Abstract
Patients with heparin-induced thrombocytopenia (HIT) remain at risk for recurrent thromboembolic complications despite improvements in management. HIT is caused by antibodies that preferentially recognize ultralarge complexes (ULCs) of heparin and platelet factor 4 (PF4) tetramers. We demonstrated previously that a variant PF4K50E forms dimers but does not tetramerize or form ULCs. Here, we identified small molecules predicted to bind PF4 near the dimer-dimer interface and that interfere with PF4 tetramerization. Screening a library of small molecules in silico for binding at this site, we identified 4 compounds that inhibited tetramerization at micromolar concentrations, designated PF4 antagonists (PF4As). PF4As also inhibited formation of pathogenic ULCs, and 3 of these PF4As promoted the breakdown of preformed ULCs. To characterize the ability of PF4As to inhibit cellular activation, we developed a robust and reproducible assay that measures cellular activation by HIT antibodies via FcγRIIA using DT40 cells. PF4As inhibit FcγRIIA-dependent activation of DT40 cells by HIT antibodies as well as platelet activation, as measured by serotonin release. PF4As provide new tools to probe the pathophysiology of HIT. They also may provide insight into the development of novel, disease-specific therapeutics for the treatment of thromboembolic complications in HIT.
Introduction
Heparin-induced thrombocytopenia (HIT) is a serious complication of heparin therapy. HIT develops in ∼ 1% to 3% of patients treated with unfractionated heparin for 5 to 10 days but also in patients treated with low-molecular-weight heparin and other polysaccharide anticoagulants.1-3 It is generally accepted that the clinical manifestations of HIT are caused by antibodies that recognize a complex composed of heparin and platelet factor 4 (PF4) tetramers.4 PF4 is a 70-amino acid, lysine-rich 7.8-kDa platelet-specific CXC chemokine that is secreted in high concentrations when platelets are activated. PF4 tetramers bind avidly to heparin and to cellular glycosaminoglycans.5-7 This interaction is central to the pathogenesis of HIT.4,8-11 HIT antibodies bind preferentially to PF4 over a narrow molar ratio of reactants, that is, ∼ 1-2 tetramers: 1 molecule of heparin, average molecular mass 15 kDa.4,10,12-14 We and others have shown that at these ratios, PF4 tetramers and heparin form stable but reversible ultralarge complexes (ULCs)10 that preferentially bind HIT antibodies and activate platelets through FcRγIIA.4,10,15 Therefore, the biochemical basis for ULC formation is of importance in understanding antigen formation and potentially as a means to disrupt antigen formation and prevent or manage HIT.
The crystal structure of PF4 reveals that the β-sheets of each monomer face the tetramer interface, with the N and C termini lying on the exterior surface of the molecule.16 The lysine-rich C-terminal α-helices contribute to a circumferential band of positively charged amino acid residues17 that have been implicated in the binding of heparin.18 Importantly, the energy contributed by salt bridges, specifically involving glutamic acid 28 and lysine 50, drive PF4 tetramer formation.16 In solution, PF4 exists in a dynamic equilibrium between monomeric, dimeric, and tetrameric forms.19 We have reported previously that when Lys50 is mutated to glutamic acid (K50E), PF4 readily forms dimers but not tetramers.10 Importantly, we also have demonstrated that ULCs are not formed when heparin is incubated with PF4K50E,10 demonstrating that tetramerization is a prerequisite for ULC formation. Therefore, inhibition of tetramer formation, which shifts the PF4 equilibrium toward dimers and monomers, provides a novel and rational approach to decreasing ULC formation.
Here, we have taken advantage of these observations and used computational chemistry to identify a potential binding site on the surface of the PF4 dimer near Glu28 and Lys50 and to screen a library of more than 1 million small molecules for potential PF4 antagonists (PF4As). We also describe the composition and activity of several of these compounds with inhibitory activities in the micromolar range that provide insight into the mechanism of autoantigen formation and cellular activation in HIT.
Methods
Unfractionated heparin was an injectable sodium salt from porcine intestinal mucosa (Sagent Pharmaceuticals). HiTrap heparin affinity columns used for protein purification were purchased from GE Healthcare. Immunochemicals used included horseradish peroxidase–conjugated sheep polyclonal anti–human PF4 from Enzyme Research Laboratory, mouse immunoglobulin G 2bκ (IgG2bκ; MOPC 141) from Sigma, and sheep anti–mouse IgG and alkaline phosphatase–conjugated goat anti–mouse IgG from Jackson ImmunoResearch Laboratories. Murine monoclonal antibodies KKO (anti–human PF4-heparin complex), RTO (anti–human PF4), and IV.3 (FcγRIIA-blocking antibody) have been described previously and were the kind gift of G. Arepally (Duke University).20 Human plasma samples were from patients with a high clinical suspicion for HIT,21 and who had positive heparin/PF4 ELISA (GTI X-HAT45; Waukesha) and serotonin release assay results. Use of these samples was approved by the University of Pennsylvania institutional review board. A bicinchoninic acid protein assay reagent kit and Bis(sulfosuccinimidyl)suberate (BS3) cross-linker were obtained from Pierce Chemical. Immulon 4 HBX microtiter plates for ELISA were from Thermo Fisher Scientific. Bovine serum albumin, ionomycin, phorbol 12-myristate 13-acetate, and p-nitrophenyl phosphate tablets were from Sigma-Aldrich, and the 3,3′,5,5′-tetramethylbenzidine liquid substrate system for ELISA was from Kirkegaard and Perry Laboratories.
Computational screening to identify potential PF4As
We used DOCK22 to screen a library of ∼ 1.1 million lead-like compounds from the ZINC database23 to identify those with the potential to bind to the PF4 dimer surface within a pocket located in the vicinity of Lys50 and Glu28. To do so, we generated a set of overlapping sphere centers to fill this site using SPHGEN24 to represent the negative image of the binding site on PF4. The coordinates of the sphere centers were used to orient ligands within the target site. Sphere-sphere distances were compared with ligand atom distances to find compatible orientations of the ligand, and a scoring grid to evaluate ligand orientations was produced using the program GRID.25 The docking calculations were run in parallel, using 56 simultaneous processes, on a 14-node compute cluster.26
Generation of human PF4 in S2 cells
cDNA encoding human PF4 was cloned into the plasmid pMT/BiP/V5-His A (Invitrogen) for expression in Drosophila Expression System (Invitrogen). Cloning was performed using BglII and AgeI cloning sites. A hexanucleotide encoding the BglII site was then eliminated by site-directed mutagenesis so that the expressed protein contained full-length wild-type (WT)–PF4 or PF4K50E with an identical sequence as their counterparts expressed in Escherichia coli.27 PF4 expression was induced by adding copper sulfate (to 0.5mM). The induced S2 cells were incubated in serum-free medium Insect-Xpress (Lonza Walkersville) for 3 to 5 days; supernatants were collected, sodium azide (0.02% final concentration) and EDTA (2.5mM final concentration) were added, and the media were filtered through an Express Plus 0.22-μm filter (Millipore).
Purification of PF4 and PF4K50E
WT-PF4 was purified from the media on a heparin HiTrap column on an ATKA Prime (GE Healthcare) at 4°C in 10mM Tris, 1mM EDTA, pH 8.0 buffer. Media were loaded in buffer containing 0.5M NaCl, and PF4 was eluted at 1.8M NaCl using a linear gradient. Fractions containing purified PF4 as detected by silver staining of 12% NuPAGE Bis-Tris gels (Invitrogen) were pooled, concentrated, and the buffer was exchanged into 50mM HEPES, 0.5M NaCl, pH ∼7.2 using an Amicon Ultra centrifugal filter (3K NMWL; Millipore). Protein was quantified using a bicinchoninic acid assay. PF4K50E was purified as WT-PF4, with the following modifications. The column buffer system used was 50mM MES, 1mM EDTA, pH 6.5. Media were loaded in buffer containing 0.3M NaCl and PF4K50E eluted at 1.3M NaCl using a linear gradient.
Measurement of PF4 oligomerization
The ability of compounds to disrupt the formation of PF4 tetramers was examined using a previously described cross-linking assay.10 PF4 (10 μg/mL) in PBS was incubated with or without various concentrations of potential antagonists for 60 minutes at room temperature, followed by addition of the chemical cross-linking reagent BS3 (0.2mM final concentration) for 30 minutes at room temperature. In other experiments, incubating PF4As with PF4 for as little as 5 minutes gave similar results. The reaction was stopped by adding NuPAGE lithium dodecyl sulfate sample buffer and denatured by heating to 70°C for 10 minutes according to the manufacturer's instructions. Aliquots (15 μL) were analyzed electrophoretically on a 12% NuPAGE Bis-Tris gel under reducing conditions, and protein was visualized by silver staining. To determine the relative amounts of variously sized oligomers, bands were quantified by densitometry using the Gel Logic 100 imaging system with Kodak molecular imaging software (Version 4.5.1). Data are expressed as percentage of total PF4 in each oligomeric state measured in at least 3 independent experiments. PageRuler Plus Pre-stained Protein Ladder (MBI Fermentas) served as the molecular weight standards.
Measurement of PF4:heparin ULCs by ELISA
ULCs were formed with PF4 that had been incubated in the presence or absence of potential PF4As. In brief, PF4 (7.5 μg/mL) was preincubated with various concentrations of PF4As for 1 hour at 37°C before adding heparin and the mixture (concentration of PF4, 5 μg/mL; heparin, 0.2 U/mL) was incubated for an additional 15 minutes at 37°C. In other experiments, incubating PF4As with PF4 for 5 minutes gave similar results. The resulting complexes were diluted to a final PF4 concentration of 0.1 μg/mL in buffer containing PF4A to maintain a constant PF4A concentration, and antibody binding was measured by ELISA, as described previously.10 In brief, ULCs were incubated in wells precoated with KKO, detected by adding horseradish peroxidase–conjugated sheep polyclonal anti–human PF4 antibody, and developed with 3,3′,5,5′-tetramethylbenzidine substrate. After stopping the enzymatic reaction with 1M H3PO4, absorbance was measured at 450 nm in a SpectraCount plate reader (Packard). To determine whether PF4As facilitate the dissociation of preformed ULCs, the ELISA was modified so that PF4 was incubated with heparin for 30 minutes before PF4As were added. Solutions were then incubated overnight at 37°C before measuring binding to KKO-coated wells. To show whether PF4As inhibit PF4:heparin antibody binding, a direct ELISA was performed as described previously.20
Measurement of FcγRIIA activation in DT40 cells
NFAT-Luc, a kind gift from Prof Arthur Weiss (University of California, San Francisco),28 contains a luciferase reporter under control of the IL-2 promoter and is activated by binding of nuclear factor of activated T-cells (NFAT). pEF6-FcγRIIA, which contains the human FcγRIIA coding sequence under the control of the human EF-1α promoter, was constructed by cloning a human FcγRIIA IMAGE clone into the multiple cloning site of pEF6c (Invitrogen).
DT40 cells (chicken B cells) were cultured in RPMI-1640 supplemented with 10% fetal bovine serum, 1% chicken serum (Invitrogen), 50μM β-mercaptoethanol, 2mM GlutaMAX (Invitrogen), 100 U/mL penicillin, and 100 U/mL streptomycin at 37°C under 5% CO2. One day before transfection, cells were split to 2.5 × 105/mL (75-cm2 flask). On the day of transfection, 2 × 106 cells were pelleted, washed twice with PBS, and then resuspended in 400 μL PBS. Cells were cotransfected with 5 μg of pEF6-FcγRIIA and 20 μg of pEF6-NFAT-Luc by electroporation in a 0.4-cm cuvette (Bio-Rad Laboratories) at 330 V, 500 μF using an Electroporator II (Invitrogen). Cells were then transferred to 1 well of a 6-well plate containing 8 mL of complete media and incubated overnight at 37°C under 5% CO2. On the following day, cells were pelleted by centrifugation and resuspended at 2 × 106 cells/mL in fresh media without serum. Cell aliquots (50 μL) were placed into each well of a 96-well plate. ULCs ± PF4As were prepared at 2 × concentration in media without serum, and 50 μL was added per well to a final well volume of 100 μL. To establish basal expression, 50 μL of media was added. As a positive control for FcγRIIA signaling, monoclonal antibody IV.3 was added (8 μg/mL) for 15 minutes at 37°C under 5% CO2 followed by 50 μL of sheep anti–mouse IgG secondary antibody at 15 μg/mL to cross-link and activate these receptors. As a positive control for an intact intracellular signaling pathway, phorbol 12-myristate 13-acetate (25 ng/mL) with ionomycin (1μM) was added. PF4 (10 μg/mL) and various concentrations of potential antagonists were coincubated for 60 minutes at 37°C under 5% CO2 followed by the addition of heparin (0.3 U/mL) for 15 minutes at 37°C in 5% CO2. KKO (20 μg/mL) or plasma from patients with serotonin release assay confirmed HIT (1:800 dilution) was then added. We added 50 μL of each mixture to the 96-well plate for 6 hours at 37°C under 5% CO2. Plates were frozen at −80°C. To measure activation, cells were thawed and lysed with 5× passive lysis buffer (Promega) for 15 minutes. Luciferase activity was measured on a MultiLumat LB 9506 luminometer (10-second readings; Berthold Technologies) using luciferase assay reagent (Promega) following the manufacturer's instructions. In other experiments, PF4As were incubated with PF4 for 5 minutes with the same results.
Serotonin release assay
The serotonin release assay, a functional assay for platelet activation in the presence of heparin, was performed by a modification of a previously described method.20,29 14C-5-Hydroxytryptamine creatinine sulfate (GE Healthcare)–labeled platelets (platelet rich plasma) from healthy volunteers were mixed with KKO (170 μg/mL) or with known platelet-activating HIT plasma in the absence (buffer control) or presence of PF4A. Assays were performed in the presence of heparin (1.0 U/mL) and in the absence of heparin (background). Negative controls without antibody were studied in parallel. Data are expressed as percentage of maximal release of radioactivity with release by the positive control plasma and 1.0 U/mL heparin defined as 100%.
Analysis of dose-response data
Dose-response data were analyzed using the indirect Hill relationship30 using Prism Version 5.03 (GraphPad Software) sigmoidal dose-response, variable slope model.
Results
Computational chemistry: in silico screening
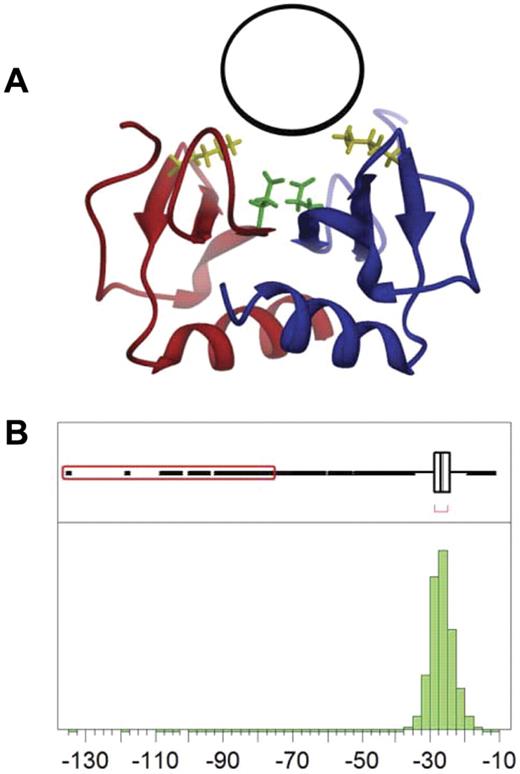
The target site used to identify potential PF4As is located on the surface of the dimer near residues Lys50 and Glu28 (Figure 1A). We screened a library of small-molecule “lead-like compounds.” Lead-like compounds are intended to serve as the nucleus of a drug and require further modification to increase affinity and specificity. Screening ∼ 1.1 million compounds required 10 days (∼ 1 compound/second) to perform. DOCK scores are empirical, with lower values representing higher predicted affinity. The docking scores obtained approximate a Gaussian distribution (Figure 1B), with a mean value of −26.6 and a standard deviation of 3.3. We identified 109 compounds that scored greater than 10 standard deviations below the mean (−60 or less), which were used as our starting point for experimental validation as PF4As.
Identification of binding site and computational screening. (A) The PF4 antagonist binding pocket (black circle) is located on the surface of the PF4 dimer (red and blue ribbons) near residues Lys50 (yellow) and Glu28 (green). (B) Distribution of DOCK scores for potential PF4 antagonists; 109 compounds have scores more than 10 SD below the mean (red box).
Identification of binding site and computational screening. (A) The PF4 antagonist binding pocket (black circle) is located on the surface of the PF4 dimer (red and blue ribbons) near residues Lys50 (yellow) and Glu28 (green). (B) Distribution of DOCK scores for potential PF4 antagonists; 109 compounds have scores more than 10 SD below the mean (red box).
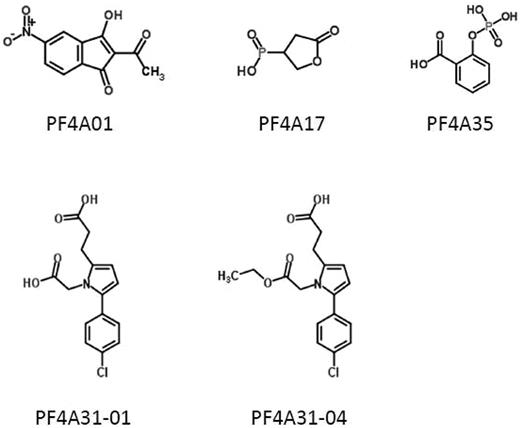
Based on availability from commercial sources, we obtained 21 of these compounds, and they were screened for their ability to inhibit PF4 tetramerization. Of these, 4 compounds were identified that inhibited PF4 tetramerization at micromolar concentrations, designated PF4A01, PF4A31-01, PF4A31-04, and PF4A35 (Figure 2). We focused our studies on these 4 compounds, as well as PF4A17 (which was predicted by our computational analysis to bind to PF4 but did not inhibit tetramerization) as a negative control.
Structures of PF4 antagonists. Chemical structures of 5 compounds characterized in this article are shown.
Structures of PF4 antagonists. Chemical structures of 5 compounds characterized in this article are shown.
Antagonism of PF4 tetramerization
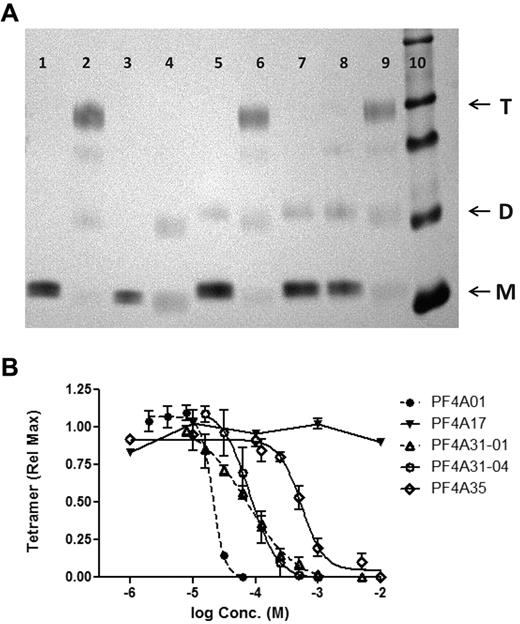
Because PF4As were designed to bind to the dimer interface of PF4 and disrupt tetramerization, our first test was to measure their ability to shift the oligomeric equilibrium of PF4 from the tetrameric toward the monomeric state. The oligomeric states of PF4 in the absence or presence of PF4As was measured by SDS-PAGE after chemical cross-linking, as described previously.10 As reported previously, tetramers are the predominant species of WT-human PF4, whereas the mutant PF4K50E is present primarily as dimers and monomers (Figure 3A). PF4As PF4A01, PF4A31-01, and PF4A31-04 each inhibited tetramerization of WT-PF4, shifting the oligomeric steady state toward the monomeric state. Neither PF4A35 (at 500μM) nor PF4A17 inhibited tetramerization (Figure 3A).
Inhibition of PF4 tetramerization. (A) SDS-PAGE of PF4 cross-linked with BS3 (or uncross-linked controls) with or without antagonists (lane 1, WT-PF4; lane 2, WT-PF4 cross-linked; lane 3, PF4K50E; lane 4, PF4K50E cross-linked; lanes 5-9, WT-PF4 cross-linked in the presence of antagonist: lane 5, 250μM PF4A01; lane 6, 500μM PF4A17; lane 7, 250μM PF4A31-01; lane 8, 250μM PF4A31-04; lane 9, 500μM PF4A35; lane 10, molecular mass markers). Arrows denote PF4 monomers (M), dimers (D), and tetramers (T). (B) Dose-response inhibition of PF4 tetramer formation by antagonists. Data are the mean ± SEM of at least 3 independent experiments. Curves represent fit of data to the indirect Hill equation.
Inhibition of PF4 tetramerization. (A) SDS-PAGE of PF4 cross-linked with BS3 (or uncross-linked controls) with or without antagonists (lane 1, WT-PF4; lane 2, WT-PF4 cross-linked; lane 3, PF4K50E; lane 4, PF4K50E cross-linked; lanes 5-9, WT-PF4 cross-linked in the presence of antagonist: lane 5, 250μM PF4A01; lane 6, 500μM PF4A17; lane 7, 250μM PF4A31-01; lane 8, 250μM PF4A31-04; lane 9, 500μM PF4A35; lane 10, molecular mass markers). Arrows denote PF4 monomers (M), dimers (D), and tetramers (T). (B) Dose-response inhibition of PF4 tetramer formation by antagonists. Data are the mean ± SEM of at least 3 independent experiments. Curves represent fit of data to the indirect Hill equation.
Four of the PF4As (including PFA35 at concentrations > 500μM) demonstrated a dose-dependent and ultimately complete inhibition of tetramer formation (Figure 3B). Analysis of these data with the indirect Hill equation revealed IC50 values for tetramer antagonism between 20 and 500μM (Figure 3B; Table 1). In contrast, PF4A17 did not inhibit tetramerization at concentrations as high as 10mM (Figure 3B). These data demonstrate that the 4 compounds that prevent spontaneous tetramerization of PF4 vary in potency.
Relative potency of antagonists
| Antagonist . | IC50, μM . | ||
|---|---|---|---|
| Tetramer . | ULC formation . | Activation . | |
| PF4A01 | 20 | 59 | 33 |
| PF4A17 | ND | ND | ND |
| PF4A31-01 | 72 | 200 | 440 |
| PF4A31-04 | 81 | 190 | 100 |
| PF4A35 | 500 | 1300 | 500 |
| Antagonist . | IC50, μM . | ||
|---|---|---|---|
| Tetramer . | ULC formation . | Activation . | |
| PF4A01 | 20 | 59 | 33 |
| PF4A17 | ND | ND | ND |
| PF4A31-01 | 72 | 200 | 440 |
| PF4A31-04 | 81 | 190 | 100 |
| PF4A35 | 500 | 1300 | 500 |
ND indicates not detectable.
Inhibiting formation and stability of PF4:heparin ULCs
We have shown previously that PF4 tetramerization is a prerequisite for formation of ULCs.10 Thus, we predicted that compounds able to inhibit PF4 tetramerization would prevent ULC formation. To test this hypothesis, we preincubated WT-PF4 with each PF4A before adding heparin. ULCs were then measured by ELISA.
As seen in Figure 4A, compounds that inhibited tetramerization (PF4A01, PF4A31-01, PF4A31-04, and PF4A35) inhibited ULC formation in a dose-dependent manner; inhibition was complete at the highest concentrations tested. PF4A17 did not inhibit ULC formation, consistent with its inability to inhibit PF4 tetramerization. Furthermore, the relative potency of these antagonists, as measured by IC50, is similar to that needed to antagonize tetramerization (Table 1). In an independent, orthogonal assessment of activity, similar results were obtained measuring ULCs by gel filtration as described previously10 (Figure 4B). These data are consistent with our hypothesis and demonstrate that PF4As that inhibit tetramer formation completely inhibit ULC formation.
Inhibition of PF4:heparin ULCs. (A) Dose response of antagonists for inhibition of ULC formation measured by ELISA. Data are the mean ± SEM of at least 3 independent experiments performed in triplicate. Curves represent fit of data to the indirect Hill equation. (B) Inhibition of ULCs as measured by gel filtration. Antagonists are at 1mM. Data are representative of 2 or more experiments. (C) Inhibition of ULCs by PF4As spans a wide range of heparin concentrations. Conditions are as in panel A, except that ULCs were formed with PF4 and various concentrations of heparin (0.05, 0.1, 0.2, 0.4, 0.8, and 1.6 U/mL from left to right in each group). (D) Incubation of KKO with PF4As (1mM) does not inhibit KKO binding to ULCs as measured by ELISA. Data are the mean ± SEM of at least 3 independent experiments performed in triplicate. (E) Dose response of antagonists for breakdown of preformed ULCs. Data are mean ± SEM of at least 3 independent experiments performed in triplicate. Concentrations of antagonists for each series (from right to left) are 2mM, 1mM, 500μM, and 250μM (125μM and 63μM for PF4A01 only).
Inhibition of PF4:heparin ULCs. (A) Dose response of antagonists for inhibition of ULC formation measured by ELISA. Data are the mean ± SEM of at least 3 independent experiments performed in triplicate. Curves represent fit of data to the indirect Hill equation. (B) Inhibition of ULCs as measured by gel filtration. Antagonists are at 1mM. Data are representative of 2 or more experiments. (C) Inhibition of ULCs by PF4As spans a wide range of heparin concentrations. Conditions are as in panel A, except that ULCs were formed with PF4 and various concentrations of heparin (0.05, 0.1, 0.2, 0.4, 0.8, and 1.6 U/mL from left to right in each group). (D) Incubation of KKO with PF4As (1mM) does not inhibit KKO binding to ULCs as measured by ELISA. Data are the mean ± SEM of at least 3 independent experiments performed in triplicate. (E) Dose response of antagonists for breakdown of preformed ULCs. Data are mean ± SEM of at least 3 independent experiments performed in triplicate. Concentrations of antagonists for each series (from right to left) are 2mM, 1mM, 500μM, and 250μM (125μM and 63μM for PF4A01 only).
Because complex formation is dependent on the relative amounts of PF4 and heparin, we examined the ability of several of these PF4As to inhibit ULC formation over a range of heparin concentrations. As seen in Figure 4C, PF4A01, PF4A31-01, and PF4A31-04 inhibit ULC formation over a range of heparin concentrations (0.05-1.6 U/mL) that span the parabolic dose-response curve described previously.8,10,31 PF4A17 shows no appreciable inhibition at any heparin concentration. These data demonstrate that inhibition of ULCs by PF4As is not dependent on the amount of heparin present.
To look at the effect of PF4As on antibody binding to intact ULCs, KKO (preincubated with PF4A or buffer) was incubated with ULCs immobilized directly onto the wells of the ELISA plate. Figure 4D demonstrates that PF4As do not directly inhibit antibody binding to preformed ULCs. These data are consistent with the mechanism for PF4A inhibition of antibody binding being decreasing ULC formation and not by competing with antibody binding.
As a more stringent test of inhibitory activity, we next examined the ability of these PF4As to facilitate the disruption of preformed ULCs. We hypothesized that this was possible because we have shown that ULCs, although relatively stable, are in a dynamic state and capable of redistributing into smaller PF4:heparin complexes.10 In these experiments, ULCs were formed and then incubated with PF4A or buffer for 24 hours at 37°C. Figure 4E shows that PF4A01, PF4A31-01, and PF4A31-04 facilitated the dissociation of preformed ULCs in a dose-dependent manner. Under the conditions examined, breakdown was incomplete (some ULCs are still present). However, for PF4A01 and PF4A31-04, less than 50% of ULCs remain compared with baseline. Of interest, PF4A35, the least potent PF4A based on inhibition of tetramer formation, was unable to promote the breakdown of preformed ULCs despite successfully preventing de novo ULC formation. As expected, PF4A17 did not inhibit the stability of preformed ULCs. These data demonstrate that a subset of PF4As that prevent ULC formation also promote ULC breakdown.
Inhibition of FcγRIIA-dependent activation by ULCs
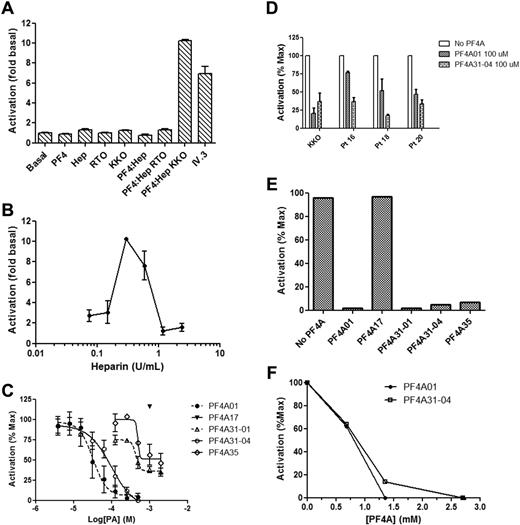
HIT antibodies bound to ULCs on platelets initiate activation by engaging FcγRIIA.32,33 We developed a model cell system to measure inhibition of FcγRIIA-mediated activation by HIT antibodies in the presence of PF4 and heparin to assess the effect of PF4As on cell activation. To do so, DT40 cells were transfected with pEF6-FcγRIIA and pEF6-NFAT-Luc (luciferase reporter under control of the IL-2 promoter). These cells are activated by the anti-FcγRIIA monoclonal antibody IV.3, as well as by the HIT-like monoclonal antibody KKO in the presence of ULCs (Figure 5A). RTO, an isotype control that binds avidly to PF4 but does not mimic the pathologic properties of HIT,20 failed to activate these cells. Importantly, KKO/ULC did not activate cells lacking FcγRIIA (transfected with pEF6-NFAT-Luc alone; data not shown), demonstrating that activation in this system is FcγRIIA-dependent. Furthermore, addition of PF4, heparin, KKO, or RTO alone did not activate the FcγRIIA-expressing cells, demonstrating the relevance of this assay to measuring the functional activity of HIT antibodies. We next examined whether FcγRIIA activation was heparin-dependent. To do so, we measured activation by KKO using a constant amount of PF4 and increasing amounts of heparin. As seen in Figure 5B, we observed the previously described bell-shaped dose dependence of heparin concentration on platelet activation.31 Taken together, the data support the use of the DT40 cell activation system as an assay of cellular activation via FcγRIIA dependent on PF4, heparin, and HIT antibody and therefore as a means to assess the effect of PF4As.
FcγRIIA-mediated activation of cells and inhibition by PF4 antagonists. (A) Activation of DT40 cells transfected with FcγRIIA and a luciferase reporter. Basal condition is a buffer only control, Hep indicates heparin, and IV.3 is the anti-FcγRIIA monoclonal antibody in the presence of anti-IgG antibody. (B) Dose response of DT40 activation by heparin in the presence of constant amounts of PF4. (C) Dose response of antagonists for inhibition of DT40 activation. Data are the mean ± SEM of at least 3 independent experiments performed in triplicate. Curves in panel C represent fit of data to the indirect Hill equation. (D) Inhibition of DT40 activation by plasma obtained from 3 HIT patients. Both antagonists completely inhibited activation by KKO and patient samples at a concentration of 500μM. Data represent the mean ± SEM of at least 2 independent experiments performed in triplicate. (E) Activation of platelets as measured by release of 14C-5-hydroxytryptamine creatinine sulfate. Data are representative of at least 2 experiments. Antagonists were present at a concentration of 2.5mM. (F) As in panel E, except that activation was by plasma from patients with HIT, and several antagonist concentrations were measured.
FcγRIIA-mediated activation of cells and inhibition by PF4 antagonists. (A) Activation of DT40 cells transfected with FcγRIIA and a luciferase reporter. Basal condition is a buffer only control, Hep indicates heparin, and IV.3 is the anti-FcγRIIA monoclonal antibody in the presence of anti-IgG antibody. (B) Dose response of DT40 activation by heparin in the presence of constant amounts of PF4. (C) Dose response of antagonists for inhibition of DT40 activation. Data are the mean ± SEM of at least 3 independent experiments performed in triplicate. Curves in panel C represent fit of data to the indirect Hill equation. (D) Inhibition of DT40 activation by plasma obtained from 3 HIT patients. Both antagonists completely inhibited activation by KKO and patient samples at a concentration of 500μM. Data represent the mean ± SEM of at least 2 independent experiments performed in triplicate. (E) Activation of platelets as measured by release of 14C-5-hydroxytryptamine creatinine sulfate. Data are representative of at least 2 experiments. Antagonists were present at a concentration of 2.5mM. (F) As in panel E, except that activation was by plasma from patients with HIT, and several antagonist concentrations were measured.
We then measured activation of DT40 cells by KKO, PF4, and heparin in the presence of increasing concentrations of PF4As (Figure 5C). PF4A01 and PF4A31-04 were the most potent inhibitors of cellular activation (Table 1), causing complete inhibition at the highest doses tested. PF4A31-01 and PF4A35 were less potent, and as expected, PF4A17 was not inhibitory. Importantly, PF4A01 and PF4A31-04 both inhibited activation by human HIT plasma with a similar potency as activation by KKO (Figure 5D).
In an independent, orthogonal assessment of activity, PF4A01, PF4A31-01, PF4A31-04, and PF4A35 (but not PF4A17) all inhibited platelet activation by KKO as measured by the serotonin release assay (Figure 5E). The rank order of potency for inhibition of cellular activation by these compounds paralleled their ability to inhibit tetramer formation. Furthermore, PF4A01 and PF4A31-04 inhibited serotonin release from platelets activated by plasma from patients with HIT (Figure 5F).
Inhibition of WT-PF4 with the nontetramerizing mutant PF4K50E
We have shown that PF4As inhibit tetramerization of PF4, and we propose that inhibition of tetramerization is the mechanistic basis for the observed effects of PF4As on ULCs and FcγRIIA antagonism. To provide independent support for this inference, we performed experiments using the nontetramerizing PF4 mutant PF4K50E as an antagonist of WT-PF4. Figure 6 shows that while keeping the total PF4 concentration constant, as the percentage of WT-PF4 mixed with PF4K50E is decreased, tetramerization, ULC formation, and FcγRIIA activation are all inhibited in a similar dose-dependent manner. These data provide independent evidence that inhibition of PF4 tetramerization leads to inhibition of FcγRIIA activation by decreasing ULC formation.
Inhibition of WT-PF4 with the nontetramerizing mutant PF4K50E. Inhibition of PF4 tetramerization (A), ULC formation (B), and FcγRIIA activation (C). PF4 concentrations were maintained constant by diluting WT PF4 with PF4K50E. Data are the mean ± SEM of at least 3 independent experiments performed in triplicate.
Inhibition of WT-PF4 with the nontetramerizing mutant PF4K50E. Inhibition of PF4 tetramerization (A), ULC formation (B), and FcγRIIA activation (C). PF4 concentrations were maintained constant by diluting WT PF4 with PF4K50E. Data are the mean ± SEM of at least 3 independent experiments performed in triplicate.
Discussion
Studies from several groups, including ours, indicate that HIT is initiated by antibodies that recognize a subset of antigenic complexes that form between heparin and PF4.4,10,14 At ratios of PF4 and heparin that maximize antibody binding and platelet activation, higher order complexes form between PF4 tetramers and heparin that are stable but reversible, bind multiple IgG molecules and more effectively activate human platelets through FcRγIIA.4,10,14 Thus, PF4 tetramerization is a prerequisite for effective platelet activation and so represents a rational mechanistic target for disruption that is both upstream of thrombin generation in the pathogenic pathway and specific for HIT rather than acting as systemic anticoagulants. In support of this mechanistic concept, other groups have recently reported that anticoagulant heparin 2-O,3-O-desulfated heparin, a heparinoid molecule, can disrupt PF4:heparin complexes and decrease platelet activation.34,35 This article describes the initial steps in the identification and characterization of small molecules that bind to specific sites on PF4 and thereby (1) inhibit PF4 tetramerization, (2) inhibit formation of PF4:heparin ULCs, (3) disrupt preformed ULCs, and (4) prevent HIT antibody and FcγRIIA-mediated platelet activation.
We also report the development of a robust and reproducible new assay to measure HIT antibodies capable of inducing cellular activation via FcγRIIA by transfecting the receptor and an NFAT-Luc reporter into DT40 cells. We have shown that these cells are activated by the HIT-like monoclonal antibody KKO, as well as by antibodies from several patients with HIT, when PF4 and heparin are present and that activation is via FcγRIIA. Activation of DT40 cells follows the expected bell-shaped heparin dose-response curve seen with HIT antibody binding, ULC formation and platelet activation.4,10,12,13 We then used this assay to measure the inhibitory capacity of PF4As on FcγRIIA-dependent cellular activation. PF4A01 and PF4A31-04, which differ structurally, both block PF4 tetramer formation and inhibit activation of DT40 cells. This is consistent with our mechanistic hypothesis that inhibition of PF4 tetramerization would decrease ULC formation, formation of large immune complexes on cell surfaces, and persistent engagement of FcRγIIA leading to cell activation. Thus, these compounds behave as predicted, affirm the role of ULCs in the pathogenesis of HIT, and they represent important new leads to study the mechanism of antigen assembly and formation of immune complexes in HIT.
PF4A31-01 is structurally similar to PF4A31-04 and has similar potency in terms of inhibiting formation of PF4 tetramers, but it was a less potent inhibitor of cell activation. The structural difference between these 2 antagonists involves esterification of an acetate group to a methyl ester in PF4A31-04, suggesting that decreased electronegativity, increased steric bulk in this position, or both may favor interaction with PF4. One possible explanation of this observation is that PF4A31-04 binds PF4 more stably compared with PF4A31-01, differentially impacting inhibition of cellular activation and tetramer formation.
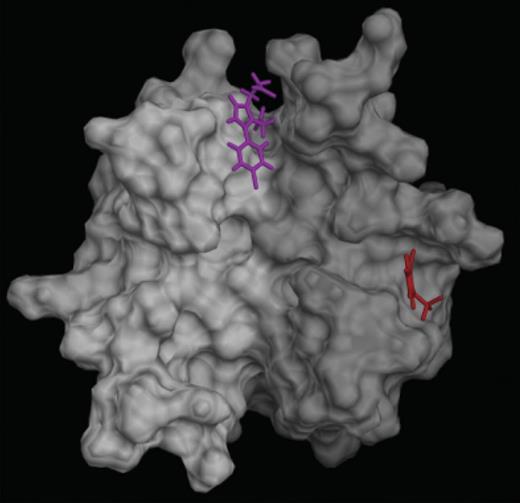
PF4A35 inhibits PF4 tetramerization and ULC formation, but it was not able to promote the breakdown of preformed ULCs and was a weak inhibitor of FcγRIIA activation in DT40 cells. Interestingly, PF4A35 is predicted to bind to a different region of the PF4 dimer interface than PF4A01 (Figure 7). A more detailed examination of the differences in the dimer-dimer interactions at these 2 sites may provide insight into the relative impotence of PF4A35. Another key distinction is that PF4A35 in solution is negatively charged because of the phosphate group, whereas PF4A01 is neutral. Preformed ULCs have negatively charged heparin on the surface that may repel the similarly charged PFA35. These factors may contribute to the relative inability of PF4A35 to inhibit ULC-driven cellular activation and lack of efficacy in the breakdown of ULCs despite the ability to inhibit PF4 tetramerization. Further studies into the mechanisms and structural basis of the activities of these compounds are required for a more definitive understanding of this phenomenon.
Predicted binding sites of PF4 antagonists. PF4 antagonists PF4A31 (magenta) and PF4A35 (red) are predicted to bind preferentially at different locations on the surface of the PF4 dimer (white-gray).
Predicted binding sites of PF4 antagonists. PF4 antagonists PF4A31 (magenta) and PF4A35 (red) are predicted to bind preferentially at different locations on the surface of the PF4 dimer (white-gray).
The computational screening methods that we applied to successfully identify a limited set of active lead compounds, were optimized to efficiently evaluate a large number of compounds and not to provide a detailed examination of the properties of each. By focusing our attention on a limited set of active compounds, it is now practical to use more sophisticated computational analyses that will comprehensibly characterize the properties of these leads, to design additional analogs, to establish structure-activity relationships, and to prioritize further experimental investigations.
In summary, we have identified several lead-like compounds through rational design that block the proximal steps in PF4 self-association and thereby downstream effects on platelet activation induced by HIT antibody. These compounds affirm the importance of PF4 tetramerization and oligomerization in antigen assembly and thereby represent important new tools to study the pathobiology of HIT. In theory, optimization of these or additional compounds provides a potential new approach to prevent or treat HIT. Similar approaches may be applicable for other disorders, such as antiphospholipid syndrome in which the antigenic target has been identified.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Craig Hughes and Anand Lokare for support in the DT40 experiments and Majd Protty for generating the FcγRIIA construct. They thank Beverly Ptashkin and May Dela Cruz from the special coagulation laboratory at the Hospital of the University of Pennsylvania for performance of serotonin release assays.
This work was supported by the National Institutes of Health grants HL078726 and HL078726-04-S1 (B.S.S.), R41HL106878 (J.J.R. and B.S.S.), and HL099973 and HL084006 (D.B.C.); The Wellcome Trust 088410; and the British Heart Foundation CH/03/003 (S.P.W.).
National Institutes of Health
Wellcome Trust
Authorship
Contribution: B.S.S., A.H.R., P.S.W., L.I.G., and J.J.R. designed experiments; A.H.R., L.I.G., J.L.H., S.V.Y., and J.J.R. performed experiments; B.S.S., A.H.R., D.B.C., S.P.W., L.I.G., J.L.H., and J.J.R. analyzed and interpreted data; and B.S.S., A.H.R., D.B.C., S.P.W., L.I.G., J.L.H., and J.J.R. wrote and edited the manuscript.
Conflict-of-interest disclosure: J.J.R. has an ownership interest in In Silico Molecular LLC, and A.H.R. has a spouse with ownership interest in In Silico Molecular LLC. The remaining authors declare no competing financial interests.
Correspondence: Bruce S. Sachais, Department of Pathology and Laboratory Medicine, Division of Transfusion Medicine and Therapeutic Pathology, University of Pennsylvania, 605A Stellar-Chance Labs, 422 Curie Blvd, Philadelphia, PA 19104; e-mail: sachais@mail.med.upenn.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal