To the editor:
Allogeneic hematopoietic stem cell transplantation (HSCT) can grant long-term control and cure of acute myeloid leukemia (AML) thanks to the antitumor effect of the transplanted immune system. Still, relapse remains an open issue: in the haploidentical setting, we and others demonstrated that more than one-third of post- transplantation relapses are due to the de novo genomic loss of patient-specific HLA in leukemic blasts, which favors immune evasion from donor T cells.1,2 Although highly relevant in the haploidentical context, HLA loss has been poorly assessed after matched unrelated donor (MUD) HSCT,3 which is 10 times more frequent in clinical practice.4
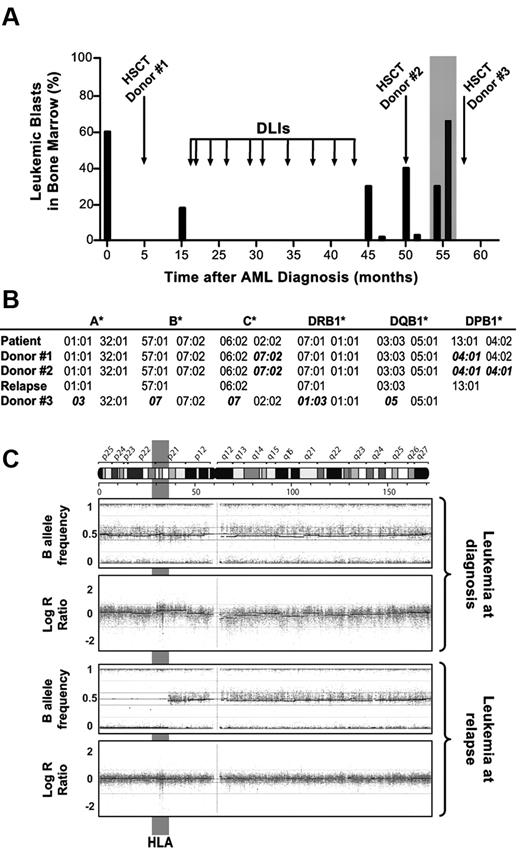
Here we report on a 36-year-old woman with normal karyotype AML, positive for the FLT3 internal tandem duplication (ITD) at diagnosis (clinical course in Figure 1A). The patient achieved complete remission after standard induction and 2 courses of consolidation chemotherapy, followed by myeloablative HSCT from a MUD (10/12 HLA matched, donor 1 in Figure 1B). Disease relapse occurred after 10 months, and was controlled for more than 2 years by salvage chemotherapy followed by serial donor lymphocyte infusions (DLIs). Thereafter, a second relapse occurred, requiring 2 cycles of chemotherapy and a second allogeneic HSCT from a different MUD (9/12 and 11/12 HLA matched with the patient and the first donor, respectively, donor 2 in Figure 1B). After 6 months from the second HSCT, a third relapse occurred (boxed in gray in Figure 1A). Interestingly, this time leukemia resulted negative for the FLT3-ITD mutation, prompting further investigation.5 HLA molecular typing and whole-genome single nucleotide polymorphism analysis were comparatively performed on leukemia at diagnosis and at third relapse, demonstrating de novo acquired uniparental disomy (UPD) of a large region of chromosome 6p. The genomic alteration spanned approximately 40 Mb and resulted in the loss of the HLA haplotype encompassing the C*02:02 and DPB1*04:02 mismatched alleles (Figure 1B and C), with the same mechanism described for relapses after haploidentical HSCT.1,2 This observation suggests that a primeval leukemic clone, even though negative for FLT3-ITD (mapped to chromosome 13), may have persisted during the course of treatments and reappeared upon the immune advantage granted by genomic HLA loss. Because T cells from the current donor are ineffective against leukemic cells that have lost the mismatched HLA,1 we proposed to our patient a third HSCT from her mother, sharing the haplotype that had been lost by UPD and therefore fully HLA-mismatched against the relapsing leukemia (donor 3 in Figure 1B). Unfortunately, the patient died in aplasia of transplant-related toxicity.
Clinical, immunogenetic, and molecular evidence of HLA loss at relapse after MUD HSCT. (A) Clinical course of the patient: histogram bars represent the percentage of leukemic blasts in the bone marrow of the patient at different time points during the posttransplantation follow-up. Boxed in gray is the disease relapse that occurred after the second MUD HSCT, when leukemic blasts were purified by fluorescence-activated cell sorting for further molecular analyses. (B) Genomic HLA typing of the patient before HSCT (and of AML blasts at diagnosis), of the 2 MUDs (donors 1 and 2), of the leukemic blasts purified at relapse after the second MUD HSCT (month 54, boxed in gray in panel A), and of the patient's mother (donor 3). HLA alleles mismatched between donors and patient are shown in bold italics. Homozygosity for HLA-DPB1*04:01 for donor 2 was inferred from univocal genomic typing. (C) Single nucleotide polymorphism (SNP) profile of chromosome 6 from purified AML blasts harvested at diagnosis (top dot plots) and at relapse after the second MUD HSCT (bottom dot plots), analyzed using the Illumina Human660W-Quad BeadChip. Top and bottom plots show the B allele frequency and the LogR ratio, indicating zygosity and gene copy numbers of each SNP, respectively. Note de novo acquired UPD of leukemic blasts at relapse in a 40-Mb region of chromosome 6p encompassing HLA (evidenced in gray).
Clinical, immunogenetic, and molecular evidence of HLA loss at relapse after MUD HSCT. (A) Clinical course of the patient: histogram bars represent the percentage of leukemic blasts in the bone marrow of the patient at different time points during the posttransplantation follow-up. Boxed in gray is the disease relapse that occurred after the second MUD HSCT, when leukemic blasts were purified by fluorescence-activated cell sorting for further molecular analyses. (B) Genomic HLA typing of the patient before HSCT (and of AML blasts at diagnosis), of the 2 MUDs (donors 1 and 2), of the leukemic blasts purified at relapse after the second MUD HSCT (month 54, boxed in gray in panel A), and of the patient's mother (donor 3). HLA alleles mismatched between donors and patient are shown in bold italics. Homozygosity for HLA-DPB1*04:01 for donor 2 was inferred from univocal genomic typing. (C) Single nucleotide polymorphism (SNP) profile of chromosome 6 from purified AML blasts harvested at diagnosis (top dot plots) and at relapse after the second MUD HSCT (bottom dot plots), analyzed using the Illumina Human660W-Quad BeadChip. Top and bottom plots show the B allele frequency and the LogR ratio, indicating zygosity and gene copy numbers of each SNP, respectively. Note de novo acquired UPD of leukemic blasts at relapse in a 40-Mb region of chromosome 6p encompassing HLA (evidenced in gray).
Our report demonstrates that immune escape by genomic HLA loss can occur not only in the haploidentical context, but also in leukemia relapses after well-matched unrelated donor HSCT. Further studies are warranted to address the frequency of this phenomenon; still, we would already encourage the use of HLA molecular typing in AML relapses also after MUD HSCT, to avoid the toxicity of inefficacious DLIs. Ultimately, novel diagnostic and therapeutic tools will be needed for early detection and targeted treatment of these peculiar variants of posttransplantation AML relapse.
Authorship
Approval was obtained from the San Raffaele Scientific Institute review board for this study. Informed consent was provided in accordance with the Declaration of Helsinki.
Acknowledgments: This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC) IG 12042, by the Italian Ministry of Health RF-FSR-2008-1202648, and by the Cariplo Foundation 2009-2665.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Katharina Fleischhauer, Unit of Molecular and Functional Immunogenetics, San Raffaele Scientific Institute, via Olgettina 60, Milano, Italy; e-mail: fleischhauer.katharina@hsr.it.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal