Abstract
Cellular differentiation is orchestrated by lineage-specific transcription factors and associated with cell type–specific epigenetic signatures. In the present study, we used stage-specific, epigenetic “fingerprints” to deduce key transcriptional regulators of the human monocytic differentiation process. We globally mapped the distribution of epigenetic enhancer marks (histone H3 lysine 4 monomethylation, histone H3 lysine 27 acetylation, and the histone variant H2AZ), describe general properties of marked regions, and show that cell type–specific epigenetic “fingerprints” are correlated with specific, de novo–derived motif signatures at all of the differentiation stages studied (ie, hematopoietic stem cells, monocytes, and macrophages). We validated the novel, de novo–derived, macrophage-specific enhancer signature, which included ETS, CEBP, bZIP, EGR, E-Box and NF-κB motifs, by ChIP sequencing for a subset of motif corresponding transcription factors (PU.1, C/EBPβ, and EGR2), confirming their association with differentiation-associated epigenetic changes. We describe herein the dynamic enhancer landscape of human macrophage differentiation, highlight the power of genome-wide epigenetic profiling studies to reveal novel functional insights, and provide a unique resource for macrophage biologists.
Introduction
Human monocyte-to-macrophage differentiation is a process involving marked morphologic, functional, and transcriptional changes that proceed in the absence of proliferation. The mechanisms controlling this transition are not well understood on the molecular level, in part because both human monocytes and macrophages are hard to manipulate without triggering defense programs that interfere with normal differentiation.
Recent global epigenetic and transcription factor profiling studies in various cell types have provided ample evidence for a tight relationship between transcription factor binding and the local deposition/removal of some epigenetic marks, including histone methylation or acetylation, the appearance of histone variants, or DNA demethylation.1 Cell type–specific epigenetic signatures are particularly evident at promoter-distal sites, where histone H3K4 monomethylation/dimethylation,2,3 histone H3K27 acetylation,3,4 the histone variant H2AZ,2 or DNA demethylation5,6 indicate the presence of poised or activated lineage-specific enhancer elements. These distal regulatory elements are often cell type–specific, are correlated with gene expression, and are bound by combinations of common and cell type–specific key regulators.1,7 For example, in the murine hematopoietic system, macrophage-specific putative enhancer elements are characterized by PU.1, C/EBPα/β, and AP-1 binding, whereas putative enhancer elements in a related blood-cell type (murine B cells) that are also characterized by PU.1 binding associate with a distinct set of B cell–specific factors, including E2A, EBF, and OCT2.8 Observations of correlating transcription factor binding and epigenetic patterns were also made in other cellular systems, including embryonic stem cells,5 adipocytes9,10 and cancer cells,11,12 and the growing body of associative and functional data suggests that cell type–specific epigenetic patterns are indeed shaped by combinations of certain key transcription factors.
We hypothesized that the strong relationship between epigenetic modifications and transcription factor binding events, in particular at promoter-distal sites, may allow the identification of key regulators of the human macrophage differentiation process based on their epigenetic “fingerprint.” In the present study, by combining global epigenetic profiling with de novo motif analysis, an approach that allows the unbiased identification of sequence motifs without a priori knowledge, we identified 2 novel sequence motifs in the macrophage-specific enhancer signature. One of these motifs was shown to bind EGR2, which likely contributes to the (H3K4me1 or H3K27ac-marked) enhancer repertoire of human monocyte-derived macrophages in combination with other lineage-relevant DNA-binding proteins, including PU.1, C/EBPβ, and AP-1 family members.
Methods
Cells
Collection of peripheral blood monocytes from healthy donors was performed in compliance with the Declaration of Helsinki. All donors signed an informed consent. The leukapheresis procedure and subsequent purification of peripheral blood monocytes was approved by the local ethical committee (reference number 92-1782 and 09/066c). Macrophages were generated by culturing monocytes in endotoxin-free RPMI 1640 medium (Biochrom) supplemented with 2% human pooled AB-group serum on Teflon foils for up to 7 days (details are provided in supplemental Methods; see the Supplemental Materials link at the top of the article).
Western analysis
Western blotting was performed using whole-cell extracts, as described previously.13 The Abs used are listed in supplemental Methods.
ChIP
ChIP experiments were carried out as described previously.13 The Abs used and quality controls are described in supplemental Methods.
High-throughput sequencing and mapping
DNA from the ChIP analysis (10-50 ng) were adapter ligated and PCR amplified according to the manufacturer's protocol (Illumina). ChIP fragments were sequenced for 36 cycles on Illumina Genome Analyzers I or II according to the manufacturer's instructions. Sequence tags were mapped to the current human reference sequence (GRCh37/hg19) using Bowtie14 and only uniquely mapped tags were used for downstream analyses. Sequence tags from different donors were combined and tag counts were normalized to 107 specifically mapped tags. Published ChIP-sequencing (ChIP-seq) data for CD133+ hematopoietic stem cells (HSCs)15 were remapped from raw sequence data to the GRCh37/hg19 assembly using Bowtie,14 and local tag counts were normalized for GC nucleotide content to match the corresponding monocyte datasets using HOMER.8 A summary of the ChIP-seq data used in this study is provided in supplemental Table 1. Sequencing data have been deposited with the National Center for Biotechnology Information Gene Expression Omnibus database under accession number GSE31621. We also generated track hubs for the entire dataset, which are available at http://www.ag-rehli.de.
Sequencing data analysis
Analysis of mapped ChIP-seq tags was performed using HOMER.8 ChIP-seq quality control, transcription factor peak finding, and motif analysis were done as described previously.8 Genomic locations of peaks were defined relative to RefSeq transcription start sites (TSSs) and a minimum distance of 3 kb from annotated TSSs was considered promoter-distal. As opposed to the 200-bp peak size used for annotation and motif analysis of transcription factors, “peaks” in epigenetic ChIP-seq datasets were considered broader (1 kb) and were not required to exceed the local surrounding background. To compare ChIP-seq peak tag counts between 2 differentiation stages, peak sets of both cell types were merged if found within 1 kb, and the distribution of log2-transformed ChIP-seq tags was plotted as a color-coded tag count density map. Peaks showing at least 4-fold increased tag counts in comparisons between cell stages were counted as cell type–specific peaks. Histograms of tag densities were calculated using position-corrected, normalized tag counts. Motif enrichment was done by comparing sequences of cell type–specific peaks (± 100 bp for transcription factors, ± 500 bp for histone mark peaks) to 50 000 randomly selected genomic fragments of the same size, matched for GC content, and autonormalized to remove bias from lower-order oligo sequences. Motif enrichment was calculated using the cumulative hypergeometric distribution by considering the total number of target and background sequence regions containing at least one instance of the motif. De novo motif discovery was divided into 2 phases starting with a global, exhaustive scan of all oligos for their enrichment, followed by a second local optimization of motif probability matrices using the best oligos from the first phase as the initial seeds for the optimization. As motifs were discovered, their instances were masked from the input sequence to avoid convergence of multiple motifs on the same highly enriched sequence elements. The motifs with the lowest hypergeometric P values were considered the top motifs. Because of the numerous enrichment tests made during the motif discovery procedure and the vast search space, corrections for multiple hypothesis testing had to be carried out empirically by randomizing the target and background assignments and repeating the motif discovery procedure. One hundred randomizations (which were performed for each individual motif search) failed to yield motifs with enrichment P < 10−19, implying that the false discovery rate for motifs with P < 10−19 reported in this study was < 1%. To identify candidate transcription factors (or families of transcription factors) that potentially bound the identified sequences, de novo–derived motifs were compared against a library of known motifs consisting of motif matrices from the JASPAR database (http://jaspar.genereg.net/), a comprehensive library of motifs derived from published transcription factor ChIP-seq data, as well as published motifs not represented in the known motif library. Similarity was defined using matrix correlation coefficients (R2). The enrichment of Gene Ontology terms was calculated using DAVID tools.16 Hypergeometric P values for Gene Ontology term enrichment were corrected for multiple hypothesis testing using the Benjamini-Hochberg procedure. To test for associations between enhancer regions and known risk polymorphisms, the genomic locations of enhancers were compared with the positions of variants in the Genome-Wide Association Study (GWAS) catalog.17 For each GWAS study, single-nucleotide polymorphisms (SNPs) associated with increased risk of disease were assigned to enhancer regions if they were found within 2 kb of the enhancer locations. The P value of this association was calculated using the cumulative hypergeometric distribution by dividing the genome into 2-kb sections and assigning them to SNPs, enhancers, or both, and assumes that the positions of disease-risk SNPs and enhancers are distributed independently throughout the genome. Only GWAS studies with more than 5 risk SNPs were considered in this analysis. False discovery rates (q values) were calculated empirically by randomizing risk SNP positions 1000 times and recalculating the significance of their overlap with enhancer locations.
Results
Specific sequence motifs mark dynamic enhancer signatures and are correlated with potential key regulators of human macrophage differentiation
The differentiation of primary human blood monocytes into mature macrophages can be recapitulated in vitro and is accompanied by marked morphologic and functional changes. This transition proceeds in the absence of proliferation, requires the expression of novel sets of genes, and is clearly distinct from monocytic lineage commitment.18,19 The regulatory network characterizing the human macrophage differentiation program is not well understood, and it is not entirely clear which transcription factors drive the process, in particular the transition from human monocytes to macrophages. Because functional assays involving nucleic acids that are often used to study gene functions in other cellular systems (eg, RNA interference) induce defense programs and interfere with normal monocyte differentiation, systems biology approaches using comprehensive global datasets might present an alternative option to characterize the differentiation process. We hypothesized that the activation and DNA binding of key transcription factors associated with the differentiation process would be linked to local epigenetic changes and that these dynamically marked sites would be enriched for sequence motifs corresponding to such key regulators. A de novo search for such motifs in RefSeq-annotated promoter regions of differentially regulated genes did not reveal significant macrophage-specific motif signatures, so we extended our analysis to include promoter-distal regulatory elements as putative drivers of lineage differentiation. Using ChIP coupled with next-generation sequencing, we globally mapped and analyzed the distribution of the “poised” enhancer mark histone H3 lysine 4 monomethylation (H3K4me1), the “active” enhancer mark H3 lysine 27 acetylation (H3K27ac), and the mark for “open” chromatin (H2AZ) in freshly isolated human blood monocytes and monocyte-derived macrophages. We also included published data (H3K4me1 and H2AZ) from human CD133+ HSCs15 into our analysis.
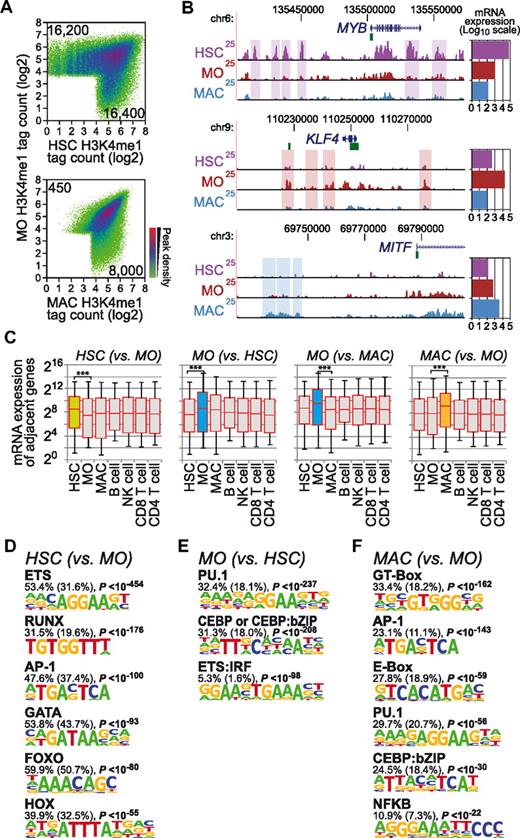
As shown in Figure 1A, the distribution of H3K4me1 between HSCs and monocytes and between monocytes and macrophages showed considerable differentiation-dependent dynamics at promoter-distal sites, defined herein as being ± 3 kb away from RefSeq-annotated gene TSSs. Tracks for 3 example genes encoding transcription factors (ie, MYB, KLF4, and MITF) that show cell stage–specific, promoter-distal H3K4me1 deposition that is correlated with their transcriptional activity are shown in Figure 1B (see corresponding bar charts to the right of the tracks). On the global level, the dynamic deposition of H3K4me1 at distal sites at a given developmental stage was correlated with higher mRNA expression levels of adjacent genes, suggesting that cell stage–dependent H3K4me1 deposition reflects a differentiation-dependent enhancer signature (Figure 1C).
Characterization of putative enhancer regions marked by cell stage–specific H3K4me1 during macrophage differentiation. (A) H3K4me1 ChIP-seq tag counts for peak regions are compared between macrophage differentiation stages (HSC, CD133+ HSCs; MO, monocytes; MAC, macrophages) in a density plot. The colors represent the relative density of peaks in each location within the density plot. Numbers in corners refer to the number of cell type–specific promoter-distal H3K4me1 sites (± 3 kb from RefSeq-annotated TSS). (B) Genome browser tracks for 3 transcription factors genes with cell stage–specific expression patterns. Boxes indicate promoter distal (based on RefSeq gene annotation), cell stage–specific H3K4me1-marked regions representing putative enhancer regions. Bar charts on the right show microarray-based mean expression values (log10 scale) for each gene in each cell type. Coloring indicates cell types (HSCs, purple; MOs, dark red; MACs, blue). (C) Box plots showing the distribution of mRNA expression levels (HSC, CD34+ HSCs; MO, monocytes; MAC, macrophages; and lymphoid cell types as indicated) for RefSeq genes adjacent to differentiation stage-specific promoter distal H3K4me1 peak regions. Cell types showing cell-stage specificity are indicated by colored boxes (HSCs, green; MOs, blue; MACs, orange). Solid bars of boxes display the interquartile ranges (25%-75%) with an intersection as the median; whiskers, 5th and 95th percentiles. Pairwise comparisons of mRNA expression levels for the indicated cell types are significant (***P < .001 by Student t test, paired, 2-sided). (D-F) De novo–extracted sequence motifs associated with differentiation stage–specific H3K4me1 peak regions. Motifs were assigned to transcription factors or transcription factor families based on similarity with known motif matrices. In addition, the fraction of H3K4me1 regions (1 kb) containing at least 1 motif instance, the expected frequency of the motif in random sequences (in parentheses), and P values (hypergeometric) for the overrepresentation of each motif are given.
Characterization of putative enhancer regions marked by cell stage–specific H3K4me1 during macrophage differentiation. (A) H3K4me1 ChIP-seq tag counts for peak regions are compared between macrophage differentiation stages (HSC, CD133+ HSCs; MO, monocytes; MAC, macrophages) in a density plot. The colors represent the relative density of peaks in each location within the density plot. Numbers in corners refer to the number of cell type–specific promoter-distal H3K4me1 sites (± 3 kb from RefSeq-annotated TSS). (B) Genome browser tracks for 3 transcription factors genes with cell stage–specific expression patterns. Boxes indicate promoter distal (based on RefSeq gene annotation), cell stage–specific H3K4me1-marked regions representing putative enhancer regions. Bar charts on the right show microarray-based mean expression values (log10 scale) for each gene in each cell type. Coloring indicates cell types (HSCs, purple; MOs, dark red; MACs, blue). (C) Box plots showing the distribution of mRNA expression levels (HSC, CD34+ HSCs; MO, monocytes; MAC, macrophages; and lymphoid cell types as indicated) for RefSeq genes adjacent to differentiation stage-specific promoter distal H3K4me1 peak regions. Cell types showing cell-stage specificity are indicated by colored boxes (HSCs, green; MOs, blue; MACs, orange). Solid bars of boxes display the interquartile ranges (25%-75%) with an intersection as the median; whiskers, 5th and 95th percentiles. Pairwise comparisons of mRNA expression levels for the indicated cell types are significant (***P < .001 by Student t test, paired, 2-sided). (D-F) De novo–extracted sequence motifs associated with differentiation stage–specific H3K4me1 peak regions. Motifs were assigned to transcription factors or transcription factor families based on similarity with known motif matrices. In addition, the fraction of H3K4me1 regions (1 kb) containing at least 1 motif instance, the expected frequency of the motif in random sequences (in parentheses), and P values (hypergeometric) for the overrepresentation of each motif are given.
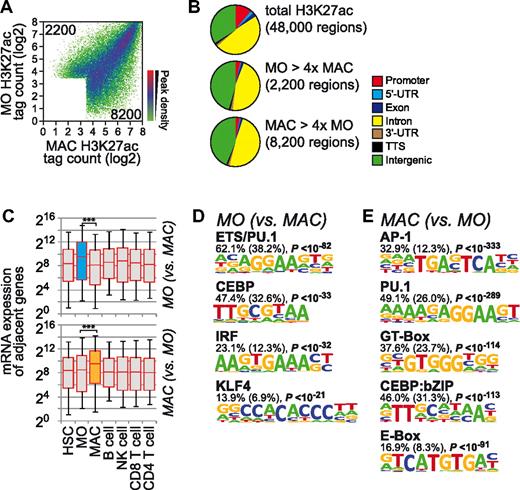
We next applied de novo motif analyses and searched for sequence motifs in differentially H3K4me1-marked sites. As detailed in “Methods,” we used HOMER, a novel motif-discovery algorithm,8 to extract motifs de novo from each set of cell type–specific peaks against a large set of nonoverlapping random sequences showing a similar nucleotide composition. Figure 1D through F show de novo–derived motifs that were significantly enriched in cell type–specific, H3K4me1-marked regions. The HSC-specific motif signature contained sequence motifs that were identified previously as binding sites for transcription factors important for HSCs, including ETS, RUNX, and GATA, an AP1-like putative basic leucine zipper transcription factor (bZIP)–binding site, as well as putative HOX or FOXO-like motifs (Figure 1D). The predominant de novo–extracted monocyte signature included motifs for PU.1, C/EBP family factors, and a composite ETS:IRF (EIRE) motif (Figure 1E). Only few sites actually lost H3K4me1 during monocyte-to-macrophage differentiation, and de novo motif analysis did not reveal any significant enrichment of specific sequences (data not shown). Macrophage-specific H3K4me1 regions were characterized by a distinct motif composition, including GT box, an AP1-like motif, an E-box element, the consensus PU.1 motif, a composite C/EBP:bZIP element,20 and a NFκB motif (Figure 1F). Similar observations were made for the distribution of 2 other enhancer-associated marks, H2AZ and H3K27ac (Figure 2 and supplemental Figure 1), with 2 main exceptions: (1) differentiation-associated presence of H2AZ was not associated consistently with mRNA expression changes in neighboring genes (supplemental Figure 1D), and (2) a larger proportion of regions lost H3K27ac marks during monocytic differentiation that were associated with motifs corresponding to PU.1/ETS, CEBP, and IRF consensus sites and a consensus sequence for KLF family factors (Figure 2D). Consistent with previous studies and supporting the enhancer nature of the studied histone modifications, one-third of randomly selected H3K27ac- or H3K4me1-marked sites showed enhancer activity in reporter assays (supplemental Figure 2).
Global distribution of H3K27 acetylation during macrophage differentiation. (A) H3K27ac ChIP-seq tag counts for peak regions are compared between monocytes and macrophages in a density plot. The colors represent the relative density of peaks in each location within the density plot. (B) Genomic distribution of total and cell stage–specific (at least 4-fold different) H3K27ac marked regions relative to RefSeq genes. (C) Box plots show the distribution of mRNA expression levels (HSC, CD34+ HSCs; MO, monocytes; MAC, macrophages; and lymphoid cell types as indicated) for RefSeq genes adjacent to differentiation stage-specific H3K27ac peak regions, as described in Figure 1. Significance of pairwise comparisons of mRNA expression levels for the indicated cell types are indicated (***P < .001 by Student t test, paired, 2-sided). (D-E) De novo–extracted sequence motifs associated with differentiation stage–specific H3K27ac peak regions. Motifs were assigned to transcription factors or transcription factor families based on similarity with known motif matrices. In addition, the fraction of H3K27ac regions (1 kb) containing at least 1 motif instance, the expected frequency of the motif in random sequences (in parentheses), and P values (hypergeometric) for the overrepresentation of each motif are given.
Global distribution of H3K27 acetylation during macrophage differentiation. (A) H3K27ac ChIP-seq tag counts for peak regions are compared between monocytes and macrophages in a density plot. The colors represent the relative density of peaks in each location within the density plot. (B) Genomic distribution of total and cell stage–specific (at least 4-fold different) H3K27ac marked regions relative to RefSeq genes. (C) Box plots show the distribution of mRNA expression levels (HSC, CD34+ HSCs; MO, monocytes; MAC, macrophages; and lymphoid cell types as indicated) for RefSeq genes adjacent to differentiation stage-specific H3K27ac peak regions, as described in Figure 1. Significance of pairwise comparisons of mRNA expression levels for the indicated cell types are indicated (***P < .001 by Student t test, paired, 2-sided). (D-E) De novo–extracted sequence motifs associated with differentiation stage–specific H3K27ac peak regions. Motifs were assigned to transcription factors or transcription factor families based on similarity with known motif matrices. In addition, the fraction of H3K27ac regions (1 kb) containing at least 1 motif instance, the expected frequency of the motif in random sequences (in parentheses), and P values (hypergeometric) for the overrepresentation of each motif are given.
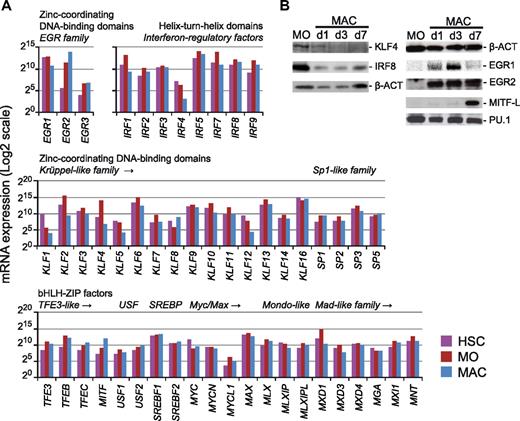
To narrow down transcription factor candidates that might correspond to the motifs discovered in H3K27ac- or H3K4me1-marked sites, we defined members of candidate transcription factor gene families that showed considerable gene expression in at least 1 of the 3 cell types using own published microarray expression data.21 Whereas mRNA expression alone is not equivalent for binding or activity (which may be regulated by the presence of ligands, posttranscriptionally or posttranslationally), it provides first clues about which factors are present and/or regulated during differentiation. Figure 3A summarizes gene-expression profiles for expressed members of the bHLH-ZIP, IRF, and zinc finger families (expression profiles for the bZIP, ETS, HOX, E2F, RUNX, and GATA families are provided in supplemental Figure 3).
Expression of motif-corresponding candidate transcription factors. (A) Shown are mRNA expression profiles for members of the bHLH-ZIP, IRF, and zinc finger gene families based on published microarray data for HSCs, MOs, and MACs.21 (Expression profiles for the bZIP, ETS, HOX, E2F, RUNX, and GATA gene families are found in supplemental Figure 3.) Classification of transcription factors is based on the properties of their DNA-binding domains (http://www.edgar-wingender.de/TFclass.html). Only genes with detectable expression are shown. Cell types are indicated by coloring (HSCs, purple; MOs, dark red; MACs, blue). (B) Western blot analysis of candidate transcription factor protein expression during human monocytic differentiation in vitro.
Expression of motif-corresponding candidate transcription factors. (A) Shown are mRNA expression profiles for members of the bHLH-ZIP, IRF, and zinc finger gene families based on published microarray data for HSCs, MOs, and MACs.21 (Expression profiles for the bZIP, ETS, HOX, E2F, RUNX, and GATA gene families are found in supplemental Figure 3.) Classification of transcription factors is based on the properties of their DNA-binding domains (http://www.edgar-wingender.de/TFclass.html). Only genes with detectable expression are shown. Cell types are indicated by coloring (HSCs, purple; MOs, dark red; MACs, blue). (B) Western blot analysis of candidate transcription factor protein expression during human monocytic differentiation in vitro.
Consistent with the de novo–derived motif signatures, the ETS-factor ERG, RUNX1, and various HOXA, E2F, and GATA family members are down-regulated in monocytes compared with HSCs, whereas PU.1, several bZIP factors, and IRF factors are induced in blood monocytes (Figure 3A and supplemental Figure 3). The Krueppel-like family member KLF4, a known key regulator in human monocytes,22,23 was most strongly and specifically up-regulated in monocytes and down-regulated during macrophage differentiation at both the mRNA and protein levels (Figure 3A and B, respectively) and thus represents a good candidate for the monocyte-derived KLF motif. The composite element EIRE, which was overrepresented in monocytes, was shown previously to bind PU.1 and IRF8.24,25 Whereas the latter is not drastically regulated on mRNA level, it is down-regulated on protein level during monocyte-to-macrophage differentiation (Figure 3B), suggesting that IRF8 may be part of the monocyte-specific enhancer signature.
Candidate transcription factors corresponding to the novel macrophage-specific motifs included the family of b-ZIP transcription factors, which are known to bind E-box elements. The macrophage-specific E-box element is similar to the M-box identified previously as the consensus binding site for the b-ZIP transcription factor microphthalmia (MITF).26 This factor was also highly induced during late macrophage differentiation on the mRNA and protein levels (Figure 3A-B), suggesting that it might represent a reasonable candidate for the macrophage-specific sequence motif. The second macrophage-specific element (the GT box) showed some similarity to the consensus site of EGR family zinc finger transcription factors. EGR2 was specifically induced in macrophages on both the mRNA and protein levels (Figure 3), whereas EGR1 was only expressed transiently during the early phase of monocyte differentiation, suggesting that both factors may contribute to early enhancer signatures (which were not studied here), whereas only EGR2 remains present in mature macrophages.
PU.1 and C/EBPβ binding are associated with the human macrophage-specific epigenetic signature
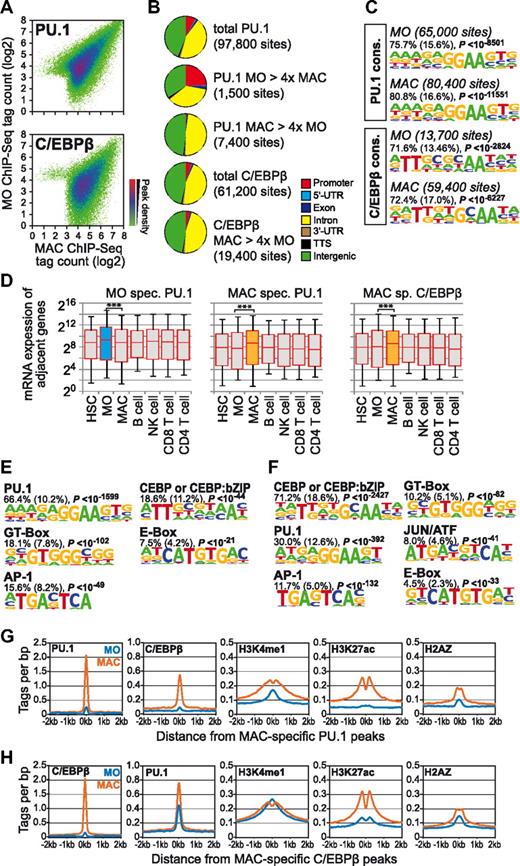
Because both HSC and monocyte epigenetic signatures revealed the “expected” motifs, the macrophage-specific motif signature likely reflected the appearance of key regulators driving monocyte-to-macrophage differentiation. The corresponding candidate transcription factors for some motifs present in the macrophage-specific enhancer signature were deducible, such as PU.1 and C/EBPβ, which were shown previously to be involved in human macrophage-specific gene regulation.13,27 To determine their overlap with macrophage-specific H3K4me1/H3K27ac-marked regions, we initially mapped global DNA binding of PU.1 and C/EBPβ. Whereas the latter factor is induced on the protein level during human monocyte-to-macrophage differentiation,13 cellular levels of PU.1 remain fairly constant in monocytes and macrophages (Figure 3), and its presence in the macrophage-specific enhancer signature was somewhat surprising. Figure 4 shows basic features of sites that were bound by each of the 2 factors. PU.1 binding was detectable in monocytes and macrophages, but displayed some degree of dynamics (Figure 4A). It was specifically lost at several promoter regions during macrophage differentiation, whereas the sites gained in macrophages were mostly located in promoter-distal regions (Figure 4B). The de novo–derived consensus motif, however, was almost identical at both differentiation stages (Figure 4C left panel). In contrast, C/EBPβ binding was strongly induced during differentiation (Figure 4A). In monocytes, C/EBPβ was actually only detected in 1 of 3 monocyte samples, which is consistent with our previous observation that C/EBPβ is phosphorylated and translocated into the nucleus during the differentiation process.13 The genomic distribution of total and macrophage-specific sites was similar (Figure 4B). The de novo–derived consensus motif were similar and represented a classic CEBP motif (in monocytes) or a mixture of motifs for C/EBP dimers and C/EBP:bZIP heterodimers20 in macrophages (Figure 4C). Dynamic binding of PU.1 in monocytes or macrophages and induction of C/EBPβ were correlated significantly with higher mRNA expression levels of adjacent genes (Figure 4D), suggesting that differentiation stage–dependent transcription factor binding is functionally relevant.
Dynamics of PU.1 and C/EBPβ binding during monocyte-to-macrophage differentiation. (A) ChIP-seq tag counts of each of the indicated transcription factors are compared for peak regions between monocytes (MO) and macrophages (MAC) in a density plot as described in the legend to Figure 1. (B) Pie charts depicting the genomic distribution of total transcription factor–bound sites or differentially bound regions (defined as having at least a 4-fold tag count difference in peak regions). For C/EBPβ, only a small fraction of monocyte-specific peaks were detected and are not included. (C) De novo–extracted consensus motifs for PU.1 and C/EBPβ bound sites. Motifs were assigned to transcription factors or transcription factor families based on similarity with known motif matrices. In addition, the fraction of transcription factor–bound regions containing at least 1 motif instance, the expected frequency of the motif in random sequences (in parentheses), and P values (hypergeometric) for the overrepresentation of each motif are given. (D) Box plots showing the distribution of mRNA expression levels for genes adjacent to differentiation stage-specific transcription factor peak regions as described in the legend to Figure 1. (E) De novo–extracted sequence motifs associated with macrophage-specific promoter-distal (according to RefSeq annotation) PU.1 peak regions. Motifs were assigned to transcription factors or transcription factor families based on similarity with known motif matrices. In addition, the fraction of PU.1-bound regions (200 bp) containing at least 1 motif instance, the expected frequency of the motif in random sequences (in parentheses), and P values (hypergeometric) for the overrepresentation of each motif are given. (F) Corresponding data for macrophage-specific C/EBPβ-bound regions are shown. (G) Histograms for genomic distance distributions of monocytes (MO) and macrophage (MAC) PU.1, C/EBPβ, H3K4me1, H3K27ac, and H2AZ tag counts centered across macrophage-specific PU.1-bound sites across a 4-kb genomic region. (H) Corresponding data for macrophage-specific C/EBPβ-bound regions.
Dynamics of PU.1 and C/EBPβ binding during monocyte-to-macrophage differentiation. (A) ChIP-seq tag counts of each of the indicated transcription factors are compared for peak regions between monocytes (MO) and macrophages (MAC) in a density plot as described in the legend to Figure 1. (B) Pie charts depicting the genomic distribution of total transcription factor–bound sites or differentially bound regions (defined as having at least a 4-fold tag count difference in peak regions). For C/EBPβ, only a small fraction of monocyte-specific peaks were detected and are not included. (C) De novo–extracted consensus motifs for PU.1 and C/EBPβ bound sites. Motifs were assigned to transcription factors or transcription factor families based on similarity with known motif matrices. In addition, the fraction of transcription factor–bound regions containing at least 1 motif instance, the expected frequency of the motif in random sequences (in parentheses), and P values (hypergeometric) for the overrepresentation of each motif are given. (D) Box plots showing the distribution of mRNA expression levels for genes adjacent to differentiation stage-specific transcription factor peak regions as described in the legend to Figure 1. (E) De novo–extracted sequence motifs associated with macrophage-specific promoter-distal (according to RefSeq annotation) PU.1 peak regions. Motifs were assigned to transcription factors or transcription factor families based on similarity with known motif matrices. In addition, the fraction of PU.1-bound regions (200 bp) containing at least 1 motif instance, the expected frequency of the motif in random sequences (in parentheses), and P values (hypergeometric) for the overrepresentation of each motif are given. (F) Corresponding data for macrophage-specific C/EBPβ-bound regions are shown. (G) Histograms for genomic distance distributions of monocytes (MO) and macrophage (MAC) PU.1, C/EBPβ, H3K4me1, H3K27ac, and H2AZ tag counts centered across macrophage-specific PU.1-bound sites across a 4-kb genomic region. (H) Corresponding data for macrophage-specific C/EBPβ-bound regions.
We next studied the binding behavior of PU.1 or C/EBPβ at cell stage–specific, distal H3K4me1 and H3K27ac marked regions. Macrophage-specific “enhancer” regions were increasingly bound by both factors in macrophages, suggesting that PU.1 and C/EBPβ contribute significantly to the macrophage-specific enhancer repertoire, whereas monocyte-specific regions were only enriched for PU.1 binding in monocytes (supplemental Figure 4). The few sites that lost PU.1 binding at distal regions in macrophages were associated with the CTCF and bZIP motifs (supplemental Figure 5A). The loss of PU.1 was associated with an average loss of local H3K4me1 and H3K27ac, suggesting that this factor is linked to the deposition of these marks at these (and likely other) sites (supplemental Figure 5B).
Regions that were bound specifically by PU.1 or C/EBPβ in macrophages were co-enriched for the same motifs that were identified in distal regions gaining H3K4me1 or H3K27ac during monocyte differentiation (Figure 4E-F). On average, PU.1 recruitment was correlated with increased H3K4me1, H3K27ac, H2AZ deposition, and C/EBPβ binding (Figure 4G), whereas C/EBPβ mainly resulted in the differentiation-dependent deposition of H3K27ac (Figure 4H). Factor binding generally induced an altered distribution of histone marks in the vicinity of the peak center, which has been observed previously for transcription factor–bound sites8,28 and suggests the differentiation-dependent positioning of adjacent nucleosomes (Figure 4G-H). Therefore, the cell stage–specific binding of either factor was strongly correlated with a cell stage–specific enhancer signature.
Novel transcription factors associated with the human macrophage-specific enhancer signature
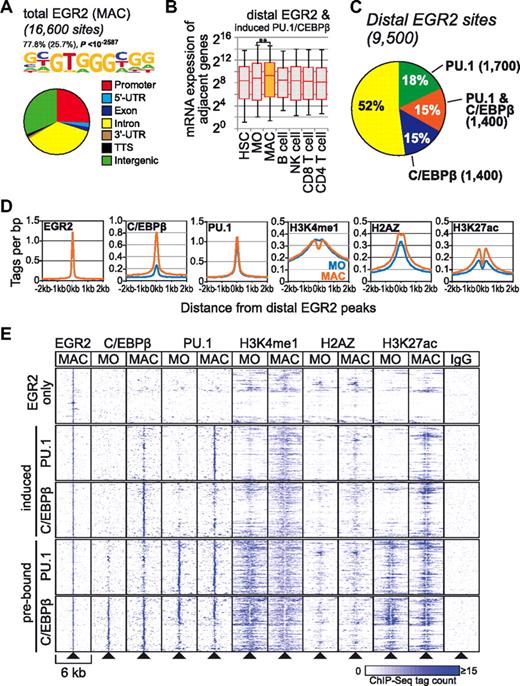
Whereas we were unsuccessful in studying global occupancy of one candidate transcription factor (MITF) in macrophages using ChIP assays (none of the tested commercially available Abs efficiently enriched previously identified MITF target sites, data not shown), we were able to map the global binding patterns of the second candidate, EGR2, in human macrophages using ChIP-seq. The ChIP-derived consensus motif of EGR2 was found to be identical to the GT box associated with macrophage-specific epigenetic signatures (Figure 5A). Co-associated motifs at distal sites included PU.1, C/EBP, and bZIP, confirming the frequent association of these factors in macrophages (supplemental Figure 6A). Consistent with its GC-rich recognition site, EGR2 showed a stronger association with promoters compared with PU.1 or C/EBPβ (Figure 5A). Genes adjacent to all EGR2-bound sites were modestly but significantly up-regulated in macrophages (supplemental Figure 6B). The average increase in macrophage gene expression was more pronounced at sites that also gained either PU.1 and/or C/EBPβ (Figure 5B), suggesting that EGR2 indeed participates in the regulation of macrophage-specific genes. Almost half of all distal EGR2 peaks had PU.1 or C/EBPβ peaks nearby (Figure 5C), and binding of the latter factors increased at distal EGR2 peaks during differentiation (Figure 5D). Whereas H3K27ac and H2AZ increased around EGR2 peaks, H3K4me1 deposition at EGR2-bound sites showed no average increase; however, the shift from a unimodal to a bimodal distribution indicated the differentiation-dependent positioning of adjacent nucleosomes (Figure 5D). Separation of EGR2 sites into those prebound by PU.1 or C/EBPβ, exhibiting induced binding of PU.1 or C/EBPβ, or showing no nearby PU.1 or C/EBPβ revealed that the increased enhancer marking and nucleosome positioning were associated primarily with induced PU.1 or C/EBPβ binding. Prebound PU.1 or C/EBPβ sites appeared to lose some of the H3K4me1 signal, but showed increases in the nucleosome-depleted area in the vicinity of the peak center, whereas only a smaller number of isolated EGR2 peaks was associated with a gain in enhancer mark density (Figure 5E).
EGR2 is associated with the macrophage-specific epigenetic enhancer signature. (A) De novo–extracted consensus motif for EGR2-bound sites and genomic distribution of total transcription factor–bound sites depicted as a pie chart. (B) Box plots showing the distribution of mRNA expression levels for genes adjacent to the 1800 EGR2 peak regions that also gained PU.1 or C/EBPβ in macrophages (indicated by orange). Solid bars of boxes display the interquartile ranges (25%-75%) with an intersection as the median; whiskers, 5th and 95th percentiles. Pairwise comparisons of mRNA expression levels for the indicated cell types are significant (**P < .01 by Student t test, paired, 2-sided). (C) Pie chart depicting the overlap of EGR2, PU.1, and C/EBPβ peaks within 200 bp of centered EGR2 ChIP-seq peaks. (D) Histograms for genomic distance distributions of tag counts for macrophage (MAC) EGR2 and monocyte (MO) and MAC PU.1, C/EBPβ, H3K4me1, H3K27ac, and H2AZ centered across macrophage-specific EGR2-bound sites across a 4-kb genomic region. (E) Promoter distal (according to RefSeq annotation) EGR2 peaks were subdivided into groups that showed induced binding for PU.1 and C/EBPβ, were already preoccupied by 1 of the 2 factors in monocytes, or were not co-bound by PU.1 or C/EBPβ. Regions (6-kb-wide, 500 of each group) centered on EGR2-bound peaks were clustered according to their C/EBPβ, PU.1, H3K4me1, H2AZ, and H3K27ac ChIP-seq profiles in monocytes and macrophages and results are presented as heat maps.
EGR2 is associated with the macrophage-specific epigenetic enhancer signature. (A) De novo–extracted consensus motif for EGR2-bound sites and genomic distribution of total transcription factor–bound sites depicted as a pie chart. (B) Box plots showing the distribution of mRNA expression levels for genes adjacent to the 1800 EGR2 peak regions that also gained PU.1 or C/EBPβ in macrophages (indicated by orange). Solid bars of boxes display the interquartile ranges (25%-75%) with an intersection as the median; whiskers, 5th and 95th percentiles. Pairwise comparisons of mRNA expression levels for the indicated cell types are significant (**P < .01 by Student t test, paired, 2-sided). (C) Pie chart depicting the overlap of EGR2, PU.1, and C/EBPβ peaks within 200 bp of centered EGR2 ChIP-seq peaks. (D) Histograms for genomic distance distributions of tag counts for macrophage (MAC) EGR2 and monocyte (MO) and MAC PU.1, C/EBPβ, H3K4me1, H3K27ac, and H2AZ centered across macrophage-specific EGR2-bound sites across a 4-kb genomic region. (E) Promoter distal (according to RefSeq annotation) EGR2 peaks were subdivided into groups that showed induced binding for PU.1 and C/EBPβ, were already preoccupied by 1 of the 2 factors in monocytes, or were not co-bound by PU.1 or C/EBPβ. Regions (6-kb-wide, 500 of each group) centered on EGR2-bound peaks were clustered according to their C/EBPβ, PU.1, H3K4me1, H2AZ, and H3K27ac ChIP-seq profiles in monocytes and macrophages and results are presented as heat maps.
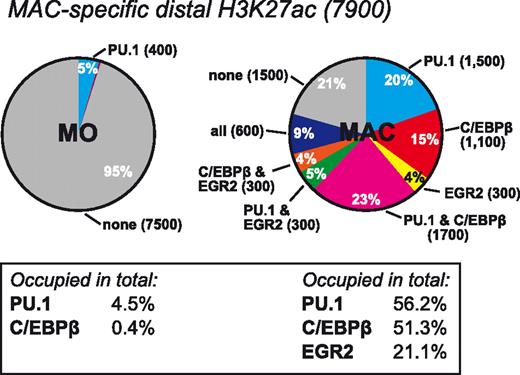
The global distribution of differentiation-associated transcription factor binding shows that the appearance of H3K4me1 or H3K27ac during differentiation is clearly associated with induced and combinatorial binding of PU.1, C/EBPβ, or EGR2. This was particularly evident for macrophage-specific H3K27ac sites, in which the overlap with PU.1, C/EBPβ, or EGR2 transcription factor peaks increased from 5% in monocytes to 79% in macrophages (Figure 6; corresponding data for H3K4me1 and monocyte-specific peaks are given in supplemental Figure 7). This result suggests that these 3 factors are key factors in establishing the macrophage-specific epigenetic signature.
Overlap between macrophage-specific enhancer marking and transcription factor binding in monocytes and macrophages. (A) Pie chart depicting the overlap of EGR2, PU.1, and C/EBPβ peaks within 1000 bp of centered macrophage-specific distal H3K27ac ChIP-seq peaks in monocytes (MO, left chart) and macrophage (MAC, right chart). The fraction of peaks bound by each factor in total is given below each chart. Corresponding charts for monocytes and H3K4me1 for both cell types are provided in supplemental Figure 7.
Overlap between macrophage-specific enhancer marking and transcription factor binding in monocytes and macrophages. (A) Pie chart depicting the overlap of EGR2, PU.1, and C/EBPβ peaks within 1000 bp of centered macrophage-specific distal H3K27ac ChIP-seq peaks in monocytes (MO, left chart) and macrophage (MAC, right chart). The fraction of peaks bound by each factor in total is given below each chart. Corresponding charts for monocytes and H3K4me1 for both cell types are provided in supplemental Figure 7.
Epigenetic enhancer signatures and disease-associated genetic variants
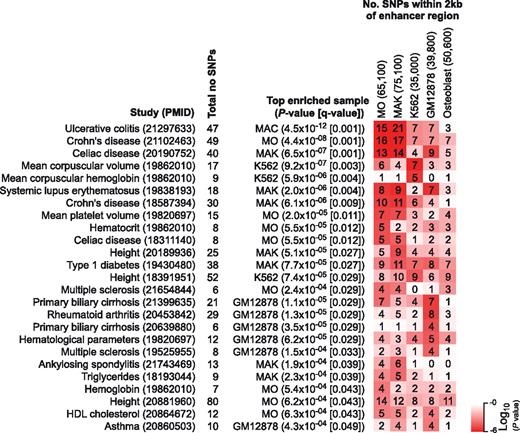
Monocytes and macrophages represent important components of our innate immune system and are important for tissue homeostasis and wound healing. They also play important roles in various diseases including infections and cancer. Whereas GWAS continue to provide us with genetic variants that are associated with diseases, the majority of these variants are found within the noncoding genome, where the functional consequences of the variation are mostly unknown. One functional consequence of an intergenic or intragenic noncoding variant could be the inactivation or aberrant activation of an enhancer. We therefore explored the association of candidate enhancer regions with disease-associated variants extracted from a comprehensive GWAS catalog.17 In the present study, we focused on promoter-distal regions marked by H3K4me1 and/or H3K27ac. H2AZ was not included because it is a marker for open chromatin (which also marks boundary elements), is less associated with cell type–specific gene regulation, and is frequently associated with developmental genes (supplemental Figure 8). Given a set of disease-risk SNPs identified by an individual GWAS, we hypothesized that the more SNPs are found in the vicinity of a set of enhancers, the greater the likelihood that the set of enhancers contributes to the phenotype of the disease. To control for cell type–specificity, we extracted corresponding putative enhancer marks from publicly available ChIP-seq datasets for an erythroleukemia line (K562 cell line), a lymphoblastoid cell line (GM12878), and primary osteoblasts. As shown in Figure 7, we identified several studies with disease-risk SNPs that were strongly enriched in candidate enhancer regions. SNPs associated with ulcerative colitis, Crohn disease, systemic lupus erythematosus, and celiac disease showed a strong correlation with the enhancer signature of human monocytes and macrophages. Consistent with their erythroid origin, K562 candidate enhancers were enriched for SNPs associated with mean corpuscular volume and hemoglobin, whereas candidate enhancers of the lymphoblastoid cell line GM12878 were correlated with SNPs from several autoimmune disorders (eg, primary biliary cirrhosis and rheumatoid arthritis). Within the monocyte/macrophage-enriched SNP set, we also identified cases in which disease-associated SNPs changed a motif instance for a relevant transcription factor (supplemental Figure 9), suggesting that the modulation of cell type–specific enhancer function by sequence variants may contribute significantly to disease.
Enhancer signatures and disease-associated genetic variants. Overlap of H3K4me1 and/or H3K27ac-marked promoter-distal regions (± 3 kb from RefSeq-annotated TSS) with disease-associated sequence variants (SNPs) from the GWAS catalog.17 Only studies that identified at least 5 SNPs were included in the analysis and shown are all studies with a false discovery rate < 5%. P values for enrichment are presented as a heat map where red indicates significance values. Numbers of total enhancer regions for each cell type are given in brackets above the heat map.
Enhancer signatures and disease-associated genetic variants. Overlap of H3K4me1 and/or H3K27ac-marked promoter-distal regions (± 3 kb from RefSeq-annotated TSS) with disease-associated sequence variants (SNPs) from the GWAS catalog.17 Only studies that identified at least 5 SNPs were included in the analysis and shown are all studies with a false discovery rate < 5%. P values for enrichment are presented as a heat map where red indicates significance values. Numbers of total enhancer regions for each cell type are given in brackets above the heat map.
Discussion
In the present study, we analyzed dynamic enhancer signatures during the transition of primary human blood monocytes into monocyte-derived macrophages, a naturally occurring postproliferative differentiation process that is accompanied by marked phenotypical changes.18,19 Our study includes the generation and interpretation of genome-wide distribution maps of 3 histone marks, H3K4me1, H3K27ac, and H2AZ, which have been implicated previously in enhancer biology,2-4,7,11 as well as maps of several transcription factors by ChIP-seq. In addition, we integrated published HSC ChIP-seq data15 and transcriptome data for major blood cell types21 to describe comprehensively the epigenetic enhancer signatures associated with monocyte differentiation.
Cell type–specific epigenetic fingerprints associate with sequence motifs for key regulators
Differentiation-associated epigenetic enhancer signatures (H3K4me1, H3K27ac, and, to a lesser degree, H2AZ) contained specific sequence motifs at all 3 cell-differentiation stages (ie, HSCs, monocytes, and macrophages) that correspond to consensus motifs for known key transcriptional regulators for each cell stage. The HSC-specific H3K4me1-derived enhancer fingerprint included, for example, ETS, RUNX, and GATA motifs that correspond to consensus sequences for known stem-cell regulators (eg, ERG, FLI1, RUNX1, and GATA2) found previously to co-occupy regulatory regions in blood stem cells,29 as well as an AP1-like motif and a motif for HOX-family transcription factors that are also known to regulate stem cell functions.30 Compared with HSCs, the blood monocyte–associated epigenetic enhancer signature shifted toward sequence motifs for the myeloid lineage-determining ETS-family factor PU.1, C/EBP family members, and a composite element (EIRE), which is known to bind a heterodimer of PU.1 and IRF8 (also called ICSBP) in human monocytes/macrophages.24 These factors are all known to be crucial for monocyte/macrophage biology. PU.1 and C/EBPα/β are master regulators of the myeloid differentiation program and are able to reprogram nonmyeloid cells toward the macrophage lineage,31-33 whereas IRF8 mutations were shown recently to impair monocyte and dendritic-cell development in humans.34
Compared with HSC-to-monocyte differentiation, which is a process involving a stepwise lineage restriction through several cell cycles, human monocyte-to-macrophage differentiation proceeds without proliferation and the cell types are closely related. Nevertheless, monocyte-to-macrophage differentiation has a significant impact on the epigenetic enhancer signature, with primarily macrophages gaining additional H3K27ac and H3K4me1 sites. In monocytes, cell stage–specific H3K27ac signatures associated with ETS, C/EBP, an EIRE-like motif, and a KLF-motif that likely corresponds to KLF4, a member of the Kruppel-like factor family, which has been implicated previously in monocyte differentiation22,23 and is strongly down-regulated during macrophage differentiation.
Similar to the motif composition in murine macrophages,8,35 the human macrophage-specific epigenetic enhancer signature contained PU.1, AP-1-like bZIP, and a composite C/EBP:bZIP motif.20 In addition, the macrophage-specific H3K4me1 signature contained a motif for NFκB, which likely represents a fraction of poised enhancers that become activated upon macrophage stimulation.35
Interestingly, human macrophage enhancer regions were also enriched for 2 “novel” motifs, including a GT-box and an E-box element, which were not identified previously in mouse macrophages.8,35 Both novel motifs were also enriched around macrophage-specific PU.1- or C/EBPβ-binding sites, suggesting that the motif-corresponding factors may contribute significantly to the establishment and/or maintenance of human macrophage–specific epigenetic patterns equivalent to the findings in murine macrophages for PU.1.8
Comparison with known consensus sites and gene-expression data revealed reasonable candidates for both novel motifs. The E-box resembled a motif that has been described as a binding site for MITF/TFE transcription factors.36 Two family members (TFEC and MITF) have been implicated previously in the biology of monocyte-derived cells,37-40 and the long isoform of MITF is induced during human macrophage differentiation (on both the RNA and protein levels). However, we were unable to analyze the MITF-binding pattern in macrophages and it is equally possible that other factors bind the macrophage-enriched E-box motif.
Based on gene expression and motif similarity, EGR2 presented a good candidate for the GT box and its ChIP-seq–derived binding motif, and global binding distribution confirmed that the macrophage-specific GT-box element is in fact bound by EGR2. Approximately 20% of the “active” human macrophage-specific enhancer signature showed EGR2 binding, suggesting that EGR2 contributes significantly to the human macrophage-specific enhancer repertoire. The transient up-regulation of EGR1 during macrophage differentiation suggests that the GT box may be bound by both family members at early differentiation stages. Consistent with our data, EGR factors were implicated recently in the transcriptional network shaping a macrophage-like state in the human myeloid leukemia cell line THP-1.41 The role of Egr transcription factors in murine myeloid cell differentiation has been controversial. Whereas some studies proposed an essential, nonredundant function of Egr1/2 in macrophage differentiation,42,43 another study using primary knockout cells concluded that Egr transcription factors are neither specific to nor essential for murine monocyte/macrophage differentiation.44 However, as opposed to primary murine macrophages, EGR2 expression is clearly induced and maintained during monocyte-to-macrophage differentiation, raising the possibility that it may have a species-specific role in human macrophages. A growing body of evidence suggests that both DNA binding and function of several “common” transcription factors at least partly depend on the cell type–specific pool of transcription factors.8,11,35,45-47 Therefore, the fact that EGR family factors are expressed ubiquitously and induced by adherence or growth factor stimulation in many cell types may not necessarily argue against their function in monocyte/macrophage differentiation, as has been suggested.44
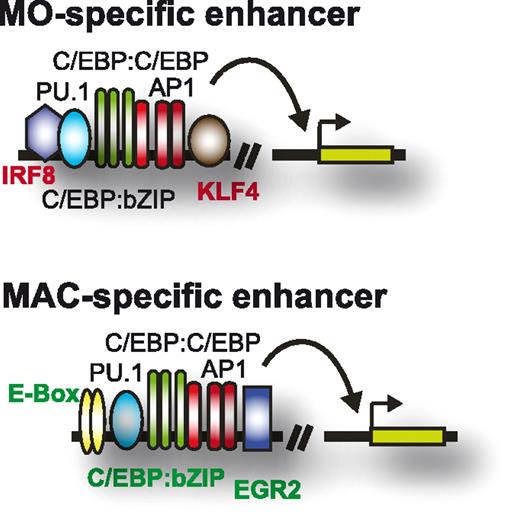
The factors comprising the cell stage–specific enhancer repertoire of primary human monocytes or macrophages are summarized schematically in Figure 8.
Enhancer signatures in human monocytes and macrophages. Schematic depicting the transcription factor repertoire-shaping enhancers in human monocytes and macrophages.
Enhancer signatures in human monocytes and macrophages. Schematic depicting the transcription factor repertoire-shaping enhancers in human monocytes and macrophages.
Enhancer signatures and their relevance for understanding disease-associated variants
Having defined candidate enhancer regions (marked by H3K4me1 and/or H3K27ac) in 2 immune cell types that have been implicated in many diseases, we also studied a possible relation between monocyte/macrophage enhancer elements and disease-associated variants that were extracted from a comprehensive GWAS catalog.17 The strong correlation between the monocyte/macrophage enhancer signature with SNPs associated with ulcerative colitis, celiac disease, Crohn disease, or systemic lupus erythematosus is entirely consistent with the proposed role of human monocytes and macrophages in these diseases14,48 and also confirms a previous, similar approach analyzing several cell lines.49 Linking GWAS data with epigenetic enhancer signatures of different cell types could thus present an important step toward a functional annotation of noncoding disease variants. The overlap of SNPs with putative transcription factor–binding sites is still limited, but this could be because of the limited coverage of current SNP array platforms. Future GWAS studies may be able to use available enhancer signatures of relevant cell types to target their SNP profiling (eg, using targeted enrichment of enhancer regions and subsequent high-throughput sequencing). As opposed to current approaches covering the most common (but not necessarily relevant) variants, such targeted sequencing approaches could be better suited to explain the mechanistic basis of disease-associated variants, because they would detect all variants at “relevant” sites.
Conclusions
In the present study, we describe differentiation stage–specific enhancer signatures and identify one novel transcription factor, EGR2, that likely contributes to the human macrophage-specific enhancer repertoire. Whereas it is clear that the conclusions derived from the computational analysis of global mapping data are based on associations, our findings are consistent with previous work in human cell lines and mouse models and provide novel insights into how combinations of common and differentiation-associated transcription factors specify the enhancer repertoire during cellular differentiation.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dagmar Glatz, Ireen Ritter, and Sabine Pape for excellent technical assistance and Sven Heinz for fruitful discussions and critically reading the manuscript.
This work was funded by a grant from the Deutsche Forschungsgemeinschaft (Re1310/7) to M.R.
Authorship
Contribution: T.-H.P., M.L., L.S., and Y.H. performed the experiments; C.B. and R.A. provided important tools; T-H.P., C.B., and W.C. analyzed the results; and M.R. designed the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for M.L. is Institute of Biomedical Research, College of Medical and Dental Sciences, University of Birmingham, Birmingham, United Kingdom.
Correspondence: Michael Rehli, Department of Hematology, University Hospital Regensburg, F-J-Strauss-Allee 11, D-93042 Regensburg, Germany; e-mail: michael.rehli@ukr.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal