We report on a forward RNAi screen in primary human hematopoietic stem and progenitor cells, using pooled lentiviral shRNA libraries deconvoluted by next generation sequencing. We identify MAPK14/p38α as a modulator of ex vivo stem cell proliferation and show that pharmacologic inhibition of p38 dramatically enhances the stem cell activity of cultured umbilical cord blood derived hematopoietic cells. p38 inhibitors should thus be considered in strategies aiming at expanding stem cells for clinical benefit.
Introduction
Umbilical cord blood (CB) is a highly promising and increasingly used therapeutic source of hematopoietic stem and progenitor cells (HSPCs) for transplantation regimens in a variety of malignant and nonmalignant hematologic diseases. However, the relatively low number of HSPCs present in single CB units poses major constraints and intense efforts are therefore being devoted to develop conditions that would enable the ex vivo amplification of transplantable stem cells.1,–3 Here, we have developed a forward RNAi screen to identify modifiers of primary human HSPCs that may be targeted to promote expansion of undifferentiated cells.
Methods
Cell culture
CB CD34+ cells (see supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) were cultured in SFEM serum-free expansion medium (StemCell Technologies) supplemented with cytokines (Peprotech): stem cell factor (SCF), thrombopoietin (TPO), and FLT3-ligand (FLT3L), each at 100 ng/mL (STF conditions). For p38 inhibition, the following compounds reconstituted in DMSO were added: 10μM SB203580 (Sigma-Aldrich), 1μM Vx702 (Tocris Biosciences), 400nM BIRB796, and 100nM Ly2228820 (Selleck Chem).
Screening procedure
CD34+ cells were lentivirally transduced as described elsewhere4 and subsequently passaged in STF conditions. Genomic DNA from cells collected at day 3 and at day 20 after CD34 enrichment (see supplemental Methods) was isolated with the High Pure PCR Template Preparation Kit (Roche Applied Science). Provirus integration sites were PCR amplified and sequenced (Illumina Genome Analyzer) at the High Throughput Sequencing Facility of the Partners Healthcare Center for Personalized Genetic Medicine (PCPGM; Cambridge, MA). The number of reads for all shRNAs was normalized to the total number of annotated shRNA sequences for each sample. For validation experiments, shRNAs were sub-cloned through the Nde1/Spe1 restriction sites to a modified version of the pLKO1 vector (Sigma-Aldrich), where GFP replaced the puromycin resistance marker.
Human engraftment assay
The cultured equivalent of 30 000 input CD34+ cells was intravenously injected into 12- to 16-week-old NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice (The Jackson Laboratory). Human contribution was assessed by anti–human CD45 antibody (HI30) and lympho/myeloid distribution was evaluated with anti-CD33 (P67.6), anti-CD15 (HI98), and anti-CD19 (HIB19) antibodies. All animal experiments were reviewed and approved by the Lund/Malmö Local Ethical committee. Western blot, assays for cell division history, apoptosis, and reactive oxygen species. See supplemental Methods for details.
Results and discussion
We previously reported on the utility of lentiviral shRNA libraries to screen for modifiers of primary human HSPCs using the limited persistence of HSPCs ex vivo as a basis for functional selection of shRNAs promoting maintenance or expansion of undifferentiated HSPCs.5 Here, we developed this screening approach further by taking advantage of next generation sequencing to track shRNA-transduced HSPCs, allowing thousands of perturbations to be tested in parallel in a single pool of cells.
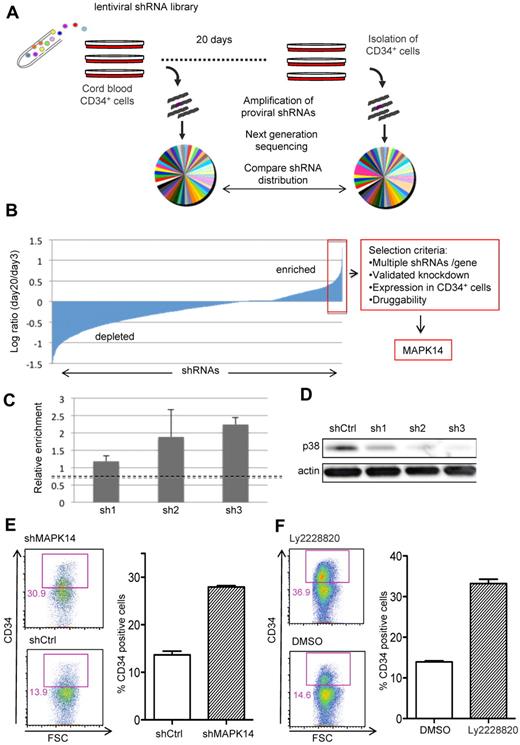
We used a pooled subset of the RNAi consortium (TRC) lentiviral shRNA library6 targeting around 1000 genes with 2 to 5 shRNAs per gene. The library was selected to include a high proportion of “druggable” genes (mainly kinases and phosphatases), enabling exploitation of any hits from the screen using small molecules and pharmacologic inhibition. CB-derived CD34+ cells (1-2 × 106 cells per replicate screen) were transduced with the pooled library at a transduction rate allowing for mostly single copy integrations (30%-35%), and cultured in serum-free medium supplemented with growth factors. Candidate target genes for HSPC expansion were identified by shRNAs conferring sustained CD34 expression during the culture. We assessed the distribution of all shRNAs in the library by next generation sequencing of integrated proviruses amplified from genomic DNA of CD34+ cells, 3 days after transduction and after 20 days of culture (Figure 1A). This analysis generated over 15 million annotated shRNA sequence tags. The relative changes in contribution of all shRNAs during the culture period, calculated from 3 independent replicate screens, are shown in Figure 1B. Candidate target genes were then identified from the most enriched fraction (top 5%) of shRNAs (Figure 1B). We have previously shown that shRNAs have a high tendency to generate false-positive hits through off-target effects5 and consequently only considered genes for which 2 or more shRNAs showed enrichment in the screen as well as a documented knockdown efficiency above 70% (supplemental Table 1). Many of the top-ranked genes were kinases involved in regulation of cell proliferation and we also identified genes with known tumor-suppressive functions, for example, LATS1 and WT1 (see supplemental Table 1 for details). Since our primary goal was to identify genes that could be manipulated in expansion protocols, we decided to first focus on MAPK14 as it is readily druggable by several pharmacologic inhibitors and has distinct expression in CD34+ cells. MAPK14 encodes p38α, a member of p38 family of MAP kinases, previously implicated in maintenance of the hematopoietic stem cell pool in mice.7 The screen revealed an enrichment of 3 independent shRNAs targeting MAPK14 (Figure 1C), displaying a direct correlation between level of enrichment and knockdown efficiency (Figure 1D). Validation experiments showed that either MAPK14 silencing by lentiviral shRNA (Figure 1E) or p38α inhibition by a small molecule inhibitor (Figure 1F) enhanced the frequency of CD34+ cells during culture, introducing MAPK14/p38α as a promising target to manipulate ex vivo HSPC activity.
A forward RNAi screen identifies MAPK14 as a regulator of human HSPCs. (A) CB CD34+ cells were transduced with a pooled lentiviral library and cultured for 20 days. Cell samples harvested after 3 and 20 days were analyzed by next generation sequencing of proviral shRNAs. (B) Relative changes in shRNA distribution calculated as the normalized number of shRNA sequence reads at day 20 divided by the number of reads day 3. The mean from 3 independent screens is plotted in ascending order. (C) The relative enrichment in CD34+ cells at day 20 for 3 independent shRNAs (sh1 to sh3) targeting MAPK14. The mean value of all shRNAs in the screen is shown as a dotted line. (D) Western blot of p38 expression in CD34+ cells transduced with MAPK14-targeting shRNAs (sh1 to sh3) or control shRNA targeting β-galactosidase (shCtrl). (E) CD34 expression after 17 days of culture of CB CD34+ cells transduced with MAPK14-targeting shRNA or control shRNA (n = 3). (F) CD34 expression after 14 days of culture of CB CD34+ cells treated with the P38α inhibitor Ly2228820 or vehicle (DMSO; n = 3). Error bars represent SEM.
A forward RNAi screen identifies MAPK14 as a regulator of human HSPCs. (A) CB CD34+ cells were transduced with a pooled lentiviral library and cultured for 20 days. Cell samples harvested after 3 and 20 days were analyzed by next generation sequencing of proviral shRNAs. (B) Relative changes in shRNA distribution calculated as the normalized number of shRNA sequence reads at day 20 divided by the number of reads day 3. The mean from 3 independent screens is plotted in ascending order. (C) The relative enrichment in CD34+ cells at day 20 for 3 independent shRNAs (sh1 to sh3) targeting MAPK14. The mean value of all shRNAs in the screen is shown as a dotted line. (D) Western blot of p38 expression in CD34+ cells transduced with MAPK14-targeting shRNAs (sh1 to sh3) or control shRNA targeting β-galactosidase (shCtrl). (E) CD34 expression after 17 days of culture of CB CD34+ cells transduced with MAPK14-targeting shRNA or control shRNA (n = 3). (F) CD34 expression after 14 days of culture of CB CD34+ cells treated with the P38α inhibitor Ly2228820 or vehicle (DMSO; n = 3). Error bars represent SEM.
We next addressed the utility of p38 inhibition to enhance stem cell expansion protocols. As reference conditions we used serum-free medium supplemented with SCF, TPO, and FLT3L (STF conditions), which has been shown by us and others to maintain or modestly expand HSPC activity in short-term cultures (5-8 days) as measured by engraftment potential in immunodeficient mice (C.K. and J.L., unpublished observations, November 2010).1,8,9 We measured the levels of p38 and its active phosphorylated form during a 5-day culture period and found high levels of active p38 especially during the first 3 days of culture (Figure 2A). Addition of a p38 inhibitor rapidly reduced the levels of phosphorylated p38 (Figure 2A). We next tested 4 independent p38 inhibitors under these culture conditions and monitored expression of CD34 and CD90 as recent findings from our laboratory show that the SCID repopulating activity of cultured HSPCs is exclusively contained within a small fraction of cells that coexpress CD34 and CD90 (A.B., C.K., and J.L., Expression of CD90 marks the SCID repopulating activity of cultured human hematopoietic stem cells, manuscript in preparation). While cells treated with each of the 4 inhibitors showed a modest reduction in total cell number (supplemental Figure 1A), we observed a marked increase in both frequency and number of CD34+CD90+ cells after 5 days of culture (Figure 2B-C). To further determine the content of functional HSPC in these cultures, we transplanted cells to sublethally irradiated Nod Scid Gamma (NSG) mice. Remarkably, cells treated with the p38 inhibitors exhibited a substantial increase in the long-term human reconstitution levels, compared with cells grown in STF conditions alone (Figure 2D and supplemental Figure 2). The engrafted cells showed robust repopulation of both the myeloid and lymphoid lineage (Figure 2E). Thus, modulating MAPK14/p38α in ex vivo expansion cultures dramatically enhances the output of HSPCs with in vivo reconstitution ability.
Pharmacologic inhibition of MAPK14/p38 enhances the HSPC activity of cultured CB CD34+ cells. CB CD34+ cells (70 000-100 000 cells per well) were cultured in STF medium supplemented by the specified MAPK14/p38 inhibitors or vehicle (DMSO) during 5 days and were then analyzed by FACS or transplanted to sublethally irradiated NSG mice. (A) Time course expression analysis of total p38 (t-P38) and phosphorylated p38 (p-p38). Actin expression was used as loading control (act). D indicates days. (B) FACS analysis for CD34 and CD90 expression. (C) Numbers of CD34+90+ cells (data from a representative experiment with 3 independent replicates). Error bars represent SEM (D) Percentage of human (CD45+) cells in peripheral blood of NSG recipients (3-4 animals per group) at 6, 10, and 16 weeks after transplantation. (E) Lympho-myeloid potential assessed by FACS analysis of human CD19 (B-lymphoid cells) and CD33/CD15 (myeloid cells) in bone marrow of NSG recipient mice, 17 weeks after transplantation. Each plot represents an animal from the experiment presented in panel D. Numbers indicate the percentage of positive cells in whole bone marrow. (F) ROS staining (DCFDA probe) assessed by FACS and presented as fluorescence mean intensity (fmi) in CD34+ cells. Pooled data from 3 independent experiments. Error bars represent SEM. D indicates DMSO; SB, SB203580; Vx, Vx702; B, BIRB796; and Ly, Ly2228820.
Pharmacologic inhibition of MAPK14/p38 enhances the HSPC activity of cultured CB CD34+ cells. CB CD34+ cells (70 000-100 000 cells per well) were cultured in STF medium supplemented by the specified MAPK14/p38 inhibitors or vehicle (DMSO) during 5 days and were then analyzed by FACS or transplanted to sublethally irradiated NSG mice. (A) Time course expression analysis of total p38 (t-P38) and phosphorylated p38 (p-p38). Actin expression was used as loading control (act). D indicates days. (B) FACS analysis for CD34 and CD90 expression. (C) Numbers of CD34+90+ cells (data from a representative experiment with 3 independent replicates). Error bars represent SEM (D) Percentage of human (CD45+) cells in peripheral blood of NSG recipients (3-4 animals per group) at 6, 10, and 16 weeks after transplantation. (E) Lympho-myeloid potential assessed by FACS analysis of human CD19 (B-lymphoid cells) and CD33/CD15 (myeloid cells) in bone marrow of NSG recipient mice, 17 weeks after transplantation. Each plot represents an animal from the experiment presented in panel D. Numbers indicate the percentage of positive cells in whole bone marrow. (F) ROS staining (DCFDA probe) assessed by FACS and presented as fluorescence mean intensity (fmi) in CD34+ cells. Pooled data from 3 independent experiments. Error bars represent SEM. D indicates DMSO; SB, SB203580; Vx, Vx702; B, BIRB796; and Ly, Ly2228820.
To understand the basis for the increase in stem cell activity, we assayed p38-inhibited cells with respect to cell cycling and survival rate. Since we previously observed a reduction in total cell number (supplemental Figure 1A), we first asked whether the increased regenerative ability might result from the maintenance of quiescent stem cells throughout the expansion period. However, although p38 inhibited cells overall had undergone fewer divisions, all cells had divided at least once at the transplantation time point (supplemental Figure 1B). We then assessed apoptosis as well as levels of reactive oxygen species (ROS), which are known to negatively impact stem cell maintenance.10 While no difference in apoptotic rate was observed (supplemental Figure 1C), we detected significantly reduced levels of reactive oxygen species (ROS) on p38 inhibition (Figure 2F), implicating reduced oxidative stress as one possible mechanism behind the enhanced stem cell output.
In summary, using a functional forward genetic screen, we identified MAPK14/p38α as a highly promising target to enhance hematopoietic stem cell activity in ex vivo expansion settings. It will be of considerable interest to test p38 inhibitors in combination with other recently identified molecules that have been suggested to promote human HSPC expansion such as Notch2, Pleiotrophin, Aryl hydrocarbon receptor antagonists or angiopoietin-like proteins.3,8,11,12 Our findings further support the feasibility of pooled RNAi screens in conjunction with next generation sequencing to identify genes and pathways that regulate primary human stem cell populations.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Nir Hacohen for providing the lentiviral shRNA library and Oleg Iartchouk for assistance with Solexa sequencing.
This work was supported by grants from the Swedish Research Council, the Swedish Cancer Foundation, the Swedish Pediatric Cancer Foundation and an EU project grant (STEMEXPAND) to J.L.
Authorship
Contribution: J.L. conceptualized and designed the study together with A.B.; A.B. performed all experiments with assistance from M.S.T., C.K., and M.M.; R.G. assisted with data analysis; and A.B. and J.L. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jonas Larsson, Molecular Medicine and Gene Therapy, BMC A12, 221 84, Lund, Sweden; e-mail: jonas.larsson@med.lu.se.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal