Abstract
Th17 cells play an essential role in the pathogenesis of autoimmune and inflammatory diseases. Most of our current understanding on Th17 cell differentiation relies on studies carried out in mice, whereas the molecular mechanisms controlling human Th17 cell differentiation are less well defined. In this study, we identified gene expression changes characterizing early stages of human Th17 cell differentiation through genome-wide gene expression profiling. CD4+ cells isolated from umbilical cord blood were used to determine detailed kinetics of gene expression after initiation of Th17 differentiation with IL1β, IL6, and TGFβ. The differential expression of selected candidate genes was further validated at protein level and analyzed for specificity in initiation of Th17 compared with initiation of other Th subsets, namely Th1, Th2, and iTreg. This first genome-wide profiling of transcriptomics during the induction of human Th17 differentiation provides a starting point for defining gene regulatory networks and identifying new candidates regulating Th17 differentiation in humans.
Introduction
Naive CD4+ T cells differentiate into functionally distinct lineages in response to environmental cues and interaction with APCs. The nature of invading pathogens determines the cytokine environment in which the cognate Ag recognition by TCR takes place, subsequently influencing the phenotype of differentiating CD4+ Th cells. Classically, presentation of intra- or extracellular pathogens to naive T cells leads to either a Th1 response or a Th2 response, respectively.1 Today, new functionally distinct subtypes of CD4+ have been identified.2 Since the original identification of IL17-secreting T cells,3 further research has led to the definition of an effector Th17 cell lineage.4-6 Shortly after these findings, Th17 cells were characterized also in humans by using peripheral blood T cells and T-cell clones derived from gut tissue of patients having Crohn disease.7-9 Human Th17 cells express CCR6, CCR4, IL23R, and CD161 on their cell membrane.9,10 The characteristic cytokine secreted by these cells is IL17A (also referred to as IL17). IL17A stimulates the secretion of wide range of proinflammatory chemokines and cytokines. As its receptor is widely expressed, many cells of the immune system as well as other cell types can respond to it.11 In addition to IL17A, cytokines IL17F, IFNγ, IL22, and IL26 have been shown to be secreted by human Th17 cells.7 Proper function of Th17 cells is needed for eradication of extracellular bacterial and fungal infections.11
CD4+ cells isolated from peripheral or cord blood have been used to examine Th17 polarization in human. In several studies, differentiation of naive cells from peripheral blood has not succeeded, or the IL17A production has been markedly less efficient than detected by memory cells. IL17A secretion of polarized cord blood cells is also modest.12 Human Th17 cells have also been shown to originate from CD161+ precursor cells,10 which represent a small percentage of CD4+ cells present in cord blood. Recently, it has been shown that the expression of CD161 is developmentally regulated, and Th17 cells can differentiate from CD161− cells of preterm infants.13 Based on the early findings it seemed that the cytokine requirements of human and mouse Th17 cells are fundamentally different. For example, while initially the species specific role of TGFβ in the process was actively discussed, it is becoming widely accepted that the human and mouse Th17 cell development is dependent on similar factors. Nevertheless, there is still an ongoing debate about the combinations of cytokines and cell activation needed for driving the process. TGFβ, IL23, IL1β, and IL6 are most often reported to be effective in in vitro differentiation of CD4+ cells toward Th17 phenotype at various concentrations and in combination with other cytokines.12 In addition, prostaglandin E2 and aryl hydrocarbon receptor ligands have been shown to modulate the process.14,15 To summarize, differentiation and function of Th17 cells is controlled by a complex interplay of various factors playing a role at the distinct stages of the process and regulating different characteristics of cells secreting IL17A. To improve the understanding of Th17 differentiation as a whole, it would be important to move from a factor- or a gene-centric view to a holistic one revealing pathways and gene regulatory pathways controlling the process.
The aim of the present study was to identify new candidate genes to be tested for their role in regulating Th17 cell polarization. Th17 cells have been extensively studied by using mouse differentiation and disease models. As there is a clear link between many autoimmune diseases and IL17A similarly in humans, it is crucial to identify the factors controlling differentiation and function of Th17 subset. Interestingly, the dysregulation of Th17 responses is associated with certain forms of asthma, which is classically thought to be mediated by Th2 cells. Th17 cells have also been connected to regulation of immune reactions against cancer cells.16 In this study, by using CD4+ T cells isolated from human umbilical cord blood, we report, for the first time, genome-wide profiling of gene expression throughout human Th17 cell priming. We demonstrate that gene expression at the very early stage of human Th17 differentiation is highly dynamic. We further validated the differential expression of the selected genes at the protein level and analyzed their expression in Th1, Th2, and inducible regulatory T cell (iTreg) Th subsets. The results indicate that KDSR, ATP1B1, CXCR5, and IL2RB are selectively regulated in Th17 condition during the early priming toward Th17 phenotype. On the other hand, CD52, VDR, and CTSL1 were highly regulated also in response to initiation of some of the other Th programs. In summary, our study not only provides an overview of genes and pathways regulated in response to induction of Th17 differentiation in humans, but also provides several candidates for modulation of Th17 responses, which will be of interest for further studies.
Methods
Human CD4+ T-cell isolation and culture
CD4+ T cells were purified from umbilical cord blood of healthy neonates (Turku University Central Hospital, Turku, Finland). Mononuclear cells were isolated (Ficoll-Paque PLUS; GE Healthcare) after which CD4+ cells were collected (Dynal CD4 Positive Isolation Kit; Invitrogen). Cells were activated with plate-bound anti-CD3 (750 ng/24-well culture plate well; Immunotech) and soluble anti-CD28 (1 μg/mL; Immunotech) in a density of 0.5 × 106 cells/mL of X-vivo 20 serum-free medium (Lonza) supplemented with 2mM L-glutamine (Sigma-Aldrich), and 50 U/mL penicillin and 50 μg/mL streptomycin (Sigma-Aldrich) and cultured at 37°C in 5% CO2. Th17 polarization was initiated with IL6 (20 ng/mL; Roche), IL1β (10 ng/mL) and TGFβ (10 ng/mL) in the presence of neutralizing anti-IFNγ (1 μg/mL) and anti-IL4 (1 μg/mL). Control Th0 cell were cultured in a medium containing only the neutralizing Abs. Th1, Th2, and iTreg differentiation was initiated with IL12 (2.5 ng/mL), IL4 (10 ng/mL), and TGFβ (10 ng/mL), respectively. All cytokines and neutralizing Abs were from R&D Systems unless otherwise stated. Comparisons between the different polarization conditions were always done with the same pool of aliquoted cells. The usage of blood of unknown donors was approved by the Finnish Ethics Committee.
Transcriptional profiling
Samples were collected at 0-, 0.5-, 1-, 2-, 4-, 6-, 12-, 24-, 48-, and 72-hour time points of culture. Total RNA from 3 cultures was DNase treated (RNase-Free Dnase Set; QIAGEN) during the isolation (RNeasy Kit; QIAGEN), processed, and hybridized on Illumina Sentrix HumanHT-12 Expression BeadChip Version 3.
The microarray data were analyzed using a Bioconductor package beadarray. The time-series microarray data were filtered by choosing to analyze only the probes with detection P values < .05 at least in 1 time point at 1 cell type. In addition, only the probes having an SD > 0.15 over all the samples were used in the analysis. The data were normalized using quantile normalization. For each time point, differentially expressed genes were identified using the Bioconductor limma package with moderated t-statistic with false discovery rate (FDR) < 0.1. Annotations of genes were gathered using Ingenuity Pathways Analysis (Ingenuity Systems, www.ingenuity.com). Clusters were identified using k-means clustering.
Quantitative RT-PCR
RNA was isolated (RNeasy Mini Kit; QIAGEN) and treated in-column with DNase (RNase-Free Dnase Set; QIAGEN) for 15 minutes. The removal of genomic DNA was ascertained by treating the samples with DNase I (Invitrogen) before cDNA synthesis either with SuperScript II Reverse Transcriptase (Invitrogen) or with Transcriptor First Strand cDNA Synthesis Kit (Roche). Quantitative RT-PCR (qPCR) was performed using Universal ProbeLibrary probes (Roche Applied Science) or with FAM (reporter), TAMRA (quencher) double-labeled probes in a 10-μL reaction volume. Reaction mix used was Absolute QPCR ROX Mix (Thermo Scientific) and amplification was monitored with Applied Biosystems 7900HT Fast Real-Time PCR System (15 minutes enzyme activation and 40 cycles of 15 seconds 95°C, 1 minute 60°C). The cycle threshold (Ct) values of the transcripts studied were normalized against the signal acquired with EF1α.17 The primers and probes are listed in supplemental Table 1 (see the Supplemental Materials link at the top of the article).
Western blotting
Samples were lysed in Triton-X sample buffer (50mM Tris-HCl, pH 7.5; 150mM NaCl; 0.5% Triton-X-100; 5% glycerol; 1% SDS), containing proteinase (Roche) and phosphate inhibitors (Roche) and sonicated (Bioruptor UCD-200; Diagenode). Sonicated samples were centrifuged at maximum speed for 20 minutes at 4°C and supernatants were collected. Samples were quantified (DC Protein Assay; Bio-Rad) and boiled with 6× loading dye (330mM Tris-HCl, pH 6.8; 330mM SDS; 6% β-ME; 170μM bromophenol blue; 30% glycerol). Samples were loaded on 10% or 12% SDS-PAGE gels, transferred to nitrocellulose membranes, and probed with Abs listed in supplemental Table 2.
Flow cytometry
CCR6 detection was done at day 3. Cells were washed with PBS and staining was done in 0.5% FBS/0.1% atzide/PBS for 15 minutes at 4°C. Cells were fixed with 1% paraformaldehyde, and an LSRII flow cytometer (BD Biosciences) was used in data acquisition. Living cells were gated for analysis based on forward and side scattering. CD52, IL2RB, and CXCR5 detections were done as CCR6 detection except the Ab incubation lasted 30 minutes. Cells were fixed with 4% paraformaldehyde before analysis of ITM2A, and in addition permeabilized with methanol before analysis of LMNA. Thirty-minute incubation with primary Ab was followed with 15-minute incubations with secondary Ab if needed. The stainings were controlled with isotype Ab or by staining the cells only with the secondary Ab. The Abs used are listed in supplemental Table 2.
IL17A secretion
Differentiation of CD4+ T cells toward the Th17 phenotype was characterized by detection of IL17A production in cell-culture supernatant using the Millipore Human Cytokine/Chemokine, 96-Well Plate Assay on day 3. IL17A secretion was normalized with the number of living cells detected based on forward and side scattering in flow cytometric analysis (LSRII flow cytometer; BD Biosciences).
Ethical aspects
The usage of blood of unknown donors was approved by the Ethics Committee of the Hospital District of Southwest Finland.
Accession numbers
Microarray data can be found at the NCBI Gene Expression Omnibus with accession number GSE35103.
Results
Genome-wide transcriptional regulation at the early stage of human Th17 differentiation
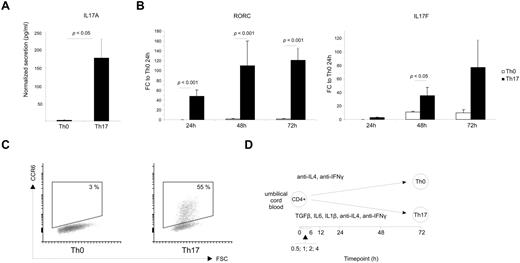
Because the essential role of Th17 cells in the pathogenesis of inflammation, autoimmunity, and cancer has been well documented by several studies in recent years,16 it has been of great interest to define the molecular mechanisms involved in the fate decision of naive CD4+ T cells to acquire the highly inflammed Th17 phenotype. To address this question, we performed a genome-wide gene expression profiling of in vitro–cultured human CD4+ cells during the initiation of Th17 polarization. First, we evaluated the polarization conditions by analyzing IL17A secretion (Figure 1A), IL17F, RORC, and CCR6 expression (Figure 1B-C). The up-regulation of these marker genes showed that the cell population was successfully biased toward Th17 phenotype. We were interested in the initiation of Th17 differentiation and hence focused our study on the early changes in transcriptomics during human Th17 polarization. CD4+ T cells isolated from umbilical cord blood were activated with anti-CD3 and anti-CD28 in the presence of a cytokine cocktail containing IL1β, IL6, and TGFβ as well as neutralizing Abs against IL4 and IFNγ. The samples for microarray analysis were collected at 9 time points during the culture starting from 0.5 hours to 72 hours (Figure 1D).
In vitro differentiation of cord blood CD4+ cells for Th17 gene expression profiling. (A) IL17A secretion after 72 hours of culture. The detection was done directly from the culture supernatant with fluorescent beads. The values are normalized with the number of living cells determined based on cell size and granularity detected with flow cytometer. The data shown are the average of 6 cultures with the SEM and statistical significance was determined with the Student t test. (B) The expression of IL17F and RORC in the cells cultured toward Th17 phenotype or left as control (Th0). The data are presented as fold change over the expression level in Th0 control cells at 24 hours. The data show the average expressions and the SEM. Statistical significance of the results has been determined with the Student t test. (C) Representative CCR6 expression after 72 hours of culturing. (D) Schematic presentation of the microarray profiling showing the culturing conditions used and the sampling time points.
In vitro differentiation of cord blood CD4+ cells for Th17 gene expression profiling. (A) IL17A secretion after 72 hours of culture. The detection was done directly from the culture supernatant with fluorescent beads. The values are normalized with the number of living cells determined based on cell size and granularity detected with flow cytometer. The data shown are the average of 6 cultures with the SEM and statistical significance was determined with the Student t test. (B) The expression of IL17F and RORC in the cells cultured toward Th17 phenotype or left as control (Th0). The data are presented as fold change over the expression level in Th0 control cells at 24 hours. The data show the average expressions and the SEM. Statistical significance of the results has been determined with the Student t test. (C) Representative CCR6 expression after 72 hours of culturing. (D) Schematic presentation of the microarray profiling showing the culturing conditions used and the sampling time points.
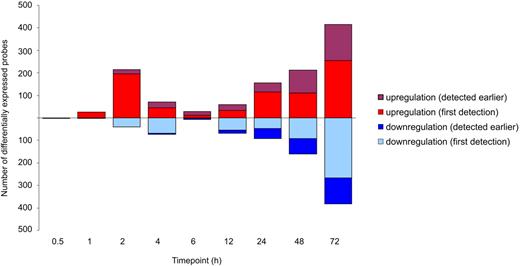
The overall gene expression changes revealed 2 waves of transcriptional changes in response to the Th17 stimulation used compared with TCR activation alone; the first one took place within the first 4 hours of culture, and the second one started after 6 hours (Figure 2). The selected time points captured the earliest changes in the overall transcriptome, because there were only very few genes differentially expressed at the earliest 0.5-hour time point after which the number of changes increased (supplemental Table 3). After 1 hour of stimulation, 28 probes representing 25 genes were already differentially expressed in the cells cultured under the Th17 polarizing condition compared with the cells cultured in the Th0 condition (Figure 1D). The first peak of changes of gene expression was observed after 2 hours of culture, 256 probes representing 228 genes were differentially expressed during the maximal primary transcriptional response after the initiation of polarization. Among the differentially expressed genes, most genes were up-regulated, suggesting many signaling pathways to be turned on in response to the stimulation by the Th17 polarization inducing cytokine cocktail. From 6 hours to 72 hours, the number of differentially expressed genes increased, resulting in up to 799 differentially expressed probes at the 72-hour time point (Figure 2). Within them, 416 probes were up-regulated and 383 were down-regulated, suggesting that Th17 polarization is regulated both by actively up-regulating the signaling pathways driving the differentiation and by shutting down the interfering gene expression.
The genes regulated during the early stages of human Th17 differentiation. Three biologic replicates of time-series data of Th17 polarized or control cord blood CD4+ cells were hybridized to Illumina Sentrix Human HT-12 Expression Version 3 BeadChips. The differential expression analysis was performed for the probes having a detection P < .05 at least in 1 time point at 1 cell type and SD > 0.15 over all the samples. The differentially expressed genes between Th17 and Th0 conditions were identified with the false discovery rate (< 0.1). The differentially expressed probes are classified according to the appearance of the difference.
The genes regulated during the early stages of human Th17 differentiation. Three biologic replicates of time-series data of Th17 polarized or control cord blood CD4+ cells were hybridized to Illumina Sentrix Human HT-12 Expression Version 3 BeadChips. The differential expression analysis was performed for the probes having a detection P < .05 at least in 1 time point at 1 cell type and SD > 0.15 over all the samples. The differentially expressed genes between Th17 and Th0 conditions were identified with the false discovery rate (< 0.1). The differentially expressed probes are classified according to the appearance of the difference.
Dynamic changes of gene expression during the early stage of human Th17 cell differentiation
Interestingly and importantly, a considerable number of the differentially expressed genes were enzymes, kinases, and transcription regulators. Statistical enrichment of cytokines (Table 1) and localization of the differentially expressed genes preferentially on the cell surface or on extracellular space (Table 2) indicate that, even at the early stages during the polarization process, cells start to focus on activating the pathways needed for signaling with the neighboring cells. Nevertheless, acquisition of fully competent phenotype continues actively beyond the first 3 days of polarization as evidenced by the appearance of new differentially expressed genes at all time points studied (Figure 2).
Functional classification of the differentially expressed genes
| . | 0.5 h . | 1 h . | 2 h . | 4 h . | 6 h . | 12 h . | 24 h . | 48 h . | 72 h . |
|---|---|---|---|---|---|---|---|---|---|
| Cytokine | 0 | 0 | 1 | 1 | 0 | 3 | 9* | 12* | 18* |
| Enzyme | 1 | 4 | 43 | 20 | 2 | 10 | 34 | 52 | 120 |
| G-protein coupled receptor | 0 | 0 | 3 | 4 | 1 | 4 | 7 | 12* | 11 |
| Growth factor | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Ion channel | 0 | 0 | 1 | 0 | 0 | 1 | 2 | 4 | 5 |
| Kinase | 0 | 0 | 17 | 9 | 2 | 12 | 16 | 14 | 34 |
| Ligand-dependent nuclear receptor | 0 | 0 | 1 | 0 | 1 | 2 | 2 | 3 | 4 |
| MicroRNA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Other | 1 | 13 | 107 | 62 | 13 | 57 | 110 | 159 | 370 |
| Peptidase | 0 | 0 | 6 | 2 | 3 | 6 | 10 | 13 | 19 |
| Phosphatase | 0 | 0 | 4 | 0 | 0 | 0 | 5 | 5 | 15 |
| Transcription regulator | 1 | 5 | 27 | 15 | 6 | 7 | 11 | 24 | 52 |
| Translation regulator | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 4 |
| Transmembrane receptor | 0 | 0 | 3 | 6 | 3 | 3 | 7 | 7 | 10 |
| Transporter | 0 | 3 | 12 | 7 | 0 | 7 | 9 | 18 | 29 |
| . | 0.5 h . | 1 h . | 2 h . | 4 h . | 6 h . | 12 h . | 24 h . | 48 h . | 72 h . |
|---|---|---|---|---|---|---|---|---|---|
| Cytokine | 0 | 0 | 1 | 1 | 0 | 3 | 9* | 12* | 18* |
| Enzyme | 1 | 4 | 43 | 20 | 2 | 10 | 34 | 52 | 120 |
| G-protein coupled receptor | 0 | 0 | 3 | 4 | 1 | 4 | 7 | 12* | 11 |
| Growth factor | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Ion channel | 0 | 0 | 1 | 0 | 0 | 1 | 2 | 4 | 5 |
| Kinase | 0 | 0 | 17 | 9 | 2 | 12 | 16 | 14 | 34 |
| Ligand-dependent nuclear receptor | 0 | 0 | 1 | 0 | 1 | 2 | 2 | 3 | 4 |
| MicroRNA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Other | 1 | 13 | 107 | 62 | 13 | 57 | 110 | 159 | 370 |
| Peptidase | 0 | 0 | 6 | 2 | 3 | 6 | 10 | 13 | 19 |
| Phosphatase | 0 | 0 | 4 | 0 | 0 | 0 | 5 | 5 | 15 |
| Transcription regulator | 1 | 5 | 27 | 15 | 6 | 7 | 11 | 24 | 52 |
| Translation regulator | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 4 |
| Transmembrane receptor | 0 | 0 | 3 | 6 | 3 | 3 | 7 | 7 | 10 |
| Transporter | 0 | 3 | 12 | 7 | 0 | 7 | 9 | 18 | 29 |
The number of unannotated probes: 25.
Enrichment of the functional class (false discovery rate < 0.05).
Location of the differentially expressed genes
| . | 0.5 h . | 1 h . | 2 h . | 4 h . | 6 h . | 12 h . | 24 h . | 48 h . | 72 h . |
|---|---|---|---|---|---|---|---|---|---|
| Extracellular space | 0 | 1 | 5 | 3 | 2 | 11* | 20* | 30* | 51* |
| Plasma membrane | 0 | 3 | 31 | 24* | 6 | 31* | 49* | 72* | 95* |
| Cytoplasm | 1 | 8 | 74 | 38 | 11 | 34 | 70 | 100 | 242 |
| Nucleus | 1 | 10 | 56 | 29 | 7 | 16 | 41 | 51 | 148 |
| Unknown | 1 | 3 | 59 | 34 | 5 | 20 | 42 | 72 | 156 |
| . | 0.5 h . | 1 h . | 2 h . | 4 h . | 6 h . | 12 h . | 24 h . | 48 h . | 72 h . |
|---|---|---|---|---|---|---|---|---|---|
| Extracellular space | 0 | 1 | 5 | 3 | 2 | 11* | 20* | 30* | 51* |
| Plasma membrane | 0 | 3 | 31 | 24* | 6 | 31* | 49* | 72* | 95* |
| Cytoplasm | 1 | 8 | 74 | 38 | 11 | 34 | 70 | 100 | 242 |
| Nucleus | 1 | 10 | 56 | 29 | 7 | 16 | 41 | 51 | 148 |
| Unknown | 1 | 3 | 59 | 34 | 5 | 20 | 42 | 72 | 156 |
The number of unannotated probes: 25.
Enrichment of the cellular compartment (false discovery rate < 0.05).
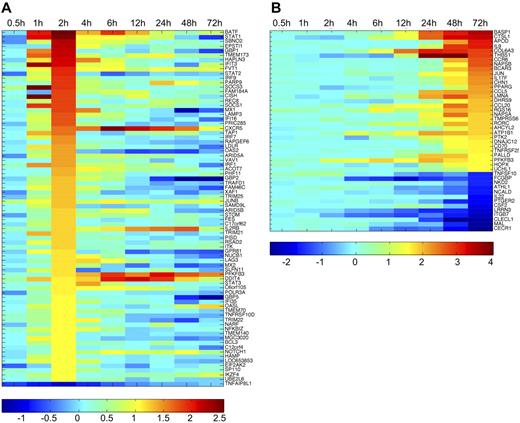
CD4+ cells vigorously regulate the gene expression pattern during the early polarization toward Th17 differentiation. This is clearly demonstrated by following the gene expression throughout the selected timeframe; for example, most of the genes differentially expressed at the 2-hour time point are significantly differentially regulated only at that time point. Similarly, there are many differentially expressed genes only regulated at the 72-hour time point (supplemental Table 3). The expression pattern of the genes with the highest signal log-ratio between Th17 and Th0 culturing condition at 2 and 72 hours are visualized in Figure 3A and B, respectively. Among the differentially expressed genes, there are several genes known to be preferentially expressed in Th17 cells, such as CCR6, IL17F, RORC, RORA, IL9, IL23R, BATF, and VDR9,18-21 which validates the cultures and the analysis done in this study. Of note, the differential expression of IL17A is not detectable with the probe on the microarray used while its expression was clearly up-regulated in response to Th17-inducing cytokines in our samples when measured with other methods. Most importantly, our microarray study also revealed novel genes in this context.
Dynamic regulation of the genes in cord blood CD4+ cells during Th17 differentiation. (A) Heatmap presentation of the expression kinetics of the differentially expressed genes between Th17 and Th0 control cells at 2 hours. The genes with signal log ratio > 1 or < −1 are presented. (B) Heatmap presentation of the expression kinetics of the differentially expressed genes at 72 hours. Signal log ratio > 1.5 or < −1.5 was used as cutoff. Gene up-regulation in Th17 culturing condition is shown with red, and down-regulation with blue as indicated with the scales below the heatmaps.
Dynamic regulation of the genes in cord blood CD4+ cells during Th17 differentiation. (A) Heatmap presentation of the expression kinetics of the differentially expressed genes between Th17 and Th0 control cells at 2 hours. The genes with signal log ratio > 1 or < −1 are presented. (B) Heatmap presentation of the expression kinetics of the differentially expressed genes at 72 hours. Signal log ratio > 1.5 or < −1.5 was used as cutoff. Gene up-regulation in Th17 culturing condition is shown with red, and down-regulation with blue as indicated with the scales below the heatmaps.
Next, we clustered the genes identified to be differentially expressed between our Th17 and Th0 culturing conditions, based on their gene expression difference to the freshly isolated unactivated CD4+ T cells (Figure 4, supplemental Table 4). The clustering reveals 3 main expression patterns: up-regulation, down-regulation, and somewhat steady expression throughout the analyzed timeframe after exposure to the Th17-polarizing cytokines. In addition to these predominant patterns, the level of expression categorizes the genes into different clusters. Compared with the differential expression between Th17 and Th0 control cells (Figure 3, supplemental Table 3), the regulation in Th17 cells compared with the freshly isolated CD4+ cells is more straightforward. On average, there is only some fluctuation in expression during the very first time points and after this, the gene expression follows the selected direction. This indicates that the differential expression in Th17 polarized cells compared with Th0 cells is not only because of either active up- or down-regulation of the genes, but also counterregulation to the stimulus caused by activation without polarizing cytokines.
Gene expression profiles of the identified Th17 differentiation–associated genes. The differentially regulated genes between Th17 polarized and Th0 control cord blood CD4+ cells were clustered based on their expression profile during Th17 polarization over the analyzed timeframe. For each gene, the probe found to be differentially expressed for the first time was used in the analysis. The number of genes belonging to each cluster is marked to the figure. Clustering was done with the k-means method. Gene expression patterns were drawn by using cluster center values.
Gene expression profiles of the identified Th17 differentiation–associated genes. The differentially regulated genes between Th17 polarized and Th0 control cord blood CD4+ cells were clustered based on their expression profile during Th17 polarization over the analyzed timeframe. For each gene, the probe found to be differentially expressed for the first time was used in the analysis. The number of genes belonging to each cluster is marked to the figure. Clustering was done with the k-means method. Gene expression patterns were drawn by using cluster center values.
Validation of selected differentially expressed genes
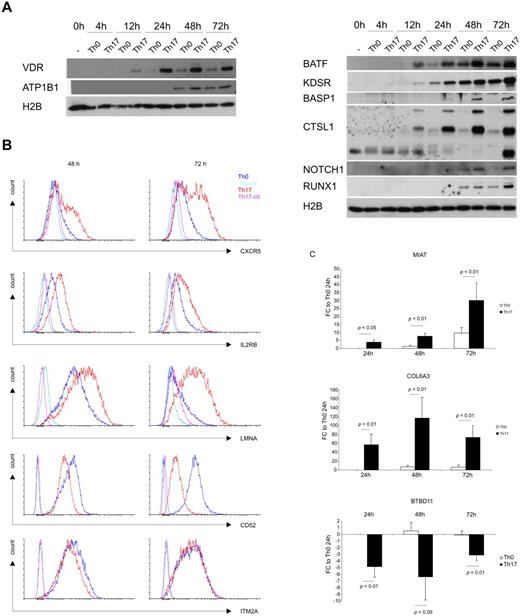
We selected some of the genes with the highest and long-lasting gene expression difference between Th17 and Th0 conditions to be studied further at protein level (supplemental Figure 1). Previously known function of the genes was also used as a criterion in selecting the targets for validation. Western blotting results showed that BASP1, CTSL1, RUNX1, BATF, KDSR, NOTCH1, VDR, and ATP1B1 were highly expressed in Th17 cells compared with their expression in Th0 cells at various time points during the first 3 days of polarization (Figure 5A). CXCR5, IL2RB, and LMNA were up-regulated in CD4+ T cells cultured under Th17-polarizing conditions compared with Th0 cells in our microarray study, and this difference is reflected to protein level (Figure 5B). On the other hand, the flow cytometric detection of CD52 and ITM2A at 48 and 72 hours showed down-regulation of these 2 proteins in CD4+ T cells cultured under a Th17-polarizing condition compared with cells cultured under a Th0 condition (Figure 5B), which is also consistent with the patterns seen in the microarray data. The microarray data from this study showed the up-regulation of COL6A3 and MIAT genes, and down-regulation of the BTBD11 gene in Th17-polarizing cells. These findings were confirmed with RT-PCR detections at 24-, 48-, and 72-hour time points (Figure 5C).
Validation of the selected genes. (A) Representative Western blot detection of RUNX1, CTSL1, BATF, BASP1, KDSR, NOTCH1, VDR, and ATP1B1 at 0, 4, 12, 24, 48, and 72 hour time points after culturing of cord blood CD4+ cells in Th17-polarizing medium or Th0 control condition. Histone H2B was analyzed to confirm equal loading. (B) Flow cytometric detection of CD52, IL2RB, CXCR5, LMNA, and ITM2A after culturing for 48 and 72 hours. Analysis of the stained cells was done with an LSRII flow cytometer (BD Biosciences). (C) RT-PCR detection of MIAT, BTBD11, and COL6A3 at 24-, 48-, and 72-hour time points. EF1α was used as an endogenous normalization control. Fold changes were calculated by comparing the normalized expression values to the corresponding expression in Th0 control samples at the 24-hour time point. SEM, statistical significance were determined with Student t test, and the average fold change is shown in the figure.
Validation of the selected genes. (A) Representative Western blot detection of RUNX1, CTSL1, BATF, BASP1, KDSR, NOTCH1, VDR, and ATP1B1 at 0, 4, 12, 24, 48, and 72 hour time points after culturing of cord blood CD4+ cells in Th17-polarizing medium or Th0 control condition. Histone H2B was analyzed to confirm equal loading. (B) Flow cytometric detection of CD52, IL2RB, CXCR5, LMNA, and ITM2A after culturing for 48 and 72 hours. Analysis of the stained cells was done with an LSRII flow cytometer (BD Biosciences). (C) RT-PCR detection of MIAT, BTBD11, and COL6A3 at 24-, 48-, and 72-hour time points. EF1α was used as an endogenous normalization control. Fold changes were calculated by comparing the normalized expression values to the corresponding expression in Th0 control samples at the 24-hour time point. SEM, statistical significance were determined with Student t test, and the average fold change is shown in the figure.
Selective expression of genes during Th-subset differentiation
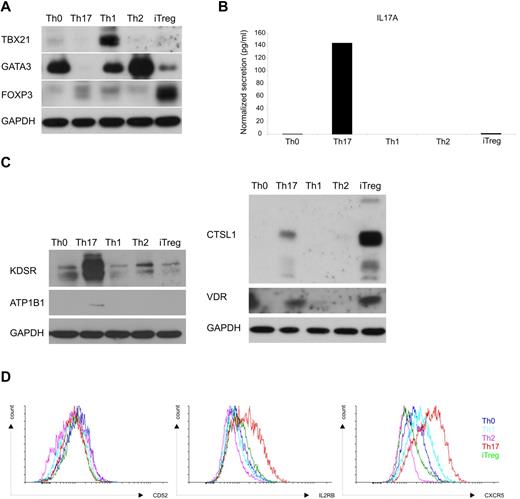
To further characterize the selectivity of the differentially expressed genes in the Th17-culturing condition, cord blood CD4+ T cells were cultured under Th0, Th1, Th2, iTreg, or Th17 conditions. Cells were harvested at 72 hours and the expression of selected proteins was analyzed with Western blotting or flow cytometry. As shown in Figure 6A, the expression of TBX21, GATA3, and FOXP3 the key transcription factors for Th1, Th2, or iTreg differentiation were detected preferentially expressed in the corresponding subset of helper T cells. IL17A was also selectively expressed in the cells polarized to Th17 direction (Figure 6B). Based on the kinetic profiling study, the expression of KDSR, CTSL1, ATP1B1, BASP1, NOTCH1, VDR, RUNX1, IL2RB, and CXCR5 was increased after Th17 polarization compared with the cells cultured under a Th0 condition. In contrast, the expression of CD52 was reduced in the cells cultured under a Th17 condition compared with those cultured in a Th0 condition. Consistent with the findings from the microarray study as well as the validation (Figure 5A-B), ATP1B1, KDSR, IL2RB, and CXCR5 were expressed at a higher level in the cells cultured under a Th17 condition compared with Th0 cells. In addition, the expression of these proteins was higher after Th17 polarization than after Th1, Th2, or iTreg induction (Figure 6C-D). However, although the expression of VDR was higher in cells under Th17-polarizing conditions than in Th0 cells, which was consistent with the results from the microarray study, it was also highly expressed in the cells cultured toward the iTreg (Figure 6C) phenotype. Again, the detection of CTSL1 confirmed the result from the gene profiling study, but the highest expression of CTSL1 was detected in the cells cultured under the iTreg condition (Figure 6C). The down-regulation of the expression of CD52 by a Th17-culturing condition was observed in the gene expression profiling study. However, the expression of CD52 was found to be lowest in Th2 cells (Figure 6D). Clear cell-type specificity could not be determined for BASP1, RUNX1, or NOTCH1 (data not shown). In summary, further analysis revealed that some of the identified genes regulated during Th17 polarization are selectively expressed during the differentiation of this particular lineage. However, some of the genes were not specific for Th17 cells as the differential expression was also observed in other Th subset–polarizing conditions.
Expression of the validated proteins throughout Th-cell subsets. CD4+ T cells isolated from umbilical cord blood were activated with plate-bound anti-CD3 and soluble anti-CD28. The only activated cells cultured with neutralizing anti-IL4 and anti-IFNγ were used as Th0 control. Cells were stimulated with IL12 (2.5 ng/mL) for initiation of Th1, IL4 (10 ng/mL) for Th2, TGFβ (10 ng/mL) for iTreg, and IL1β (10 ng/mL) + IL6 (20 ng/mL) + TGFβ (10 ng/mL) + anti-IL4 (1 μg/mL) + anti-IFNγ (1 μg/mL) for Th17 differentiation. Cells were harvested after 72 hours of culture. (A) Polarization toward different Th-cell subtypes was examined by analyzing the expression of key transcription factors of each subtype; TBX21 for Th1, GATA3 for Th2, and FOXP3 for iTreg cells with Western blotting. (B) Cytokine secretion was used to validate the selective expression of IL17A in the cells polarized toward the Th17 phenotype. The representative result from 2 experiments is shown. (C) The expression of CTSL1, ATP1B1, KDSR, and VDR was analyzed with Western blotting. (D) CD52, IL2RB, and CXCR5 were detected with flow cytometry. GAPDH detection was used as a loading control in Western blotting analysis.
Expression of the validated proteins throughout Th-cell subsets. CD4+ T cells isolated from umbilical cord blood were activated with plate-bound anti-CD3 and soluble anti-CD28. The only activated cells cultured with neutralizing anti-IL4 and anti-IFNγ were used as Th0 control. Cells were stimulated with IL12 (2.5 ng/mL) for initiation of Th1, IL4 (10 ng/mL) for Th2, TGFβ (10 ng/mL) for iTreg, and IL1β (10 ng/mL) + IL6 (20 ng/mL) + TGFβ (10 ng/mL) + anti-IL4 (1 μg/mL) + anti-IFNγ (1 μg/mL) for Th17 differentiation. Cells were harvested after 72 hours of culture. (A) Polarization toward different Th-cell subtypes was examined by analyzing the expression of key transcription factors of each subtype; TBX21 for Th1, GATA3 for Th2, and FOXP3 for iTreg cells with Western blotting. (B) Cytokine secretion was used to validate the selective expression of IL17A in the cells polarized toward the Th17 phenotype. The representative result from 2 experiments is shown. (C) The expression of CTSL1, ATP1B1, KDSR, and VDR was analyzed with Western blotting. (D) CD52, IL2RB, and CXCR5 were detected with flow cytometry. GAPDH detection was used as a loading control in Western blotting analysis.
Discussion
Th17 cells play essential roles in the pathogenesis of both autoimmune and allergic inflammatory diseases. There is also accumulative evidence linking Th17 cells to cancer biology. In the past few years, studies focusing on this subset of Th cells have thus drawn great attention in the field of immunology. However, compared with the comprehensive studies carried out in mouse models, the studies on molecular mechanisms and signaling pathways involved in regulating human Th17 differentiation from naive CD4+ T-cell progenitors have not been as thorough. The strength of our study is the usage of CD4+ T cells isolated from umbilical cord blood of newborn babies. As the majority of these cells are naive T cells, cord blood is the best material available for large profiling studies done with human primary cells without prior expansion or cloning. Of note, there is a possibility, albeit remote, that non-naive Th17 precursors present in small numbers in cord blood could influence the results. To capture and characterize the upstream molecular mechanisms of Th17 fate decision, we followed the detailed kinetic changes of gene expression starting from 0.5 hours to 72 hours after the cells were exposed to TGFβ, IL6, and IL1β. We identified the immediate target genes responding to Th17-polarizing stimulation peaking at 2 hours. More and more genes were regulated after 24 hours, indicating increasing deviation of the Th17 polarized lineage from the control cells and accumulation of primary and secondary target genes of the polarizing cytokines. As such, the presented dataset is the first comprehensive, genome-wide transcriptomics study of the first steps of human Th17 differentiation.
Comparing the selectively expressed genes in Th17 cells from our study with the published studies done with mouse cells indicates that a large amount of genes are similarly regulated in both species, for example, IL23R, RORA, BATF, RUNX1, and VDR.9,18-21 This indicates that the studies characterizing the function of these genes done with mouse models are likely to be relevant and applicable in medical research. However, findings should be validated in both systems and extrapolation directly from one organism to another should be carefully considered.22 This is clearly a challenge because of genetic variation and restricted availability of human cells compared with cells from inbred model organisms. An additional level of intricacy arises from complex interactions of cytokines promoting Th17 cell development. The known cytokines promoting Th17 cell differentiation provide all specific features to a differentiating cell. Single-cell analysis have also shown that cells expressing IL17A are heterogeneous for their cytokine expression profile and cytokines associated with Th17 cells can be secreted without IL17A. An additional level of complexity comes from the fact that Th-cell differentiation is known to be flexible leading to plasticity of the acquired phenotype.2 Although a lot of effort has been put on identification of an optimal cytokine cocktail for in vitro differentiation of human Th17 cells, the conditions needed for robust IL17A secretion remain elusive.12 Despite challenges, identification of genes regulated during human Th17 priming is an important step needed to understand, and to be able to modulate, this convoluted process in humans.
In this study, profiling of human Th17 differentiation revealed > 1000 probes differentially expressed in response to the Th17 culture condition compared with the Th0 control condition. In addition, we took a step forward in identifying new candidate genes putatively important for Th17 differentiation by validating the differential expression of the selected genes at the protein level. We show that the expression of transcription regulators BASP1, NOTCH1, RUNX1, and VDR is increased after TGFβ, IL6, and IL1β stimulation in cord blood CD4+ cells. The function of BASP1 in T cells is not known, but there are indications that it might be involved in modulation of the transcriptional program during T-cell apoptosis.23 In contrast to BASP1, the role of NOTCH1, RUNX1, and VDR in T cells has been intensively studied. Recently, it has been reported that NOTCH1, along with HIF-1α, controls human Th17 cell survival and apoptosis.24 It has also been shown that inhibition of Notch signaling directly, and indirectly via regulation of RORC, turns down the production of IL17A.25 RUNX1 interacts with FOXP3 and RORC, and is needed for Treg and Th17 cell function, respectively.26,27 Calcitriol-activated VDR constrains IL17A transcription by inhibiting NFAT, recruiting HDAC, and sequestrating RUNX1.28 We showed that BTBD11 is down-regulated during Th17 polarization. BTBD11 is a retinoic acid–inducible gene in the neuroblastoma cell line.29 As retinoic acid inhibits TGFβ- and IL6-driven Th17 cell differentiation and promotes induction of regulatory T cells,30 BTBD11 may be a novel transcriptional regulator determining the selection between Th17 and iTreg lineages.
In addition, we validated the up-regulation of membrane proteins ATP1B1, IL2RB, and CXCR5, and down-regulation of ITM2A and CD52 in Th17 cells compared with control cells. The ATP1B1 subunit regulates the assembly of the Na+/K+-ATPase protein pump, and eventually the number of functional protein complexes on the plasma membrane. Interestingly, apart from its classic function, ATP1B1 has been shown to interact with the E2A transcription factor regulating its nuclear localization.31 In epithelial cells, Na+/K+-ATPase subunits have been shown to play a role in regulation of tight junctions, cell polarity, actin dynamics, cell movement, and cell signaling.32 It remains to be shown whether similar atypical functions can also be found in T cells. IL2RB is instead one of the 3 chains forming a receptor for IL2. It is also a part of IL15 receptor complex. Interestingly, IL15 has been shown to increase IL17A production by the T-cell lines derived from synovial fluid of rheumatoid arthritis patients.33 CXCR5 is one of the markers of the newly identified lineage of follicular helper T cells providing help for B cells and adaptive immunity.34 CXCR5+ cells in human blood contain heterogeneously different Th-cell subsets capable of producing Th1, Th2, and Th17 marker cytokines. In the systemic autoimmune disease juvenile dermatomyositis, CXCR5+ Th cells were biased toward Th2 and Th17 phenotypes and this correlated with disease activity.35 It has been shown that activated CD4+ T cells can express CXCR5,36 but importantly our data show that during in vitro polarization toward the Th17 phenotype, cord blood Th cells are skewed to up-regulate CXCR5. ITM2A is a membrane protein suggested to regulate chondrocyte differentiation.37 In T cells, ITM2A expression is induced by activation and it down-regulates CD8 expression during positive selection.38 Alemtuzumab, also known as Campath-1, an Ab specifically recognizing CD52, has been extensively studied and used in cancer therapy. Recently, this humanized Ab has also been examined for treatment of multiple sclerosis patients.39 There are indications that CD52 is involved in costimulation of T cells and induces the differentiation of Treg cells.40 However, precise function of this glycoprotein, mainly expressed on mature T and B lymphocytes, remains obscure.
Enzymes CTSL1 and KDSR were found to be up-regulated in Th17 cells. CTSL1 is a ubiquitously expressed protease, which has been linked to the regulation of immune responses at the level of MHC complex maturation and Ag presentation influencing differentiation of CD4+ cells and autoimmune reactions.41 Recent studies have shown that CTSL1 also has T cell–specific functions; cathepsin L inhibition prevents the cytotoxic CD8+ T-cell response in autoimmune diabetes NOD mice.42 We found 3-ketodihydrosphingosine reductase, KDSR, to be up-regulated already after an hour of culture in the Th17-polarizing condition. KDSR regulates sphingolipid biosynthesis by reducing 3-ketodihydrosphingosine to dihydrosphingosine.43 Sphingolipids are expressed on the plasma membranes ubiquitously, and are involved in structural organization of lipid rafts, regulation of apoptosis, differentiation, and proliferation. The role of the T-cell sphingolipid metabolism on the Th17 differentiation or function has not been explored. Interestingly though, altered sphingolipid metabolism is associated with pathogenesis of multiple sclerosis,44 and externally added sphingolipids augment Th17 differentiation.45
We also validated the Th17 cell–associated up-regulation of long noncoding RNA MIAT, mRNA of structural protein COL6A3 encoding α-3 chain of the type VI collagen, and LMNA which is one of the nuclear lamina proteins. Previous studies have indicated that MIAT plays a role in the development of the nervous system,46 retinal cells,47 and regulation of pluripotency,48 but so far, there has not been implication on its function in the immune system cells. COL6A3 is an important organizer of the extracellular matrix proteins. Mutations in the type VI collagen genes are associated with Bethlem myopathy and Ullrich congenital muscular dystrophy. In humans, COL6A3 expression is increased with body mass index and it contributes to adipose tissue inflammation.49 Mutations in LMNA lead to severe laminopathies. Lmna−/− mice display reduced thymus and spleen size, but so far no cell-intrinsic immune defect related to this gene has been found.50
The present study was carried out by using primary CD4+ T cells isolated from cord blood. The dataset describing the detailed kinetic changes at a very early stage of human Th17 differentiation provides a valuable resource for characterization of a variety of molecular mechanisms and signal networks involved in polarization of human Th17 cells. Although further functional analysis is needed to show whether the candidate genes identified in this study play a specific role in the Th17 differentiation process and function, the study provides a valuable starting point for identifying new pharmaceutical targets possibly regulating the process. This knowledge is essential for tackling the Th17 cell–mediated human diseases.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all voluntary blood donors and the personnel of the Department of Obstetrics and Gynaecology, Maternity Ward, Turku University Hospital (Hospital District of Southwest Finland) for the cord blood collection. Sumedha Gattani and Omid Rasool are acknowledged for their technical help. Microarray hybridizations were done at Finnish Microarray and Sequencing Center, Turku, Finland.
This work was supported by the Academy of Finland (Center of Excellence in Molecular Systems Immunology and Physiology Research, 2012-2017, Decision no. 250114, and grants 207490 SYSBIO, 116639, 115939, 140019), the European Commission Seventh Framework grants (EC-FP7-SYBILLA-201106, EC-FP7-NANOMMUNE-214281 and EC-FP7-DIABIMMUNE-202063), the JDRF, the Sigrid Juselius Foundation, a Turku University Hospital grant, and the Turku University Foundation.
Authorship
Contribution: S.T., Z.C., B.S., H.L., and R.L. designed research; V.S. and B.G. set up Th17 culturing; S.T., V.S., S.K.T., B.G., and L.O. performed experiments; S.T., V.S., S.K.T., Z.C., K.L., T.Ä., H.L., and R.L. analyzed and interpreted data; B.S. provided expertise and guidance; and S.T., Z.C., and R.L. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Riitta Lahesmaa, Turku Centre for Biotechnology, University of Turku and Åbo Akademi University, PO Box 123, FIN-20521 Turku, Finland; e-mail: riitta.lahesmaa@btk.fi.
References
Author notes
S.T., V.S., and S.K.T. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal