The role of platelets in hemostasis and thrombosis is clearly established; however, the mechanisms by which platelets mediate inflammatory and immune pathways are less well understood. Platelets interact and modulate the function of blood and vascular cells by releasing bioactive molecules. Although the platelet is anucleate, it contains transcripts that may mirror disease. Platelet mRNA is only associated with low-level protein translation; however, platelets have a unique membrane structure allowing for the passage of small molecules, leading to the possibility that its cytoplasmic RNA may be passed to nucleated cells. To examine this question, platelet-like particles with labeled RNA were cocultured with vascular cells. Coculture of platelet-like particles with activated THP-1, monocytic, and endothelial cells led to visual and functional RNA transfer. Posttransfer microarray gene expression analysis of THP-1 cells showed an increase in HBG1/HBG2 and HBA1/HBA2 expression that was directly related to the transfer. Infusion of wild-type platelets into a TLR2-deficient mouse model established in vivo confirmation of select platelet RNA transfer to leukocytes. By specifically transferring green fluorescent protein, we also observed external RNA was functional in the recipient cells. The observation that platelets possess the capacity to transfer cytosolic RNA suggests a new function for platelets in the regulation of vascular homeostasis.
Introduction
Platelets are anucleate cells generated from megakaryocytes and released in the bloodstream where they play an active role in hemostasis, inflammation, and immune responses.1,,–4 They also are involved in tumor progression and metastasis, with complex molecular mechanisms that continue to be elucidated.5,,,–9 Recent studies have focused on analyzing platelet RNA content including microRNA (miRNA) and messenger RNA (mRNA).10,11 Although lacking nuclei, platelets retain megakaryocyte-derived cytoplasmatic RNA and may translate small amounts of mRNAs as well as process miRNAs.12,–14 Platelets express high levels of miRNAs, and profiling studies have shown expression changes. Platelet transcript variability also has been described in a large community-based cohort study with inflammatory RNA significantly correlated with specific cardiovascular risk factors, notably increased body mass index.15 The platelet transcriptome mirrors the protein expression profile, but only 69% of proteins have been found at the mRNA level leaving open the question about the functionality of the remaining RNA.16,–18
Cell-cell communication can be achieved not only through direct contact but also through exchange of soluble factors or vesicles containing bioactive molecules. Recent studies have shown that microparticles and exosomes, released from different cell types, transfer not only proteins and lipids but also nucleic acids to recipient cells.19,20 Activated platelets shed microparticles that represent the most abundant microparticles in the bloodstream.21 Microparticles have been associated with hemostasis and thrombosis; they are actively involved in cancer metastasis, angiogenesis, and immune diseases.22,23
Taken together, these observations suggest that platelet transcript content may play a biologic role beyond being remnant RNA derived from the megakaryocyte. To test the hypothesis that platelet-derived RNA is transferred, we have developed an in vitro transfer model using MEG-01 cells, a human megakaryoblastic cell line known to release platelet-like particles (PLPs) with characteristics similar to human platelets.24,–26 These PLPs were then examined for potential transfer of nucleic acids to recipient cells. In this study, we show not only that PLPs are able to transfer RNA to THP-1 cells, a monocytic cell line, and human umbilical vein endothelial cells (HUVECs), but also that the transferred RNA is functional.
Methods
Cell cultures
MEG-01 cells and THP-1 cells (ATCC) were grown in suspension in RPMI 1640 (Invitrogen) supplemented with l-glutamine, 10% heat-inactivated fetal bovine serum (Invitrogen), and 1% antibiotic/antimycotic (Invitrogen). HUVECs were obtained from Lonza Walkersville, cultured in endothelial growth medium (Lonza Walkersville) and used between second and fifth passages.
PLP isolation
PLPs were isolated from MEG-01 cultures treated with 100 ng/mL recombinant human thrombopoietin (TPO; R&D Systems) for 48 hours. Preparations were centrifuged at 150g for 15 minutes, and sediments were discarded and centrifuged again at 750g for 15 minutes. Supernatants thus obtained were centrifuged at 1600g for 15 minutes, and sediments containing PLPs were washed and resuspended in culture medium.27 In selected experiments, PLPs were treated with 1 U/mL RNase A/T128 (Invitrogen) or RNase ONE (Promega) for 1 hour at 37°C. Then, the PLPs were then inhibited with 10 U/mL SUPERase·In RNAse (Invitrogen) inhibitor and 0.2M potassium phosphate buffer, respectively. PLPs were washed in culture medium after this treatment.
PLP-vascular cell coincubation
THP-1 cells treated with 1 μg/mL Pam3CSK4 (PAM; InvivoGen) were coincubated for 24 hours with PLPs isolated from MEG-01 cells cultures at a ratio 1:10. PLPs were stained with 2μM PKH67 (Sigma-Aldrich) according to the manufacturer's directions or with CD41a, CD42b FITC-conjugated antibodies (eBioscience). PKH67 is a fluorescent green dye attached to long aliphatic tails that is incorporated into the cell membranes. HUVECs were treated with 0.5 U/mL thrombin (Sigma-Aldrich) for 10 minutes in endothelial basal medium (Lonza Walkersville) without phenol red, washed, and coincubated with PKH67-labeled PLPs for 30 minutes in endothelial basal medium containing 2mM CaCl2 (Sigma-Aldrich) at the same ratio. After coincubation, both cell types were washed in phosphate-buffered saline (PBS; Invitrogen) and analyzed by flow cytometry and confocal microscopy.
BrUTP transfection and transfer
MEG-01 cells were transfected with 0.1mM 5-bromouridine-5′-triphosphate (BrUTP; Invitrogen) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's directions. Cells were treated with 100 ng/mL TPO for 24 hours, and then PLPs were collected and cocultured with THP-1 cells, as described in “PLP-vascular cell coincubation,” with time points 0, 3, 6, 12, and 24 hours. HUVECs were coincubated with PLPs for 1 hour in the same conditions described in “PLP-vascular cell coincubation.” HUVECs and THP-1 cells were analyzed for BrUTP positivity by flow cytometry using a BrdU Flow kit (BD Biosciences) according to the manufacturer's protocol.
GFP transfection and transfer
MEG-01 cells were transfected with a plasmid encoding green fluorescent protein (pmaxGFP; Lonza Walkersville) using a Nucleofector II device and Nucleofector kit C (Lonza Walkersville) optimized for MEG-01 cells according to the manufacturer's suggestions. Cells were allowed to recover after nucleofection and grow for 48 hours with 100 ng/mL TPO treatment. PLP collection and coincubations were performed as described in the preceding paragraphs. THP-1 cells were analyzed after 24 hours and then washed to eliminate residual PLPs and cultured for additional 24 hours. HUVECs were coincubated with PLPs for 1 hour and then washed and allowed to grow for 24 hours. GFP fluorescence was recorded by flow cytometry.
Animal models and in vivo platelet RNA transfer
C57BL/6J (WT) and B6.129-Tlr2tm1Kir/J (TLR2−/−) mice were purchased from The Jackson Laboratory and housed in the animal facility at the University of Massachusetts Medical School. All procedures were approved by the Institute Animal Care and Use Committee at University of Massachusetts Medical School.
Platelets were isolated from wild-type (WT) mice by centrifugation following a previously described method.29 In brief, whole blood was collected into syringes containing citrate-phosphate-dextrose solution (15.6mM citric acid anhydrous, 89.4mM sodium citrate, 18.5mM sodium phosphate, and 2.56% dextrose, pH 7.35). Samples were centrifuged at 450g for 7 minutes. The top layer of plasma was removed and diluted by 2.33 times the volume with platelet wash buffer (10mM sodium citrate, 150mM sodium chloride, 1mM EDTA, and 1% dextrose, pH 7.4). Samples were centrifuged at 300g for 4 minutes. The resulting pellet contains peripheral blood mononuclear cells (PBMCs). The supernatant was further diluted with 3 times the volume with platelet wash buffer and centrifuged at 3500g for 10 minutes. The resulting pellet contains platelets. Approximately 5.5 × 108 platelets resuspended in citrate-phosphate-dextrose solution were injected into TLR2−/− mice via tail-vein injection (total volume, 165 μL). After 1 hour, TLR2−/− mice were injected with 5 μg/g lipopolysaccharide (LPS; InvivoGen) or saline intraperitoneally. Blood samples were collected into citrate-phosphate-dextrose solution (1:5) at 3 and 6 hours after LPS injection. Whole blood samples were tested for the percentage of heterotypic aggregates (platelet-monocyte aggregates) and TLR2-positive monocytes by flow cytometry. Isolated platelets and PBMCs also were processed for RNA isolation and real-time RT-PCR as described in “Real-time RT-PCR.”
Flow cytometry
THP-1 cells were stained with anti–human CD11b–PE-Cy7 and CD11b-APC, and HUVECs were stained with CD54–PE-Cy5. Anti–human CD41a-FITC and CD42b-FITC (eBioscience) antibodies were used for PLPs, and anti–human BrdU-FITC (BD Bioscience) was used to detect BrUTP-labeled RNA. Mouse platelets and monocytes were stained with anti–mouse CD41-PE and CD11b-APC (eBioscience), respectively, whereas CD282(TLR2)–FITC (eBioscience) was used for TLR2 detection. All the stainings were performed along with the corresponding isotype controls. The experiments were run on a FACScan analyzer (BD Biosciences) using CellQuest Pro Version 5.2 software (BD Biosciences).
Confocal microscopy
Samples from the experiments described thus far were analyzed by confocal microscopy. Imaging was performed on LSM 510 (Carl Zeiss) and TCS SP2 (Leica Microsystems) confocal microscopes. Images were captured using and MetaMorph Version 6.1 software (Molecular Devices) and Leica Confocal Version 2.5 software.
Microarray gene expression analysis
Total RNA was isolated from Pam3CSK4-treated THP-1 cells cultured in presence and absence of PLPs using the miRNeasy Mini kit (QIAGEN) according to the manufacturer's directions. Microarray gene expression profile was determined using a Human Genome U133 2.0 Plus chip, human transcriptome complete (Affymetrix). The Gene Expression Omnibus accession number for the data is GSE38027.
Real-time RT-PCR
Total RNA was isolated as described in “Microarray gene expression analysis.” RNA was converted to cDNA using the High-Capacity cDNA Reverse Transcription kit (Invitrogen). Gene expression of human GAPDH and HBG1/HBG2, mouse GAPDH and TLR2 (primers and probes from Invitrogen), using the TaqMan Gene Expression Master Mix (Invitrogen), was assessed with real-time PCR (Applied Biosystems 7900 HT Fast Real-Time PCR System with SDS Version 2.2.2 software; Invitrogen).
SOD2 activity and ROS measurement
THP-1 cells were cocultured for 24 hours with PLPs as described in “PLP-vascular cell coincubation” and then tested for superoxide dismutase 2 (SOD2) activity using EpiQuik Superoxide Dismutase kit (Epigentek) according to the manufacturer's protocol. Reactive oxygen species (ROS) generation was measured using 5-(and-6)-carboxy-2′,7′-difluorodihydrofluorescein diacetate (carboxy-H2DFFDA; Invitrogen) as described previously.30 In brief, cells were washed in PBS, treated with 5μM 5-(and-6)-carboxy-2′,7′-difluorodihydrofluorescein diacetate for 30 minutes at 37°C, and washed twice with PBS. The resulting fluorescence was analyzed by flow cytometry.
Data analysis
All values are expressed as the average ± SD of at least 3 different experiments. Statistical analysis was performed using Student t test and significance of P less than or equal to .05. Prism Version 5 software (GraphPad) was used for all analyses.
Results
PLPs coincubation with HUVECs and THP-1 cells
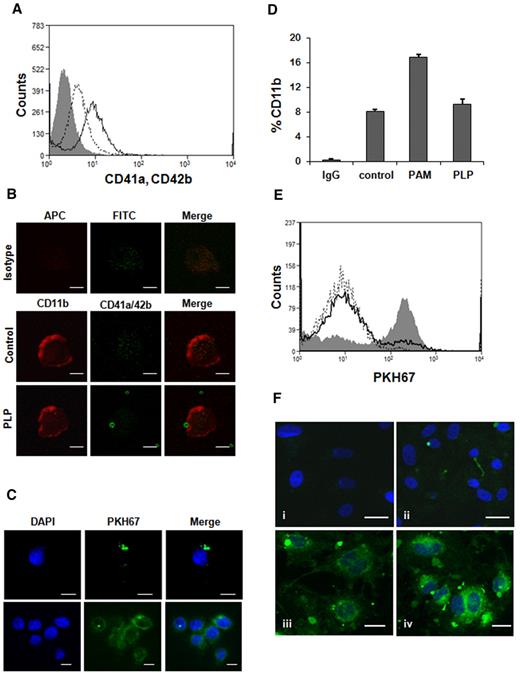
In culture, MEG-01 cells have the ability to spontaneously produce PLPs with size, structures, and characteristics very similar to human platelets.24,31 In coincubation experiments, THP-1 cells were stimulated with Pam3CSK4, a synthetic lipoprotein and TLR2 ligand, to induce interaction with PLPs. As a previous study29 showed, the concentration used for all experiments (1 μg/mL) does not induce any platelet aggregation. PAM-treated THP-1 cells, after 24 hour coincubation with PLPs, were stained with CD41a and CD42b, specific markers for human platelets (Figure 1A). Flow cytometry analysis showed that ∼ 20% of THP-1 cells were positive for the double staining, confirming the PLP binding. This result was further supported by confocal microscopy (Figure 1B-C). Untreated THP-1 cells did not significantly bind PLPs (data not shown). As shown in Figure 1B (PLP), there is colocalization between a THP-1 cell and a PLP followed by the PLP internalization, an observation confirmed using a complementary staining method (Figure 1C). Changes in THP-1 CD11b staining also were measured during coincubation experiments; as seen in Figure 1D, the staining of the adhesion molecule CD11b, in presence of PLPs, was decreased, suggesting its involvement in the binding process. Similar results were obtained with HUVECs over a shorter period of coincubation. HUVECs were activated with 0.5 U/mL thrombin to promote the adhesion to PLPs, in fact, 19% of activated HUVECs were able to bind and internalized PLPs, although less than ∼ 4% of resting cells were fluorescently positive (Figure 1E). Binding also was seen with HUVECs (Figure 1Fii), as was the progressive increase of cytosol staining from PLP internalization (Figure 1Fiii-iv).
PLPs coincubation with HUVECs and THP-1 cells. (A) THP-1 cells, treated with 1 μg/mL Pam3CSK4, were coincubated with PLPs for 24 hours and stained with CD41a-FITC, CD42b-FITC (open curve; solid line) for flow cytometry analysis. Isotype IgG (gray filled curve), control (no PLPs; open curve, dotted line). (B) The same experiment was carried out staining THP-1 cells for CD11b-APC, CD41a-FITC, and CD42b-FITC. Samples were then mounted on coverslips for confocal microscopy (PLP). IgG control (isotype) and THP-1 alone (control). Scale bar denotes 10 μm; 100× objective lens. (C) Confocal microscopy also was performed on PKH67-labeled PLPs (green fluorescence) cocultured for 24 hours with THP-1 cells to demonstrate PLP internalization. Blue nuclear staining was performed with 4,6-diamidino-2-phenylindole (DAPI). Scale bar denotes 10 μm; 60× objective. (D) The experiment was performed as described in panel A using CD11b-FITC. Untreated cells (control), activated cells (PAM), PLP (THP-1 cells cocultured with PLPs). (E) HUVECs were treated for 10 minutes with 0.5 U/mL thrombin and then coincubated with PKH67-labeled PLPs for 30 minutes (gray filled curve) and analyzed by flow cytometry. Control (open curve, dotted line), untreated HUVEC + PLPs (open curve, solid line). (F) Confocal microscopy of the experiment described in panel E with additional DAPI staining. HUVECs incubated with PKH67-labeled PLPs (ii-iv) and unlabeled PLPs (i). Scale bar denotes 20 μm; 40× objective.
PLPs coincubation with HUVECs and THP-1 cells. (A) THP-1 cells, treated with 1 μg/mL Pam3CSK4, were coincubated with PLPs for 24 hours and stained with CD41a-FITC, CD42b-FITC (open curve; solid line) for flow cytometry analysis. Isotype IgG (gray filled curve), control (no PLPs; open curve, dotted line). (B) The same experiment was carried out staining THP-1 cells for CD11b-APC, CD41a-FITC, and CD42b-FITC. Samples were then mounted on coverslips for confocal microscopy (PLP). IgG control (isotype) and THP-1 alone (control). Scale bar denotes 10 μm; 100× objective lens. (C) Confocal microscopy also was performed on PKH67-labeled PLPs (green fluorescence) cocultured for 24 hours with THP-1 cells to demonstrate PLP internalization. Blue nuclear staining was performed with 4,6-diamidino-2-phenylindole (DAPI). Scale bar denotes 10 μm; 60× objective. (D) The experiment was performed as described in panel A using CD11b-FITC. Untreated cells (control), activated cells (PAM), PLP (THP-1 cells cocultured with PLPs). (E) HUVECs were treated for 10 minutes with 0.5 U/mL thrombin and then coincubated with PKH67-labeled PLPs for 30 minutes (gray filled curve) and analyzed by flow cytometry. Control (open curve, dotted line), untreated HUVEC + PLPs (open curve, solid line). (F) Confocal microscopy of the experiment described in panel E with additional DAPI staining. HUVECs incubated with PKH67-labeled PLPs (ii-iv) and unlabeled PLPs (i). Scale bar denotes 20 μm; 40× objective.
RNA transfer from MEG-01 to HUVECs and THP-1 cells
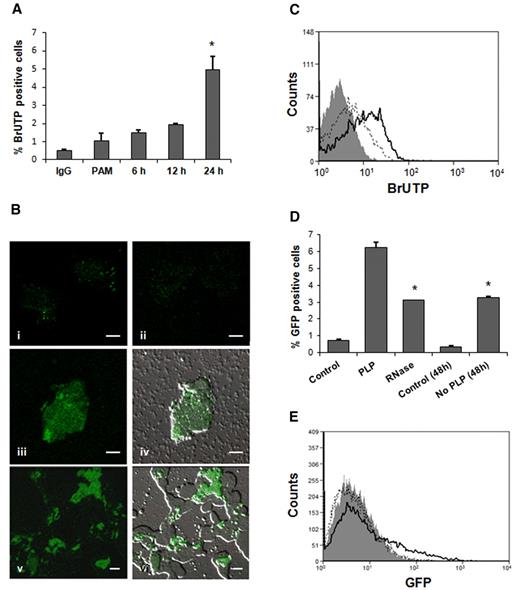
PLPs from transfected BrUTP MEG-01 cells were coincubated with THP-1 cells. After 6 hours of coincubation, labeled THP-1 cells demonstrated RNA transfer that continued gradually up to 24 hours (Figure 2A). The percentage of BrUTP-positive THP-1 cells after 24 hours was 4% to 5%. Previous studies reported MEG-01 as a difficult cell line to transfect; consequently, the amount of labeled-RNA was limited.32 Confocal microscopy showed that labeled RNA was localized outside and inside the recipient cells, indicating an ongoing transfer after 24-hour coincubation (Figure 2Biii-vi). RNA transfer also was observed using HUVECs as recipient cells; ∼ 20% of activated HUVECs were positive for labeled RNA after 1-hour coculture (Figure 2C).
RNA transfer from MEG-01 to HUVECs and THP-1 cells. (A) Flow cytometry analysis of 1 μg/mL PAM-treated THP-1 cells cultured in presence of BrUTP-labeled PLPs for 6, 12, and 24 hours. No fusion was observed at 1- and 3-hour time points (*P < .05 compared with PAM). (B) The same experiment was analyzed by confocal microscopy. THP-1 cells demonstrated BrUTP labeling after 24-hour incubation with RNA-labeled PLPs (iii-vi; iv and vi are in bright field; 100× objective). THP-1 control cells stained with IgG (i) and BrdU-FITC (ii). Scale bar denotes 10 μm; 100× objective. (C) HUVECs, treated with 0.5 U/mL thrombin, were coincubated for 1 hour with BrUTP-labeled PLPs and analyzed by flow cytometry (open curve, solid line). Isotype IgG (gray filled curve), control (no PLPs; open curve, dotted line). (D) PAM (1 μg/mL)–treated THP-1 cells cocultured for 24 hours with GFP-PLPs treated (RNase) or not with RNase (PLP). THP-1 + untreated-PLPs were washed, after 24 hours, to eliminate residual PLPs and cultured for additional 24 hours (No PLP 48 h; *P < .05 vs untreated PLPs). (E) HUVECs showed GFP fluorescence, by flow cytometry, after 1-hour coincubation with PLPs containing GFP (open curve, dotted line), cells were then washed and allowed to grow for 24 hours (open curve, solid line). Control (gray filled curve).
RNA transfer from MEG-01 to HUVECs and THP-1 cells. (A) Flow cytometry analysis of 1 μg/mL PAM-treated THP-1 cells cultured in presence of BrUTP-labeled PLPs for 6, 12, and 24 hours. No fusion was observed at 1- and 3-hour time points (*P < .05 compared with PAM). (B) The same experiment was analyzed by confocal microscopy. THP-1 cells demonstrated BrUTP labeling after 24-hour incubation with RNA-labeled PLPs (iii-vi; iv and vi are in bright field; 100× objective). THP-1 control cells stained with IgG (i) and BrdU-FITC (ii). Scale bar denotes 10 μm; 100× objective. (C) HUVECs, treated with 0.5 U/mL thrombin, were coincubated for 1 hour with BrUTP-labeled PLPs and analyzed by flow cytometry (open curve, solid line). Isotype IgG (gray filled curve), control (no PLPs; open curve, dotted line). (D) PAM (1 μg/mL)–treated THP-1 cells cocultured for 24 hours with GFP-PLPs treated (RNase) or not with RNase (PLP). THP-1 + untreated-PLPs were washed, after 24 hours, to eliminate residual PLPs and cultured for additional 24 hours (No PLP 48 h; *P < .05 vs untreated PLPs). (E) HUVECs showed GFP fluorescence, by flow cytometry, after 1-hour coincubation with PLPs containing GFP (open curve, dotted line), cells were then washed and allowed to grow for 24 hours (open curve, solid line). Control (gray filled curve).
GFP transfer from MEG-01 to HUVECs and THP-1 cells
To test mRNA functionality in the recipient cells, we transfected MEG-01 cells with a GFP vector by nucleofection. By flow cytometry, we observed that GFP was expressed in MEG-01 cells and almost 25% of PLPs collected from cultures expressed mature GFP, carried mature GFP, or both. During a coincubation experiment, both mRNA and proteins can be transferred; therefore, we examined THP-1 cells after 24-hour fusion with PLPs treated or not with RNase. To verify that PLP RNA was effectively degraded by the RNase treatment, we compared the RNA content of RNase-treated PLPs with untreated PLPs; RNase-treated PLPs contained ∼ 48% less RNA. Interestingly, more than 6% of THP-1 cells without RNase treatment were found positive for GFP, a number reduced to 3% in presence of RNase. This suggests that THP-1 cells received GFP both as a mature protein and mRNA that was then translated. When THP-1 cells cocultured with untreated PLPs were washed after 24 hours, to eliminate PLPs, and cultured for additional 24 hours, fluorescence from GFP was still detectable as a result of 2 ongoing processes: GFP-mRNA expression and transferred GFP mature protein degradation (Figure 2D). Six percent of HUVECs were fluorescent after 1-hour coincubation due to the GFP protein transfer. After 24 hours in absence of PLPs, 16.5% of cells expressed the GFP messenger, a figure assumed to include the percentage of positive cells (6%) containing the mature protein (Figure 2E).
THP-1 cell microarray gene expression analysis
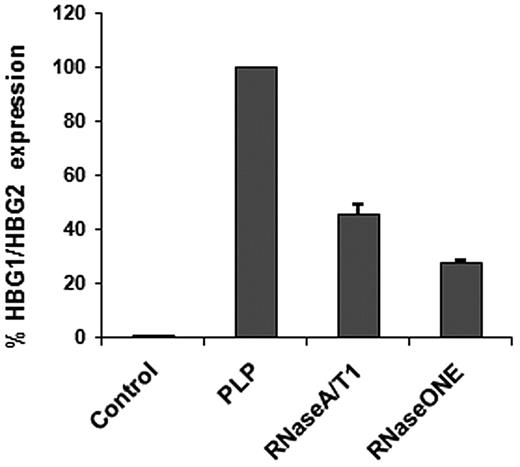
Although transfer of RNA is important, it is of greater interest if found to be functional. To investigate the possible effect of the newly transferred RNAs in the recipient cells, we carried out a gene expression analysis of THP-1 cells in presence and absence of PLPs. Microarray analysis showed that the most up-regulated genes were γ globins (HGB1/HGB2), up to 2-fold, followed by α globins (HBA1/HBA2) and ϵ globin (HBE1). Two mitochondrial genes (SOD2, MRPS15) and genes involved in transport and transfer mechanisms also were up-regulated. There were a small number of modestly down-regulated genes (< 1-fold) associated with exocytosis pathways (STXBP5L, RAB7A, GPLD1), 2 types of matrix metallopeptidase (Table 1). The most up- and down-regulated genes in the control cells (PAM-activated THP-1), noted in Table 2, also were not altered by the presence of the imported RNA. Using the γ globins (the most expressed genes), the microarray results were validated by real-time PCR verifying whether the up-regulation was due to RNA transfer. PLPs were treated with 2 different types of RNases before a coincubation and RT-PCR confirmed that the increased RNA was from outside the cells (Figure 3). The small percentage of residual RNA after RNase treatment depends on the length of treatment: a complete degradation can be achieved with longer treatments but this would have affected PLP vitality and integrity. As a negative control TPO/Pam3CSK4-treated THP-1 cells were examined for HGB1/HGB2 expression to exclude any TPO carryover effect from the previous culture. CD11B did not seem to be down-regulated, confirming that the adhesion molecule was directly involved in the PLP binding, and for this reason not accessible for antibody staining (Figure 1D).
Coincubation: THP-1 + PLPs gene expression analysis
| Up-regulated genes . | Down-regulated genes . | ||
|---|---|---|---|
| HBG1/HBG2 | HBE1 | MMP13 | MMP2 |
| HBA1 /HBA2 | CALB1 | NFYA | CD80 |
| HPGD | ACTR2 | KIF2A | STXBP5L |
| DNAJC12 | GREB1 | GPLD1 | RARA |
| TRAJ17 /TRAV20 | SOD2 | RAB7A | TNFRSF11A |
| MRPS15 | XDH | TNFRSF10D | |
| FGA | |||
| Up-regulated genes . | Down-regulated genes . | ||
|---|---|---|---|
| HBG1/HBG2 | HBE1 | MMP13 | MMP2 |
| HBA1 /HBA2 | CALB1 | NFYA | CD80 |
| HPGD | ACTR2 | KIF2A | STXBP5L |
| DNAJC12 | GREB1 | GPLD1 | RARA |
| TRAJ17 /TRAV20 | SOD2 | RAB7A | TNFRSF11A |
| MRPS15 | XDH | TNFRSF10D | |
| FGA | |||
Pam3CSK4-treated THP-1 gene expression analysis
| Up-regulated genes* . | Down-regulated genes† . | ||||
|---|---|---|---|---|---|
| ICAM1 | EPB41L3 | THBS1 | GFBP3 | NEXN | RBM35A |
| VCAM1 | ADAM28 | RTN1 | IFI44L | ANKRD55 | SYNPO2 |
| CCL2 | EBI3 | CCL4 | S100A12 | WDR49 | SERPINI2 |
| CXCL14 | RASGRP1 | CCL8 | PRR16 | CEACAM6 | CA3 |
| BCL2A1 | TRPM1 | TNFAIP6 | BIRC3 | ||
| PLA2G7 | SLAMF7 | CLEC7A | TLR8 | ||
| LUM | TRPM1 | EPYC | |||
| Up-regulated genes* . | Down-regulated genes† . | ||||
|---|---|---|---|---|---|
| ICAM1 | EPB41L3 | THBS1 | GFBP3 | NEXN | RBM35A |
| VCAM1 | ADAM28 | RTN1 | IFI44L | ANKRD55 | SYNPO2 |
| CCL2 | EBI3 | CCL4 | S100A12 | WDR49 | SERPINI2 |
| CXCL14 | RASGRP1 | CCL8 | PRR16 | CEACAM6 | CA3 |
| BCL2A1 | TRPM1 | TNFAIP6 | BIRC3 | ||
| PLA2G7 | SLAMF7 | CLEC7A | TLR8 | ||
| LUM | TRPM1 | EPYC | |||
Fold change of 2.
Fold change of 1.5.
THP-1 HBG1/HBG2 expression after PLP RNase treatment. Cells cocultured for 24 hours with RNase-treated PLPs expressed less HBG1/HBG2 (RNaseA/T1 and RNase ONE) compared with the untreated PLP coculture (PLP). HBG1/HBG2, normalized against GAPDH, was absent in the control, THP-1 cells treated with 1 μg/mL PAM and 100 ng/mL TPO.
THP-1 HBG1/HBG2 expression after PLP RNase treatment. Cells cocultured for 24 hours with RNase-treated PLPs expressed less HBG1/HBG2 (RNaseA/T1 and RNase ONE) compared with the untreated PLP coculture (PLP). HBG1/HBG2, normalized against GAPDH, was absent in the control, THP-1 cells treated with 1 μg/mL PAM and 100 ng/mL TPO.
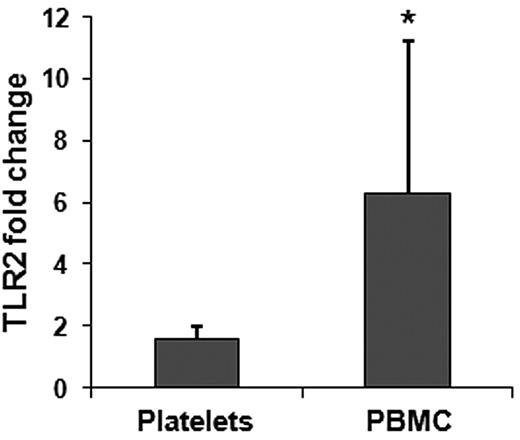
Platelet TLR2 RNA transfer in vivo
Mouse studies also were performed to test whether a specific mRNA could be transferred by platelets to monocytes in vivo. Platelets were isolated from WT mice (TLR2 RNA present) and infused into TLR2-deficient animals. Complementary to the coincubation experiments, we induced the formation of heterotypic aggregates using LPS, a bacteria LPS and TLR4 agonist. Flow cytometry results from platelet transfusion experiments performed on TLR2 knockout mice showed the presence of platelet-monocyte heterotypic aggregates 3 hours after transfusion with an increase in TLR2-positive monocytes in presence of LPS (83.60% ± 2.24%) compared with saline treatment (33.6% ± 0.33%). LPS treatment produced a modest increase in platelet-positive monocytes (12.76% ± 1.46) compared with the relative saline treatment (9.56% ± 3.46). Because coculture experiments showed RNA transfer 6 hours after coincubation, we collected platelets and PBMCs 6 hours after platelet transfusion and analyzed RNA to verify whether TLR2 mRNA was transferred to the recipient cells. A 6-fold increase in TLR2 mRNA clearly demonstrated that during the stage of heterotypic aggregates formation platelet-monocyte RNA transfer had occurred (Figure 4). This was particularly important because it established that the process occurs in vivo and with platelets as well as PLPs.
Quantitative RT-PCR of TLR2 in transfused mice.TLR2 fold changes (expressed as 2−ΔΔCt) in platelets and PBMCs. All samples were normalized against GAPDH. In each case we used 3 mice per group; the transfer of TLR2 mRNA to PBMC was considered significant (*P = .05).
Quantitative RT-PCR of TLR2 in transfused mice.TLR2 fold changes (expressed as 2−ΔΔCt) in platelets and PBMCs. All samples were normalized against GAPDH. In each case we used 3 mice per group; the transfer of TLR2 mRNA to PBMC was considered significant (*P = .05).
Relationship between SOD2 activity and ROS production in THP-1 cells
Microarray results showed a modest increase in SOD2 expression that was confirmed by assaying directly the activity in the cells (Figure 5A); there was a 13% increase in SOD2 activity in presence of PLPs compared with the control (PAM). ROS production also was increased by the presence of PLPs. PAM-activated THP-1 cells produced ∼ 30% more ROS compared with resting cells; however, in presences of PLPs, there was a further 12% increase that was consistent with the amount of SOD2 activity recorded (Figure 5B).
SOD2 activity and intracellular ROS in THP-1 cells. (A) SOD2 activity assessed in THP-1 control cells, 1 μg/mL PAM-treated and in presence of PLPs (PLP). (B) Cells coincubated with (PLP-T) or without PLPs (PAM-T) were treated with 5μM 5-(and-6)-carboxy-2′,7′-difluorodihydrofluorescein diacetate for 30 minutes and then evaluated by flow cytometry versus controls with (PLP) and without PLPs (*P < .05 compared with PLP and **P < .001 compared with NT [NT, no treatment]).
SOD2 activity and intracellular ROS in THP-1 cells. (A) SOD2 activity assessed in THP-1 control cells, 1 μg/mL PAM-treated and in presence of PLPs (PLP). (B) Cells coincubated with (PLP-T) or without PLPs (PAM-T) were treated with 5μM 5-(and-6)-carboxy-2′,7′-difluorodihydrofluorescein diacetate for 30 minutes and then evaluated by flow cytometry versus controls with (PLP) and without PLPs (*P < .05 compared with PLP and **P < .001 compared with NT [NT, no treatment]).
Discussion
Intercellular nucleic acid transfer is known to be an important mechanism regulating metastasis and stem cell communication networks.33,–35 Platelets are modestly active in translating their own mRNA,36 but few studies have examined the function of the platelet transcriptome.11,12,37 In this study, we explored the possibility that platelets influence peripheral blood and vascular cells by transferring RNA to these cells. RNA horizontal transfer was investigated by in vitro and in vivo models, shedding new light on the relationship between platelets and other vascular cells.
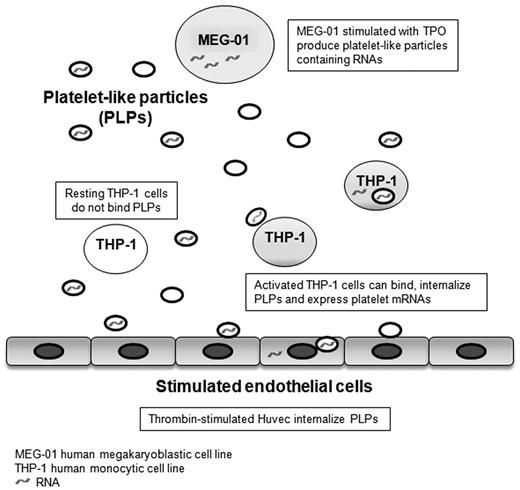
This study demonstrates that there is successful transfer of RNA from MEG-01 cells to HUVECs and THP-1 cells using PLPs as mediators of the transfer (Figure 6). Vascular cells bound and internalized PLPs with mechanisms similar to those already described for platelets and microvesicles.38,39 For example, MEG-01 cells stimulated with TPO produced PLPs carrying mainly fetal hemoglobin transcripts that were then transferred to the recipient cells having almost no baseline expression of these genes. As demonstrated in different cell lines, human TPO induces predominantly fetal and adult hemoglobin synthesis and, for the purpose of this study, were an excellent marker to validate an RNA transfer.40,–42 Interestingly, these data also suggest that RNA was not randomly transferred because MEG-01 cells may have distributed transcripts in the newly formed PLPs. This observation is supported by a recent study describing how megakayocytes selectively transfer matrix metalloproteases and their tissue inhibitors into platelets.43 PLP RNase treatments abrogated up to 75% of HGB1/HGB2 transfer after 24-hour coincubation, confirming that the mRNA recovered in THP-1 cells was from an outside source. The hemoglobin transfer was detectable after 12-hour coincubation (result not shown) in accordance with the slow process observed in THP-1 cells with the bromine-labeled RNA transfer. Many of the other gene expression changes were related to exo- and endocytosis.
In addition, the functionality of the PLP transcripts in the recipient cells was demonstrated by the GFP transfer; both the mature protein and mRNA presence were highlighted. PLPs actively interacted and transferred their RNA and GFP messenger to activated HUVECs as well; the process was faster and more efficient compared with THP-1 cells. After the fusion in THP-1 cells, we also observed an increase in ROS content that may be the result of the modest SOD2 overexpression, metabolic activity increase, or both.
Results obtained from in vitro experiments were clearly supported by the first mouse model showing that platelets can transfer TLR2 mRNA to PBMCs after an inflammatory stimulus. Considering the significant level of transcript detected 6 hours after transfusion in leukocytes, we conclude that RNA transfer in vivo is relatively fast and more efficient compared with the in vitro model. However, there are limitations to this experiment as transfer of only 1 transcript was studied.
Taken together, our observations suggest that platelets may have an interesting additional role in intercellular communication by acting as a source of transcripts. The transfer mechanism seems to require the direct contact between the cells, although the specific mechanism is not yet known. Cocultured cells have been shown to transfer short interfering RNAs with a mechanism that involves gap junctions.44 mRNAs instead were mainly identified in microparticles released from cells,18,45 indicating that size and structure prevent larger nucleic acids from being transferred through membrane channels. Our findings are unique in showing PLP internalization and GFP mRNA transfer, suggesting that platelets have a more complex regulatory role compared with microparticles.
Human platelets cannot be produced in vitro or cultured but PLPs, except for the lack of thrombotic activity, are very similar to human platelets. The RNA transfer model developed for this study suggests an intriguing in vivo scenario. The study shows that RNA was transferred and expressed in the recipient cells, and these results add a new role for the platelet transcriptome. Thus, platelet expression profiles analyzed in cancer and cardiovascular diseases46,–48 reveal transcript variations that may affect not only platelet function but also influence platelet-interacting cells and vascular homeostasis by direct transfer of genetic information.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Milka Koupenova for helpful assistance with the animal studies.
This work was supported by National Institutes of Health grants P01 A1078894 (J.E.F.) and T32 HL007224 (A.R. and L.M.B.).
National Institutes of Health
Authorship
Contribution: A.R. designed and performed experiments, analyzed data, and edited the manuscript; L.M.B. designed and performed animal studies, analyzed related data, and edited the manuscript; O.V. performed confocal microscopy shown in Figures 1B and 2B; and J.E.F. designed experiments and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jane E. Freedman, Department of Medicine, University of Massachusetts Medical School, 381 Plantation St, Biotech V, Suite 200, Worcester, MA 01605-4319; e-mail: jane.freedman@umassmed.edu.

![Figure 5. SOD2 activity and intracellular ROS in THP-1 cells. (A) SOD2 activity assessed in THP-1 control cells, 1 μg/mL PAM-treated and in presence of PLPs (PLP). (B) Cells coincubated with (PLP-T) or without PLPs (PAM-T) were treated with 5μM 5-(and-6)-carboxy-2′,7′-difluorodihydrofluorescein diacetate for 30 minutes and then evaluated by flow cytometry versus controls with (PLP) and without PLPs (*P < .05 compared with PLP and **P < .001 compared with NT [NT, no treatment]).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/119/26/10.1182_blood-2011-12-396440/5/m_zh89991293290005.jpeg?Expires=1765914920&Signature=KFDIRIJ3hRjX4j4WtEEoiYSPw5M6-xvYa1-PV0WhHkucmJSEyptQOZS5C-upqHd7LAddxAut6BVSgbvZRHfvN5VzeIK2r-uKMmMaT2Vq5wPprx2nf9LuFMtSyz0v5PzVtruvoiiVQejhlUaV2Bzjeb1f-J6eGEvbZHq~ZeJpqNPrSfmElxb2N86hFfqETv0jvy9PdNP9TxKcg1nvW0UB~Esle70loqo4N6qcqBD~ZiFWWxrObVFe4ILZ39x1qJqXOSr6PeaKpqVBpig5oog0M5AqU0wv77fkqJ07e4upWoznztPd-GQXMK-iOa-itdtUil1BRjXMolGxsoS1RyqxMA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)