the indicated time after Dox treatment. (E) Lymphoid differentiation potential of Sca-1+ cells. B-cell (left) and T-cell (right) differentiation abilities of Sca-1+ cells in the presence of Dox and cultured with IL-7 + Flt-3 ligand on OP9 (left) and OP9-DLL1 (right) stroma cell layers. Erythroid (F) and (G) megakaryocytic differentiation of Sca-1+ cells in the presence of Dox and induced by erythropoietin and thrombopoietin, respectively. FACS analysis for erythroid-specific TER119 expression, hemoglobin staining of a colony, and morphology of erythroid cells (arrow heads) are presented in panel F. A photomicrograph showing a megakaryocyte (arrowhead) is presented in panel G. (F-G) May-Grunwald Giemsa staining. Original magnifications, ×1000 (SPlan100 100×/1.25 oil 160/0.17 objective). Typical results from at least 3 independent experiments are presented in panels A through G.
We then tested the differentiation ability of the Sca-1+ cells in our culture system. Experiments were conducted 4 weeks after the infection and initiation of cultures. When Sca-1+ cells were cultured with FL and IL-7, but without Dox, on OP9 (B cell–differentiating condition) or OP9-DL1 (T cell–differentiating condition) stroma cells, there was no evidence of B- or T-cell differentiation. However, this differentiation was evident after Dox treatment; FACS analysis revealed B220 and/or CD19-positive and Thy1 and/or CD3-positive cells (Figure 6E; supplemental Figure 6E) in culture. Sca-1− cells did not show such differentiation (supplemental Figure 6E). In addition, differentiation of erythroid and megakaryocytic cells was easily induced by addition of erythropoietin and thrombopoietin, respectively, combined with Dox treatment (Figure 7F-G), thus demonstrating the potential of the Sca-1+ cell population for multilineage differentiation. Taken together, these findings suggest that AML1a expression can expand the Lin−c-kit+Sca-1+Flt3−CD34+ cell population, which has an intrinsic multilineage differentiation potential, in the cytokine-driven culture ex vivo, although AML1a expression inhibits differentiation to lineages other than the granulocytes/macrophage lineage. Nevertheless, the inhibition is relieved on silencing of AML1a expression.
Genes involved in AML1a-mediated expansion of hematopoietic cells. (A-B) BM cells infected with TetOFF-GFP-tetO-AML1a virus were sorted for GFP+ and cultured with SCF and IL-3. Sca-1+ cells in the presence (ie, without AML1a expression) or absence (ie, with AML1a expression) of Dox were used for expression microarray analysis. (A) Gene sets significantly enriched in Sca-1+ cells with AML1a expression compared with Sca-1+ cells without AML1a expression. Gene sets with FDR q < 0.25 among c2.v2. provided by the Broad Institute (http://www.broad.mit.edu/gsea) are presented in supplemental Table 1, among which gene sets related to hematopoiesis are presented along with net enrichment score (NES), FDR q-, and nominal (NOM) P values. (B) Enrichment plot for the gene sets (TAKEDA_NUP98_HOXA9_8D_UP and HSC_HSC_SHARED) and their corresponding heat maps showing genes in the leading edge. (C) Real-time PCR analysis to quantitate transcripts of the indicated genes in AML1a-expressing (Dox minus) cells relative to non–AML1a-expressing (Dox plus) cells. (D) Replating assays of AML1a-expressing cells infected with shRNA virus for the indicated genes. shRNA for luciferase served as a control. (E) Real-time PCR analyses to quantitate transcripts of the indicated genes after infection of the corresponding shRNA viruses. Results are presented as relative amounts of the transcripts compared with the control (ie, shRNA for luciferase). (F) Replating assays of AML1a-expressing cells infected with a dominant-negative form of Stat1. Vector-only infected cells served as a control. Representative data from at least 3 experiments are shown (C-F).
Genes involved in AML1a-mediated expansion of hematopoietic cells. (A-B) BM cells infected with TetOFF-GFP-tetO-AML1a virus were sorted for GFP+ and cultured with SCF and IL-3. Sca-1+ cells in the presence (ie, without AML1a expression) or absence (ie, with AML1a expression) of Dox were used for expression microarray analysis. (A) Gene sets significantly enriched in Sca-1+ cells with AML1a expression compared with Sca-1+ cells without AML1a expression. Gene sets with FDR q < 0.25 among c2.v2. provided by the Broad Institute (http://www.broad.mit.edu/gsea) are presented in supplemental Table 1, among which gene sets related to hematopoiesis are presented along with net enrichment score (NES), FDR q-, and nominal (NOM) P values. (B) Enrichment plot for the gene sets (TAKEDA_NUP98_HOXA9_8D_UP and HSC_HSC_SHARED) and their corresponding heat maps showing genes in the leading edge. (C) Real-time PCR analysis to quantitate transcripts of the indicated genes in AML1a-expressing (Dox minus) cells relative to non–AML1a-expressing (Dox plus) cells. (D) Replating assays of AML1a-expressing cells infected with shRNA virus for the indicated genes. shRNA for luciferase served as a control. (E) Real-time PCR analyses to quantitate transcripts of the indicated genes after infection of the corresponding shRNA viruses. Results are presented as relative amounts of the transcripts compared with the control (ie, shRNA for luciferase). (F) Replating assays of AML1a-expressing cells infected with a dominant-negative form of Stat1. Vector-only infected cells served as a control. Representative data from at least 3 experiments are shown (C-F).
To explore genes that are possibly linked to AML1a activity in promoting self-renewal in cytokine-driven culture, we performed an expression microarray analysis to compare Sca-1+ cells+ in the presence (ie, AML1a expression off) and absence (ie, AML1a expression on) of Dox. Dox silenced AML1a expression within 1 day (Figure 1B), whereas the disappearance of Sca-1+ cells from the culture took an additional 2 days (Figure 6D). We therefore could compare AML1a+Sca-1+ cells and AML1a−Sca-1+ cells regarding mRNA expression 2 days after Dox treatment (Figure 7). A GSEA14 (Broad Institute, http://www.broad.mit.edu/gsea/) of the data with c2.v2. gene sets (Broad Institute) revealed gene sets more enriched in AML1a+Sca-1+ than with AML1a−Sca-1+ cells (Figure 7A; supplemental Table 1). Top-ranking gene sets include those up-regulated by NUP98-HOXA9 fusion16 (NUP98-HOXA9 facilitates the self-renewal of HSCs17 ). In the leading edge, there was a prominent placement of genes suggested to be associated with the maintenance or self-renewal or proliferation of HSCs (Hoxa9, Meis1, Stat1, Evi1, Angpt1, and interferon-induced genes18-25 ; Figure 7B). Gene sets comprised genes enriched in HSCs relative to progenitors or differentiated cells26 also were listed as significantly more enriched in AML1a+Sca-1+ cells than with AML1a−Sca-1+ cells (Figure 7A; supplemental Table 1) and included genes such as Ski (Figure 7B). These changes in gene expression were not because of the nonspecific effects of Dox treatment, because Dox did not cause such changes in MIG-AML1a–infected cells (supplemental Table 2); MIG-AML1a virus allows constitutive AML1a expression regardless of Dox treatment.
Therefore, we selected Hoxa9, Meis1, Stat1, and Ski for further analysis. Quantitative PCR analysis showed an up-regulation of these genes in AML1a+Sca-1+ cells compared with AML1a−Sca-1+ cells (Figure 7C), and shRNA-mediated silencing of the expression of either one of these genes attenuated the ability of AML1a to promote self-renewal (Figure 7D-E). In addition, the expression of a dominant-negative Stat1 yielded an effect similar to that obtained by shRNA expression for Stat1 (Figure 7F). Although shRNA for Hoxa9, Meis1, and Ski attenuated HOXB4-mediated self-renewal assayed ex vivo, the extent of the attenuation seemed to be considerably less than that of AML1a-mediated self-renewal. In addition, shRNA for Stat1 and dominant-negative Stat1 did not have appreciable inhibitory effects on HOXB4-mediated self-renewal in the ex vivo assay (supplemental Figure 7). These findings suggest that AML1a influences at least several transcriptional regulators that are associated with HSCs, thus promoting self-renewal. The transcription regulators involved in AML1a-mediated self-renewal seemed, at least in part, distinct from those involved in HOXB4-mediated self-renewal, as assayed ex vivo.
Discussion
Based on our previous findings that AML1a potentiates the competitive engraftment activity of BM cells after transplantation,12 we hypothesized that AML1a expression may enable the enhanced recovery of functionally defined HSCs. Here, we could demonstrate that AML1a expression mediated the expansion of functionally defined HSCs both ex vivo and in vivo. Specifically, HSCs expanded 18-fold in 7 days in a simple ex vivo bulk culture of BM cells in the presence of SCF, IL-3, and IL-6. The expansion occurred quite rapidly, with significant HSC growth recorded in only 4 days, and the expansion of HSCs was recorded in comparison to fresh, unmanipulated BM cells. The ex vivo–expanded HSCs repopulated in the secondary recipient mice, demonstrating the retention of the self-renewal activity. Moreover, we demonstrated that AML1a expression expanded functionally defined HSCs in vivo with an efficiency that was at least 20 times greater than that of the control cells after transplantation.
Wnt/β-catenin, hedgehog, and Notch signaling pathways have been shown to expand HSCs ex vivo, but their short-term effects ex vivo and long-term effects in vivo do not seem to be consistent with each other.27-35 In contrast, the function of AML1a is consistent for both effects. Of particular note here is the finding that the in vivo HSCs expanded by AML1a expression did not exceed the normal HSC number; the recipient mouse transplanted with 7.7 CRU was highly reconstituted with GFP-positive cells (GFP ∼ 88%) 4 months after transplantation, and CRU contained in the GFP-positive fraction of BM was 17 700 (95% confidence interval [CI], 10 450%-30 200%; Figure 1C), which is within the normal range of HSC number in mouse BM.4,36 These findings suggest that AML1a-expressing HSCs remain under the control of normal hematopoiesis in a manner similar to HOXB4-expanded HSCs.4,36 Recently, HSCs that undergo extended proliferation after transplantation or challenge by lipopolysaccharide have been shown to have a propensity to return to quiescence.37 Therefore, AML1a expression conceivably confers HSCs with an enhanced self-renewal activity after transplantation or culture ex vivo, but HSCs do not continue to expand when they reach a steady state, which may be consistent with the inability of AML1a to cause leukemia in mice.
AML1/RUNX1 has been shown to be critically involved in the generation and maintenance of HSCs and progenitor cells as well as the differentiation of their progeny.8-10,38 BM cells of Aml1+/− mice exhibit increased competitiveness and a nearly doubled frequency of immature stem/progenitor cells as assessed by cobblestone area-forming cell assays, yet show diminished HSC number.39 The hematopoietic reconstitution ability of conditional Aml1−/− BM cells is maintained in the transplanted mice,9,10 but they are defective regarding the competitive repopulation capacity of HSCs.10 Interestingly, conditional Aml1−/− mice nevertheless seem to contain an increased number (∼ 9-fold) of HSCs as assessed by CRU assays.11 Our previous study showed that AML1a-expressing cells exhibit increased competitiveness, increased LTC-IC, and enhanced replating ability.12 Here, we demonstrate that AML1a expression expands functionally defined HSCs. These findings imply that although some aspects associated with forced AML1a expression mimic a reduced dosage or complete loss of Aml1, AML1a confers unique effects on stem and progenitor cell properties that cannot be achieved by reducing the dosage of the Aml1 gene. Given that HSCs forcibly expressing AML1b do not contribute to hematopoiesis after transplantation,12 the effects conferred by AML1a are isoform-specific.
We then explore the possibility of transferring the AML1a protein into HSCs, essentially in line with a strategy adopted by Krosl et al40 to deliver HOXB4 to HSCs. They employed the TAT domain of HIV as a PTD and fused it to the HOXB4 protein. The TAT-HOXB achieved a level of HOXB4 expression in cells that was comparable with that mediated by the HOXB4 retrovirus and expanded HSCs ex vivo up to 6-fold in 4 days. Our initial effort to employ a similar strategy was unsuccessful—the TAT-AML1a-fusion protein accumulated in the cytosol and could not be detected in the nuclei. We then tested a modified TAT domain, which has been reported to be more efficient in penetrating cells,15 as a PTD. We found that this modified TAT was less efficient when fused with AML1a in penetrating cells, but it was localized mainly in the nuclei. We therefore used it for further study. In our schedule of adding PTD-AML1a-fusion protein in culture, AML1a expression was constantly lower (approximately one-half to one-third) than virally expressed AML1a, but the protein nevertheless expanded HSCs up to 2.6-fold during 4 days of culture, thus demonstrating the feasibility of this cell-penetrating protein for HSC expansion. Optimizing the ex vivo conditions as well as combination with other proteins1,2,40 are needed for more effective expansion. Methodologies using vectors not involving the integration of genes into the genome would be additional options.41 We previously showed that the human cord blood lentivirally expressing AML1a contained significantly more CD34+CD38− cells and LTC-IC and cobblestone area–forming cell assays than the control cells.12 This finding suggests the potential use of AML1a expression to expand human cord blood-derived HSCs, the insufficient numbers of which limit their use in transplantation to adult patients.
Here, we further demonstrated that AML1a expression has the capacity to promote self-renewal and vigorously expand the immature progenitor population that harbors multilineage differentiation potential ex vivo in long-term culture. This remarkable ability of AML1a to expand the multipotent progenitor population yielding various blood cells ex vivo may have implications in several areas, such as transfusion medicine where blood donors are limited.
We then compared the gene expression of the phenotypically identical Lin−c-kit+Sca-1+ cells in the presence or absence of AML1a expression. The AML1a+ cells cultured for an extended period (4 weeks) ex vivo did not show an ability to engraft in mice after transplantation; thus, the analysis of such cultured cells may not reflect the property that AML1a expression confers on HSCs. However, in several instances, ex vivo culture systems are used to explore the gene expression influenced by key regulators in normal and dysregulated hematopoiesis, such as HOXB4,42-44 GATA-1,45 MLL-ENL,46 and NUP98-HOXA9,16 with considerable success in providing much information. Such an analysis suggested some shared molecular mechanisms between AML1a- and the NUP98-HOXA9–mediated expansion of stem or progenitor cells. The experimental expression of the NUP98-HOXA9 fusion gene promoted symmetric cell division (self-renewal) and expansion of stem or progenitor cells ex vivo. Takeda et al performed a microarray analysis to identify the genes regulated by NUP98-HOXA9 by culturing human cord blood cells.16 The genes enriched in NUP98-HOXA9–expressing cells were found by GSEA to be enriched in AML1a-expressing Lin−c-kit+Sca-1+ cells of our experimental system. However, unlike NUP98-HOXA9, AML1a expression did not cause leukemia in mice. Genes enriched in HSCs compared with those in more differentiated progenitors also were enriched in AML1a-expressing Lin−c-kit+Sca-1+ cells. Among the genes shared with AML1a- and NUP98-HOXA9–transduced cells or HSCs, we focused on transcriptional regulatory molecules and chose Hoxa9, Meis1, Stat1, and Ski for their functionality using shRNA. shRNA-mediated silencing of either of these genes attenuated the self-renewal activity conferred by AML1a. The effects of shRNAs that were used do not seem to be specific to AML1a-mediated self-renewal, but some, if not all, shRNAs also attenuated HOXB4-mediated self-renewal assayed ex vivo. The degree of attenuation seemed smaller for HOXB4-mediated self-renewal compared with that for AML1a-mediated self-renewal; shRNA for Stat1 as well as the expression of dominant-negative Stat1 did not appreciably inhibit HOXB4-mediated self-renewal assayed ex vivo. These findings suggest the existence of some shared and distinct molecular mechanisms between AML1a- and HOXB4-mediated self-renewal. Hoxb4 is so far the best documented intrinsic regulator capable of expanding HSCs in vivo and ex vivo, yet genes regulated by Hoxb4 have been only reported recently.42-44 Discovering molecules influenced by AML1a may lead to the elucidation of the mechanisms underlying the self-renewal activity.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Seiko Sato for assistance with animal husbandry.
This work was supported by a grant-in-aid for the second term comprehensive 10-year strategy for cancer control from the Ministry of Health and Welfare (M.S.); a grant-in-aid for scientific research from The Ministry of Education, Culture, Sports, Science and Technology (M.S. and S.T.); and a grant-in-aid for scientific research from the Japan Society for the Promotion of Science (S.T.).
Authorship
Contribution: S.T. designed and performed all experiments and wrote the paper; and M.S. contributed to the discussion.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Shinobu Tsuzuki, Division of Molecular Medicine, Aichi Cancer Center Research Institute, 1-1 Kanokoden, Chikusa-ku, Nagoya, Aichi, 464-8681, Japan; e-mail: stsuzuki@aichi-cc.jp.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal