In this issue of Blood, Müller et al show that Fanconi anemia (FA) cells are resistant, but not completely refractory, to reprogramming into induced pluripotent stem (iPS) cells.1 Genetic complementation before reprogramming restores DNA repair, rescues reprogramming, and yields iPS cells less likely to contain genetic alterations.
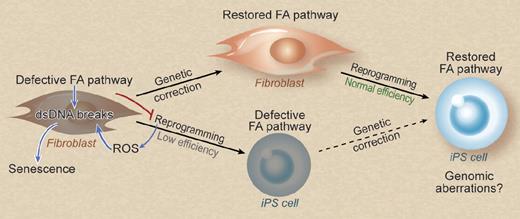
Defective DNA damage repair in FA cells induces dsDNA breaks and promotes senescence on reprogramming, thereby decreasing the efficiency of iPS cell generation. Genetic correction restores these defects and rescues the reprogramming efficiency. iPS cells derived from a previously corrected somatic cell may be less likely to harbor genomic aberrations. Professional illustration by Debra T. Dartez.
Defective DNA damage repair in FA cells induces dsDNA breaks and promotes senescence on reprogramming, thereby decreasing the efficiency of iPS cell generation. Genetic correction restores these defects and rescues the reprogramming efficiency. iPS cells derived from a previously corrected somatic cell may be less likely to harbor genomic aberrations. Professional illustration by Debra T. Dartez.
Fanconi anemia is a genetic disorder caused by loss of function of any of 14 genes in the FA pathway, a pathway coordinating cellular DNA damage repair mechanisms particularly involved in protection from DNA cross-linking agents.2 Clinically, FA manifests with bone marrow failure and increased propensity to malignancy. Genetic complementation of the FA pathway has been shown to correct the hematopoietic defects in mouse models and in cases of somatic mosaicism arising from spontaneous reversion of the mutations in humans (natural gene therapy). However, a major obstacle to translation to human gene therapy is the low yield of autologous hematopoietic stem cell (HSC) harvests from FA patients and the difficulties in their in vitro culture. Induced pluripotent stem cells offer the possibility to generate unlimited numbers of patient-specific cells and provide, from this perspective, a promising alternative therapy for FA. But pluripotent stem cell gene therapies come with their own set of problems. Reprogramming and prolonged expansion of cells may induce genomic alterations. And genetic correction should not impose additional genotoxicity.
The authors of a previous study were successful in deriving FA patient iPS cells only after the genetic defect was corrected by lentiviral transfer of the FANCA gene in the fibroblasts before reprogramming3 and not from uncorrected cells, which lead to the conclusion that restoration of the FA pathway is an absolute prerequisite for reprogramming. In contrast, in the present study, Müller and co-workers show that a defective FA pathway does not completely abolish reprogramming. The authors set to further investigate the reprogramming defect of FA cells and its mechanisms. They show that murine Fanca−/− and Fancc−/− fibroblasts have a higher incidence of dsDNA breaks—both pre-existing and induced during reprogramming—and exhibit increased reprogramming-induced senescence compared with wild-type (wt) fibroblasts (see figure). They also show that reactive oxygen species generation during reprogramming contributes to chromosome breaks and that hypoxia has a more beneficial effect in boosting reprogramming of FA than wt cells, presumably by preventing oxidative damage. Genetic correction of murine Fanca−/− fibroblasts reduces senescence and restores the reprogramming efficiency to wt levels. Similarly, genetically corrected human FA cells exhibit enhanced reprogramming compared with uncorrected fibroblasts. Importantly, the present study suggests that corrected FA iPS cell lines are less likely to contain chromosomal aberrations. These findings taken together point to a model that links the reprogramming deficit to the deficient reprogramming-induced DNA damage repair, which may lead to elimination of cells with accumulated DNA damage through senescence or other mechanisms.
This study shows that reprogramming of FA cells before genetic correction is a viable option, and raises the question, “Which should come first?” The advantage of correcting first is not only the higher reprogramming efficiency but also the potentially enhanced genomic stability of the resulting iPS cells, a very important consideration for stem cell therapies. However, the possibilities for genetic manipulation of somatic cells are limited: somatic cells are generally poorly clonable and selection of cells with the desired genetic modification or elimination of those with potentially risky transgene insertions may not be practical or even feasible. This may be particularly true with FA patient cells, which expand poorly in culture and senesce early. On the other hand, iPS cells offer a much superior platform for sophisticated genetic engineering than any somatic cell, allowing for careful selection and characterization of corrected cells, for example, by screening for safe vector integration sites or by gene targeting by homologous recombination.4,5 Furthermore, reprogramming itself may induce DNA damage, exacerbated in a background of defective DNA damage repair, but cells that harbor a great mutational load seem to be selected against (accounting for the reduced reprogramming efficiency in FA).6,7 Finally, reprogramming of FA cells also selects for genetically corrected cells that express the therapeutic transgene, as demonstrated in both the present study and the study by Raya et al,3 so it may be possible to correct and reprogram simultaneously and screen corrected iPS cell clones at the end of the process to exclude genotoxicity imposed by both the genetic modification and reprogramming at once. Which would be the preferred strategy? Further studies to address which strategy can yield iPS cells with adequate efficiency and quality together with anticipated advances in reprogramming and genetic modification methods will eventually inform the next steps toward translation to the clinic.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■
REFERENCES
National Institutes of Health

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal