The characterization and targeting of Philadelphia chromosome positive (Ph+) acute lymphoblastic leukemia (ALL)–initiating cells remains unresolved. Expression of the polycomb protein Bmi1 is up-regulated in patients with advanced stages of chronic myelogenous leukemia (CML). We report that Bmi1 transforms and reprograms CML B-lymphoid progenitors into stem cell leukemia (Scl) promoter-driven, self-renewing, leukemia-initiating cells to result in B-lymphoid leukemia (B-ALL) in vivo. In vitro, highly proliferating and serially replatable myeloid and lymphoid colony-forming cultures could be established from BCR-ABL and Bmi1 coexpressing progenitors. However, unlike in vivo expanded CML B-lymphoid progenitors, hematopoietic stem cells, or multipotent progenitors, coexpressing BCR-ABL and Bmi1 did not initiate or propagate leukemia in a limiting dilution assay. Inducible genetic attenuation of BCR-ABL reversed Bmi1-driven B-ALL development, which was accompanied by induction of apoptosis of leukemic B-lymphoid progenitors and by long-term animal survival, suggesting that BCR-ABL is required to maintain B-ALL and that BCR-ABL and Bmi1 cooperate toward blast transformation in vivo. Our data indicate that BCR-ABL targeting itself is required to eradicate Ph+/Bmi1+ B-ALL–initiating cells and confirm their addiction to BCR-ABL signaling.
Introduction
Philadelphia chromosome (Ph)+ acute lymphoblastic leukemia (ALL) represents a multiclonal evolution of heterogeneous and genomically unstable tumor-initiating and tumor-propagating cells1,,,–5 The BCR-ABL fusion protein transforms hematopoietic stem cells (HSCs) in chronic myelogenous leukemia (CML) and results in progression of the disease from the chronic phase to a poor-prognosis blast crisis (BC), in which myeloid or, more frequently, lymphoid blasts fail to differentiate.6,7 BCR-ABL may also transform granulocyte-macrophage progenitors in CML myeloid BC.8 In lymphoid BC, relapse is common despite high rates of complete response to initial therapy.9,–11 BCR-ABL+ cells appear to accumulate additional genetic mutations that result in a proliferative advantage, differentiation arrest,1,12 and the acquisition of leukemic self-renewal. Various pathways downstream of BCR-ABL have been implicated in the transformation process, including activation of transcriptional factors such as STAT5,13,–15 β-catenin,8,16,17 and MEK/ERK18 and activation of members of the Rho family of GTPases.17,19,,,–23 However, the mechanisms that are required and sufficient for the transformation of chronic-phase CML into BC are poorly defined. It is also unclear whether, once secondary mutations are triggered, newly signaling pathways become independent of BCR-ABL expression to maintain leukemogenesis.24,25
Expression of Bmi1 is up-regulated more in patients with advanced disease than in patients in chronic phase.26 Bmi1 is a polycomb family protein of transcriptional repressors that maintains normal and leukemic stem cell self-renewal.27,28 Recently Schuringa et al reported that coexpression of Bmi1 and BCR-ABL in human cord blood CD34+ cells is sufficient to induce transplantable leukemia in immunodeficient mice.29 However, the functional role of Bmi1 in Ph+ B-cell ALL (B-ALL) remains unknown. The nature of the tumor-initiating cell responsible for lymphoid transformation, whether the initiation and/or maintenance of lymphoid BC is dependent on BCR-ABL signaling, and the efficacy of BCR-ABL–targeted therapies in Ph+ lymphoblastic leukemias remain controversial.5,30,–32
In the present study, using a previously characterized stem-cell leukemia (Scl) promoter–driven and tetracycline-inducible (DOX OFF) binary transgenic (Tg) mouse model of CML (Scl/p210-BCR-ABL),21 we show that Bmi1 synergizes with BCR-ABL and is sufficient to transform chronic-phase Scl/p210 B-lymphoid progenitors, but not HSCs or multipotent progenitors (MPPs), imparting a proliferative advantage to inducing serially propagated B-ALL in vivo. Bmi1 induced the expression of HSC-specific genes such as Scl and Runx1 in Scl/p210 B-lymphoid progenitors, supporting the idea that committed progenitors can reacquire a stem-cell–associated self-renewal program to function as cancer stem cells.
Methods
Mice, transduction, and serial transplantation
The generation of Scl-tTA;TRE-p210–BCR-ABL (Scl/p210) mice has been described previously.33 FVB/N-backcrossed Scl/p210 mice (kindly provided by Dr Claudia Huettner, Harvard Medical School, Boston, MA) were crossed with C57Bl/6 mice to generate F2 Scl/p210 mice, as described previously.21 Mice fed with doxycycline (6 mg/g of food; Bioserve Biotech) and weaned from doxycycline for 3 months (DOX OFF) were used in the experiments.21 CD45.2+ C57BL/6 and CD45.1+ B6.SJLPtprca/Pep3b/BoyJ mice were purchased from The Jackson Laboratory. Animal studies were approved by the Cincinnati Children's Hospital Medical Center Institutional Animal Care and Use Committee. Lin−Sca1+cKit+ (LSK) cells were isolated from the bone marrow (BM) and spleens from Scl/p210 (DOX OFF) or nonTg control (Ntg) mice that had been back-crossed with C57Bl/6. For in vivo leukemogenesis assays, LSK cells were transduced with Bmi1-IRES-EGFP or IRES-EGFP (Mock, Mk) lentiviral vectors,34 and 1000 transduced, unsorted cells were transplanted into lethally irradiated C57Bl/6 mice. For nonlimiting dilution serial transplantation experiments, 3 × 106 splenocytes were transplanted serially into secondary and tertiary recipients. For limiting dilution transplantation experiments, unfractionated cells or HSCs (EGFP+LSK+CD34−Flk2−), MPPs (EGFP+LSK CD34+Flk2+), or B-lymphoid progenitors (EGFP+B220loCD19+CD43+/−) were isolated from primary Scl/p210;Bmi1 mice with B-ALL and input equivalent limiting doses (1 × 105, 3 × 105, 10 × 105, and 30 × 105) of donor cells were transplanted, along with 5 × 105 competitor cells, into lethally irradiated secondary recipient mice. Recipient mice were used to analyze the leukemia engraftment as EGFP+B220loCD19+CD43+IgM− leukemic cells. As described previously for BCR-ABL murine leukemias,35 recipient mice with less than 1% enhanced green fluorescent protein (EGFP) cells in the circulation on at least 2 occasions were excluded from the study because of poor hematopoietic engraftment. All mice showing more than 1% circulating EGFP+ leukemic cells had BM and/or spleen leukemic infiltration higher than 20% of the overall organ cellularity, as assessed in cytospins at the time of necropsy. An example of BM and spleen leukemic infiltration is presented in Figure 1D.
Flow cytometry analysis
To label HSCs and MPPs, BM cells were stained using a cocktail of biotin-conjugated anti–mouse lineage mAbs CD45R (B220, Clone RA3-6B2), Gr-1 (Ly6G, Clone RB6-8C5), CD4 (L3T4, Clone GK1.5), CD8a (Ly-2, Clone 53-6.7), Mac-1 (CD11b, CloneM1/70), CD3ϵ (Clone 145-2C11), and TER119 (Ly-76, all from BD Pharmingen). In a second step, the cells were incubated with streptavidin-allophycocyanin (APC)–Cy7, PE-Cy7–anti–mouse Sca-1 (Ly6A/E, clone D7, e-Bioscience); APC-anti–mouse CD117 (c-kit, Clone 2B8; BD Pharmingen); Pacific Blue–anti–mouse CD34 (RAM 3-4; Pharmingen); and phycoerythrin (PE)–anti–mouse Flk2 (BD Pharmingen) Abs. EGFP+ leukemia (B-ALL)–initiating cells were identified and sorted using APC-Cy7–anti–mouse CD45R, PE-Cy7–anti–mouse CD19, PE-anti–mouse CD43, APC–anti–mouse-CD93, and PerCP-Cy5.5–anti–mouse IgM Abs (all from BD Pharmingen). The FACSCanto flow cytometer and the FACSAria cell sorter (both BD Biosciences) were used for all analyses.
Leukemia phenotyping
Tumors arising in recipient mice of Bmi1-transduced, Scl/p210-expressing cells were analyzed for cell content of EGFP+/B220+(dim) and some were further analyzed for cell content of EGFP+B220loCD19+CD43+IgM−. Tissue histology and cytospin morphological analyses were performed with hematoxylin and eosin and Wright-Giemsa staining. Diagnosis of B-ALL was made based on the Bethesda criteria for classification of lymphoid neoplasms in mice36 ; based on those criteria, the diagnosis of precursor B-cell lymphoblastic lymphoma/leukemia was made.
Culture of freshly transduced B-lymphoid progenitors
LSK cells were isolated from Scl/p210 mice (DOX OFF) presenting with myeloproliferation and were transduced with lentiviral particles expressing Bmi1 or with empty vector control in the presence of batch selected serum (StemCell Technologies), recombinant mouse stem cell factor (rmSCF; 20 ng/mL), and rm interleukin-7 (20 ng/mL) to promote proliferation of B-lymphoid progenitors. EGFP+ cells were sorted and subjected to pro-B colony-forming unit cell (CFU-C) replating or gene expression analysis.
Serial replating analysis
LSK cells were isolated from Scl/p210 mice presenting with myeloproliferation and transduced with lentiviral or retroviral particles expressing Bmi1 or with empty vector controls. EGFP+ (lentiviral transduction34 ) or red fluorescent protein positive (SFβ1-Bmi1–IRES-RFP or IRES-RFP retroviral transduction) cells were sorted and serially replated in methylcellulose containing medium (StemCell Technologies) in myeloid (100 ng/mL of rmSCF, 100 ng/mL of recombinant human thrombopoietin, and 100 ng/mL recombinant human granulocyte colony-stimulating factor) or lymphoid (20 ng/mL of rm interleukin-7 and 20 ng/mL of rmSCF) differentiation conditions.
Reversibility of B-ALL
Scl/p210;Bmi1 mice were fed with alternative cycles of diet with or without doxycycline and leukemic blasts were analyzed by flow cytometry of peripheral blood leukocytes and of interfemoral BM aspirate at different time points.
Cell-survival analysis
The survival of EGFP+ B-lymphoid progenitors in vivo in the peripheral blood (DOX ON) were monitored at different time points using annexin V staining (BD Biosciences).
Quantitative RT-PCR
mRNA expression of Bmi1, Scl, Runx1, Cbx5, Myb, Hmgb3, p16, Ebf1, Pax5, and Ikzf1 was measured in EGFP+ cells using TaqMan reverse transcriptase (Applied Biosystems) according to the manufacturer's instructions. BCR-ABL (b3a2) mRNA expression was quantified as described previously.21
LAM-PCR
Linear amplification–mediated PCR (LAM-PCR) analysis was performed on BM and spleen cells isolated from 2 primary recipient mice in which the BM and spleen were pooled for secondary transplantation (primary): 2 secondary recipients in which the BM and spleen were pooled for transplantation into tertiary recipients (secondary), and 3 tertiary recipients (tertiary) of Scl/p210;Bmi1 cells to determine the lentiviral integration pattern, as described previously.35 Briefly, DNA proviral junction were linear amplified for 100 cycles and selectively isolated using magnetic beads (Dynal kilobase binder kit; Invitrogen). The single-strand DNA was converted to double-strand DNA using random hexamers and Klenow polymerase. The DNA was then digested with Tsp509I (New England BioLabs) and ligated to a compatible linker cassette. The DNA product was dissociated from the beads and 2 rounds of exponential amplification was carried out using nested primers specific to the viral long terminal repeat (LTR) and the linker cassette. The amplified product was analyzed using gel electrophoresis (Spreadex EL1200 gels; Elchrom Scientific AG) with Tris-acetate ethylenediaminetetraacetic acid buffer. The primers used were Lv-LTR-1(Bio): GAACCCACTGCTTAAGCCTCA; Lv-LTR-2(Bio): AGCTTGCCTTGAGTGCTTCA; LC-1: GACCCGGGAGATCTGAATTC; Lv-LTR-2(Bio): AGTAGTGTGTGCCCGTCTGT; and LC-1:GATCTGA- ATTCAGTGGCACAG.
Statistical analysis
Statistical analyses were performed using Prism Version 5.0 software (GraphPad). The Kaplan-Meier log-rank test was used for survival analysis.
Results
Bmi1 overexpression facilitates B-ALL development in Scl/p210 mice
To model and identify Bmi1-driven Ph+ CML-BC–initiating cells in vivo, we overexpressed Bmi1 using an ubiquitin C promoter–driven lentiviral bicistronic vector coexpressing EGFP as a reporter,34,37 in BCR-ABL–expressing chronic-phase CML stem cells and progenitors21 (Figure 1A). The Scl/p210 binary Tg mouse model expressing BCR-ABL has been demonstrated as a murine model of preclinical human CML used to study leukemic stem cells in vivo.21,33,38 The Scl/p210 mouse model is based on the doxycycline-inducible (DOX OFF) expression of p210–BCR-ABL driven by the Scl promoter. Inducible expression of p210–BCR-ABL is therefore circumscribed to the HSCs and progenitors (HSC/Ps). Gain of function of Bmi1 in Scl/p210-HSC/P, but not in Ntg-HSC/Ps, induced the development of B-ALL with accumulation of EGFP+/FSChi/B220lo/CD19+/CD43+/IgM− B cells, with increasing mortality by 16 weeks after transplantation (Figure 1B-C). Mice transplanted with Ntg;MOCK or Scl/p210;MOCK cells had not developed leukemia by 20 weeks after transplantation (Figure 1C). The development of B-ALL in Bmi1/Scl-p210 recipients was associated with lymphadenopathy, splenic infiltration, head and neck tumor formation, and hind-limb paralysis, all characteristics of CNS infiltration of leukemic cells, mimicking human B-ALL (Figure 1B,D). Remaining Bmi1/Scl-p210 mice maintained signs of myeloproliferative disorder without developing B-ALL when followed for more than 6 months. There was no expansion of Bmi1-expressing HSCs or MPPs in the mice developing B-ALL compared with Ntg;Mock, Ntg;Bmi1, or Scl/p210;Mock mice (Figure 1E and Table 1). B-ALL was serially transplantable into secondary and tertiary recipient mice, with a progressively shorter latency period, indicating the presence of cells with leukemic self-renewal ability (Figure 1C). Clonal analysis using LAM-PCR35 revealed that in the serially repopulated mice, the disease was initiated and propagated by a predominant clone rather than by multiple, heterogeneous clones (Figure 1F).
Bmi1 collaborates with inducible BCR-ABL to induce lymphoid BC in vivo. (A) Experimental setup. To model CML-BC–initiating cells, LSK cells were isolated from Ntg or Scl/p210 mice presenting with myeloproliferative disorder (MPD), transduced with Bmi1 or empty lentiviral vectors expressing EGFP, and serially transplanted into lethally irradiated primary, secondary, and tertiary recipient mice. (B) Representative flow cytometry (FACS) contour diagram showing the frequency of EGFP+FSChiB220loCD19+CD43+IgM− B-lymphoid blasts present in the peripheral blood, BM, spleen, and lymph nodes of Scl/p210;Bmi1 mice. (C) Cumulative survival using the Kaplan-Meier log-rank P test performed in Ntg;MOCK-, Ntg;Bmi1-, Scl/p210;MOCK-, and Scl/p210;Bmi1-transplanted primary, secondary, and tertiary recipient mice (P < .05, n = 6-12 mice per group; n = 2 independent experiments). (D) Histological evidence of leukemic cell infiltration into the BM (magnification 100×) and spleen (Magnification 100×) in Scl/p210;Bmi1 (DOX OFF) mice compared with Scl/p210;Bmi1 (DOX ON) mice. Inset pictures show tissue infiltration at higher magnification (400×) or cytospin preparation of immature lymphoblasts (magnification 1000×). (E) Representative FACS contour diagram showing the frequency of HSCs and MPPs gated on EGFP+ BM cells from Ntg;MOCK, Ntg;Bmi1, Scl/p210;MOCK, and Scl/p210;Bmi1 mice. (F) LAM-PCR amplifying lentiviral vector insertion sites in the BM and spleen of Scl/p210;Bmi1-transplanted primary, secondary, and tertiary recipient mice, demonstrating a predominantly monoclonal integration pattern. Negative control indicates mouse genomic DNA control. *Internal standard. Microphotographs were obtained with an Olympus CKX41, objectives ×10, ×40, and ×100. The images were acquired with a Moticam 2500 color camera (5.0 MPixal, USB2.0), Motich China Group Co Ltd, and processed using Motic Images Plus 2.0 software.
Bmi1 collaborates with inducible BCR-ABL to induce lymphoid BC in vivo. (A) Experimental setup. To model CML-BC–initiating cells, LSK cells were isolated from Ntg or Scl/p210 mice presenting with myeloproliferative disorder (MPD), transduced with Bmi1 or empty lentiviral vectors expressing EGFP, and serially transplanted into lethally irradiated primary, secondary, and tertiary recipient mice. (B) Representative flow cytometry (FACS) contour diagram showing the frequency of EGFP+FSChiB220loCD19+CD43+IgM− B-lymphoid blasts present in the peripheral blood, BM, spleen, and lymph nodes of Scl/p210;Bmi1 mice. (C) Cumulative survival using the Kaplan-Meier log-rank P test performed in Ntg;MOCK-, Ntg;Bmi1-, Scl/p210;MOCK-, and Scl/p210;Bmi1-transplanted primary, secondary, and tertiary recipient mice (P < .05, n = 6-12 mice per group; n = 2 independent experiments). (D) Histological evidence of leukemic cell infiltration into the BM (magnification 100×) and spleen (Magnification 100×) in Scl/p210;Bmi1 (DOX OFF) mice compared with Scl/p210;Bmi1 (DOX ON) mice. Inset pictures show tissue infiltration at higher magnification (400×) or cytospin preparation of immature lymphoblasts (magnification 1000×). (E) Representative FACS contour diagram showing the frequency of HSCs and MPPs gated on EGFP+ BM cells from Ntg;MOCK, Ntg;Bmi1, Scl/p210;MOCK, and Scl/p210;Bmi1 mice. (F) LAM-PCR amplifying lentiviral vector insertion sites in the BM and spleen of Scl/p210;Bmi1-transplanted primary, secondary, and tertiary recipient mice, demonstrating a predominantly monoclonal integration pattern. Negative control indicates mouse genomic DNA control. *Internal standard. Microphotographs were obtained with an Olympus CKX41, objectives ×10, ×40, and ×100. The images were acquired with a Moticam 2500 color camera (5.0 MPixal, USB2.0), Motich China Group Co Ltd, and processed using Motic Images Plus 2.0 software.
Comparison of complete blood counts and frequency of EGFP+ HSC/Ps and EGFP+ B-lymphoid progenitors present in the BM in mice
| Primary recipient group . | WBCs, × 103/mm3 . | ANC, × 103/mm3 . | PLTs, × 103/mm3 . | Hb, g/dL . | Spleen weight, mg . | EGFP+ LT-HSCs, % . | EGFP+ ST-HSCs, % . | EGFP+ MPPs, % . | EGFP+ B220lo, % . |
|---|---|---|---|---|---|---|---|---|---|
| Ntg;MOCK | 10.6 ± 1.9 | 2.7 ± 0.6 | 940.3 ± 101.3 | 14.4 ± 0.7 | 40 ± 10 | 0.031 ± 0.003 | 0.099 ± 0.066 | 0.098 ± 0.063 | 0.34 ± 0.10 |
| Ntg;Bmi1 | 8.7 ± 2.3 | 2.5 ± 0.7 | 931.0 ± 181.4 | 13.0 ± 1.6 | 40 ± 5 | 0.021 ± 0.003 | 0.106 ± 0.070 | 0.118 ± 0.068 | 0.42 ± 0.05 |
| Scl/p210;MOCK | 12.6 ± 3.6 | 2.7 ± 0.7 | 999.8 ± 90.8 | 13.0 ± 0.6 | 49 ± 13 | 0.038 ± 0.005 | 0.106 ± 0.108 | 0.102 ± 0.094 | 0.22 ± 0.12 |
| Scl/p210;Bmi1 (B-ALL) | 14.4 ± 2.0 | 2.8 ± 0.4 | 1048.8 ± 110.5 | 12.2 ± 2.1 | 89 ± 11* | 0.029 ± 0.009 | 0.066 ± 0.046 | 0.108 ± 0.073 | 16.5 ± 8.0* |
| Scl/p210;Bmi1 (non-B-ALL) | 11.0 ± 1.3 | 2.6 ± 1.6 | 808.0 ± 203.6 | 13.1 ± 1.9 | 41 ± 5 | 0.028 ± 0.002 | 0.084 ± 0.109 | 0.090 ± 0.014 | 0.362 ± 0.09 |
| Primary recipient group . | WBCs, × 103/mm3 . | ANC, × 103/mm3 . | PLTs, × 103/mm3 . | Hb, g/dL . | Spleen weight, mg . | EGFP+ LT-HSCs, % . | EGFP+ ST-HSCs, % . | EGFP+ MPPs, % . | EGFP+ B220lo, % . |
|---|---|---|---|---|---|---|---|---|---|
| Ntg;MOCK | 10.6 ± 1.9 | 2.7 ± 0.6 | 940.3 ± 101.3 | 14.4 ± 0.7 | 40 ± 10 | 0.031 ± 0.003 | 0.099 ± 0.066 | 0.098 ± 0.063 | 0.34 ± 0.10 |
| Ntg;Bmi1 | 8.7 ± 2.3 | 2.5 ± 0.7 | 931.0 ± 181.4 | 13.0 ± 1.6 | 40 ± 5 | 0.021 ± 0.003 | 0.106 ± 0.070 | 0.118 ± 0.068 | 0.42 ± 0.05 |
| Scl/p210;MOCK | 12.6 ± 3.6 | 2.7 ± 0.7 | 999.8 ± 90.8 | 13.0 ± 0.6 | 49 ± 13 | 0.038 ± 0.005 | 0.106 ± 0.108 | 0.102 ± 0.094 | 0.22 ± 0.12 |
| Scl/p210;Bmi1 (B-ALL) | 14.4 ± 2.0 | 2.8 ± 0.4 | 1048.8 ± 110.5 | 12.2 ± 2.1 | 89 ± 11* | 0.029 ± 0.009 | 0.066 ± 0.046 | 0.108 ± 0.073 | 16.5 ± 8.0* |
| Scl/p210;Bmi1 (non-B-ALL) | 11.0 ± 1.3 | 2.6 ± 1.6 | 808.0 ± 203.6 | 13.1 ± 1.9 | 41 ± 5 | 0.028 ± 0.002 | 0.084 ± 0.109 | 0.090 ± 0.014 | 0.362 ± 0.09 |
P < .01 comparing Scl/p210;MOCK and Scl/p210;Bmi1 mice (n = 3-5 mice per group). Scl/p210;Bmi1 mice died with features of B-ALL (hind limb paralysis, lymphadenopathy, and infiltration of lymphoblasts in different tissues).
WBC indicates white blood cells; ANC, absolute neutrophil count; PLTs, platelets; Hb, hemoglobin; LT, long-term; HSC, hematotpoietic stem cells; and ST, short-term.
Overexpression of Bmi1 collaborates with BCR-ABL to induce lineage-committed progenitor expansion, in vitro self-renewal, and Scl expression
Gain of function of Bmi1 by retroviral transduction into NTg and Scl/p210 HSC/Ps significantly maintained hematopoietic progenitor clonogenicity during serial replating on methylcellulose in vitro (Figure 2A-B). In contrast, NTg and Scl/p210 cells expressing a control vector were unable to serially replate, suggesting that Bmi1 expression alone—independent of the expression of BCR-ABL—is sufficient for hematopoietic progenitor proliferation/self-renewal in vitro (Figure 2A-B). We investigated whether overexpression of Bmi1 might also affect the expansion of HSC/Ps with or without BCR-ABL expression in a non-semisolid, liquid culture–based assay. Flow cytometry–sorted RFP+ cells were allowed to grow in presence of cytokines, and CFU-C assays were performed at different time points (Figure 2C-D). Interestingly, Bmi1 expression in Scl/p210 progenitors extended the expansion of CFU-Cs far beyond control-transduced, Scl/p210 progenitors and even Bmi1-overexpressing HSC/Ps (Figure 2B-D). These data indicate that Bmi1 expression alone can determine the proliferative capacity of both normal and leukemic stem cells and progenitors and, in conjunction with BCR-ABL expression, can maintain the expansion of the progenitor compartment. This suggests that Bmi1 augments the proliferation of BCR-ABL–expressing progenitors.
Bmi1 augments proliferation and maintains clonogenicity of Scl/p210 cells. (A) Representative fluorescence photographs showing CFU-Cs of Ntg;Mk-, Ntg;Bmi1-, Scl/p210;Mk-, and Scl/p210;Bmi1-expressing cells during serial replating on methylcellulose in vitro. Earlier, Bmi1 was subcloned into Sfβ91-IRES-RFP retroviral vector. HSC/Ps were transduced and RFP+ cells were sorted by flow cytometry (FACSAria II; BD Biosciences). (B) Frequency of CFU-Cs in the BM of Ntg;Mk-, Ntg;Bmi1-, Scl/p210;Mk-, and Scl/p210;Bmi1-expressing cells during serial replating on methylcellulose in vitro. (C) Cumulative cell number expansion of Ntg;Mk-, Ntg;Bmi1-, Scl/p210;Mk-, and Scl/p210;Bmi1-expressing cells grown in IMDM-based liquid culture supplemented with 10% FCS, 100 ng/mL of SCF, 100 ng/mL of thrombopoietin, and 100 ng/mL of G-CSF. Data represent 1 of 2 independent experiments with similar results. (D) CFU-C content (expansion) per culture of sorted RFP+ BM cells from Ntg;Mk-, Ntg;Bmi1-, Scl/p210;Mk-, and Scl/p210;Bmi1-expressing groups during liquid culture in respective time points in vitro. The CFU-C assay was performed in triplicate. Data represent 1 of 2 independent experiments with similar results. Microphotographs were obtained with a DC Imaging microscope, model BA310, with a ×20 objective (OM, ×200). The images were acquired with a Moticam 2500 color camera (5.0 MPixal, USB2.0), Motich China Group Co Ltd, and processed using Motich Image Plus 2.0 software.
Bmi1 augments proliferation and maintains clonogenicity of Scl/p210 cells. (A) Representative fluorescence photographs showing CFU-Cs of Ntg;Mk-, Ntg;Bmi1-, Scl/p210;Mk-, and Scl/p210;Bmi1-expressing cells during serial replating on methylcellulose in vitro. Earlier, Bmi1 was subcloned into Sfβ91-IRES-RFP retroviral vector. HSC/Ps were transduced and RFP+ cells were sorted by flow cytometry (FACSAria II; BD Biosciences). (B) Frequency of CFU-Cs in the BM of Ntg;Mk-, Ntg;Bmi1-, Scl/p210;Mk-, and Scl/p210;Bmi1-expressing cells during serial replating on methylcellulose in vitro. (C) Cumulative cell number expansion of Ntg;Mk-, Ntg;Bmi1-, Scl/p210;Mk-, and Scl/p210;Bmi1-expressing cells grown in IMDM-based liquid culture supplemented with 10% FCS, 100 ng/mL of SCF, 100 ng/mL of thrombopoietin, and 100 ng/mL of G-CSF. Data represent 1 of 2 independent experiments with similar results. (D) CFU-C content (expansion) per culture of sorted RFP+ BM cells from Ntg;Mk-, Ntg;Bmi1-, Scl/p210;Mk-, and Scl/p210;Bmi1-expressing groups during liquid culture in respective time points in vitro. The CFU-C assay was performed in triplicate. Data represent 1 of 2 independent experiments with similar results. Microphotographs were obtained with a DC Imaging microscope, model BA310, with a ×20 objective (OM, ×200). The images were acquired with a Moticam 2500 color camera (5.0 MPixal, USB2.0), Motich China Group Co Ltd, and processed using Motich Image Plus 2.0 software.
Bmi1 overexpression–mediated BCR-ABL B-ALL results from transformation of B-cell progenitors
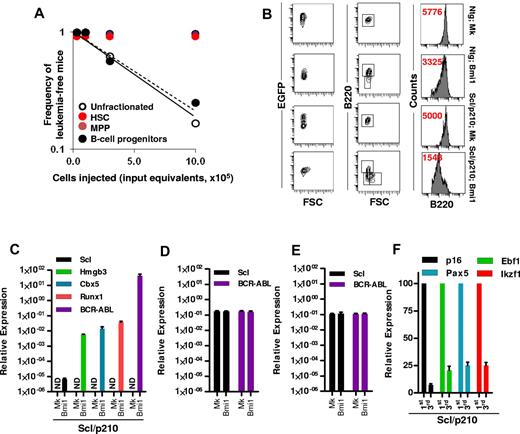
Our study was designed to characterize and quantify the leukemia-initiating cells in an in vivo model of Scl/p210;Bmi1–expressing B-ALL. Limiting dilution analysis indicated that CML-lymphoid BC was exclusively initiated by transformed B-lymphoid progenitors, because transplantation of unfractionated leukemic cells or sorted Bmi1-expressing pro-B cells into myeloablated recipients were equally able to transplant the disease, with frequencies of 1 in 700 000 cells and 1 in 765 250 cell input equivalents, respectively (Figure 3A). Neither phenotypically identified EGFP+ HSCs nor EGFP+ MPPs from leukemic mice were able to generate leukemia in the secondary recipient mice (Figure 3A), suggesting that a B-cell progenitor was responsible for the propagation of leukemias that developed in 20% of primary recipients transplanted with Scl/p210;Bmi1–expressing cells, which had developed within the first 20 weeks after transplantation. Immunophenotypic analysis further revealed that overexpression of Bmi1 in both Ntg and in Scl/p210 hematopoiesis skewed their lineage commitment toward immature B-cell differentiation with reduced expression of B220 (Figure 3B). This is in agreement with the recent finding that Bmi1 may cause lineage specification of HSC/Ps,39 thereby making the lymphoid progenitor compartment more susceptible to genetic perturbations.
Bmi-1 induces self-renewal and transforms B-lymphoid progenitors. (A) Frequency of leukemia-initiating cells in Scl/p210;Bmi1 mice in unfractionated and sorted cell fractions. Results are presented as cell input equivalents (P < .05, n = 8-10 mice per group). (B) Representative FACS contour diagram and histogram showing expression (in red, mean fluorescence intensity, MFI) of B220 on EGFP+ cells in Ntg;MOCK, Ntg;Bmi1, Scl/p210;MOCK, and Scl/p210;Bmi1 mice. (C) Relative mRNA expression (quantitative RT-PCR, 2ΔCt) analysis of Scl, Hmgb3, Cbx5, Runx1, and BCR-ABL (b3a2) in EGFP+ B-lymphoid progenitors isolated from Scl/p210;Mock (Mk) and Scl/p210;Bmi1 mice. ND indicates not detected. (D) Relative mRNA expression (quantitative RT-PCR, 2ΔCt) analysis of Scl, and BCR-ABL (b3a2) in EGFP+ HSCs isolated from Scl/p210;Mock (Mk) and Scl/p210;Bmi1 mice. (E) Relative mRNA expression (quantitative RT-PCR, 2ΔCt) analysis of Scl, and BCR-ABL (b3a2) in EGFP+ MPPs isolated from Scl/p210;Mock (Mk) and Scl/p210;Bmi1 mice. (F) Relative mRNA expression (quantitative RT-PCR, normalized) analysis of p16, Ebf1, Pax5, and Ikzf1 performed on EGFP+ B-lymphoid progenitors isolated from primary (first) and tertiary (third) Scl/p210;Bmi1 mice. Data were normalized to the leukemic cells obtained from primary (first) recipient mice.
Bmi-1 induces self-renewal and transforms B-lymphoid progenitors. (A) Frequency of leukemia-initiating cells in Scl/p210;Bmi1 mice in unfractionated and sorted cell fractions. Results are presented as cell input equivalents (P < .05, n = 8-10 mice per group). (B) Representative FACS contour diagram and histogram showing expression (in red, mean fluorescence intensity, MFI) of B220 on EGFP+ cells in Ntg;MOCK, Ntg;Bmi1, Scl/p210;MOCK, and Scl/p210;Bmi1 mice. (C) Relative mRNA expression (quantitative RT-PCR, 2ΔCt) analysis of Scl, Hmgb3, Cbx5, Runx1, and BCR-ABL (b3a2) in EGFP+ B-lymphoid progenitors isolated from Scl/p210;Mock (Mk) and Scl/p210;Bmi1 mice. ND indicates not detected. (D) Relative mRNA expression (quantitative RT-PCR, 2ΔCt) analysis of Scl, and BCR-ABL (b3a2) in EGFP+ HSCs isolated from Scl/p210;Mock (Mk) and Scl/p210;Bmi1 mice. (E) Relative mRNA expression (quantitative RT-PCR, 2ΔCt) analysis of Scl, and BCR-ABL (b3a2) in EGFP+ MPPs isolated from Scl/p210;Mock (Mk) and Scl/p210;Bmi1 mice. (F) Relative mRNA expression (quantitative RT-PCR, normalized) analysis of p16, Ebf1, Pax5, and Ikzf1 performed on EGFP+ B-lymphoid progenitors isolated from primary (first) and tertiary (third) Scl/p210;Bmi1 mice. Data were normalized to the leukemic cells obtained from primary (first) recipient mice.
In the adult hematopoietic system, the Scl promoter is active in HSCs, but not in lineage-committed B or T lymphocytes.21,33,40 Consequently, we did not detect Scl promoter–dependent BCR-ABL expression in EGFP+ B-lymphoid progenitors isolated from Scl/p210;MOCK mice (Figure 3C). Interestingly, Bmi1 overexpression up-regulated the expression of Scl and Scl-promoter–dependent BCR-ABL in EGFP+ B-lymphoid progenitors isolated from Scl/p210;Bmi1 mice, but not in those isolated from Ntg;Bmi1 mice (Figure 3C). However, the gain of function of Bmi1 did not induce expression of Scl and BCR-ABL in HSCs or MPPs isolated from Scl/p210;Bmi1 mice (Figure 3D-E). Bmi1 also induced the expression of the stem-cell–associated genes Hmgb3, Cbx5, and Runx1, and repressed the expression of the B-lymphoid differentiation genes p16, Ebf1, Pax5, and Ikzf1 in EGFP+ B-ALL–initiating cells from Scl/p210;Bmi1 mice (Figure 3C,F). To further confirm that the induction of Scl expression is a direct consequence of the gain of function of Bmi1 in Scl/p210 B-lymphoid progenitors, freshly Bmi1-transduced LSK cells isolated from Scl/p210 mice presenting with myeloproliferation were subjected to gene-expression analysis (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Bmi1 induced the expression of Scl, but not Runx1, in freshly transduced Scl/p210 B-lymphoid progenitors (supplemental Figure 1C). These results indicate that Bmi1 can transform the Scl/p210 B-lymphoid progenitor compartment into Scl-expressing, self-renewing leukemic stem cells in vivo.
Sustained BCR-ABL expression is required to maintain Bmi1-overexpressing, progenitor-initiated B-ALL
To further explore whether the initiation and maintenance of B-ALL exclusively depend on BCR-ABL signaling and whether the attenuation of BCR-ABL activity can prevent the development of Bmi1-driven B-ALL, we performed inducible genetic rescue experiments in vivo. To study leukemia initiation, mice transplanted with B-ALL–initiating cells were fed with (DOX ON) or without (DOX OFF) doxycycline immediately after transplantation (Figure 4A). To study leukemia maintenance, recipient mice that had already developed B-ALL under DOX OFF conditions were switched back to a DOX ON diet to stop further expression of BCR-ABL (Figure 4A). B-ALL failed to develop with the loss of expression of BCR-ABL expression during leukemia initiation and the mice remained alive without any detectable EGFP+ cells (Figure 4B-D). In addition, attenuation of BCR-ABL expression in mice that had already developed B-ALL completely reversed the disease and allowed these mice to survive (Figure 4B). EGFP+ B-cell progenitor clones progressively disappeared because of apoptosis (Figure 4D-F), indicating that the sustained expression of BCR-ABL is required for lymphoblast survival. Finally, we investigated whether reexpression of BCR-ABL in mice on the DOX ON diet without apparent minimal residual disease in the BM (Figure 4G), which had previously developed B-ALL, results in a relapse of leukemia. The reexpression of BCR-ABL did not result in the relapse of EGFP+ B-ALL clones in the BM of these mice (Figure 4G), which were followed for more than 4 months and maintained levels of circulating EGFP+ and BM/spleen EGFP+ content below 1% of cells. These data indicate that Bmi1-expressing clones also require BCR-ABL expression to maintain proliferation and survival.
BCR-ABL expression is indispensable for survival, initiation, and maintenance of Bmi1-expressing CML BC–initiating cells in vivo. (A) Experimental setup. To study leukemia initiation, mice transplanted with B-ALL–initiating cells were fed with (DOX ON) or without (DOX OFF) doxycycline immediately after transplantation. To study leukemia maintenance, recipient mice that had already developed B-ALL under DOX OFF conditions were switched back to a DOX ON diet to stop the further expression of BCR-ABL. (B) Representative image of Scl/p210;Bmi1 mice fed DOX-ON or DOX-OFF diet. (C) Cumulative survival of Scl/p210;Bmi1 mice during leukemia initiation and maintenance DOX-ON or DOX-OFF diets. (P < .05, n = 3-4 mice per group). (D-G) Mice fed with a DOX OFF followed by a DOX ON diet. (D) Representative FACS contour diagram showing EGFP+ B-lymphoid progenitors and annexin V+ B-lymphoid progenitors present in the peripheral blood of Scl/p210;Bmi1 mice. (E) Frequency of EGFP+ B-lymphoid progenitors present in the peripheral blood of Scl/p210;Bmi1 mice (n = 6 mice in each time point). (F) Frequency of annexin V+ B-lymphoid progenitors present in the peripheral blood of Scl/p210;Bmi1 mice (n = 6 mice in each time point). (G) Representative FACS contour diagram showing EGFP+ B-lymphoid progenitors present in the BM of Scl/p210;Bmi1 mice.
BCR-ABL expression is indispensable for survival, initiation, and maintenance of Bmi1-expressing CML BC–initiating cells in vivo. (A) Experimental setup. To study leukemia initiation, mice transplanted with B-ALL–initiating cells were fed with (DOX ON) or without (DOX OFF) doxycycline immediately after transplantation. To study leukemia maintenance, recipient mice that had already developed B-ALL under DOX OFF conditions were switched back to a DOX ON diet to stop the further expression of BCR-ABL. (B) Representative image of Scl/p210;Bmi1 mice fed DOX-ON or DOX-OFF diet. (C) Cumulative survival of Scl/p210;Bmi1 mice during leukemia initiation and maintenance DOX-ON or DOX-OFF diets. (P < .05, n = 3-4 mice per group). (D-G) Mice fed with a DOX OFF followed by a DOX ON diet. (D) Representative FACS contour diagram showing EGFP+ B-lymphoid progenitors and annexin V+ B-lymphoid progenitors present in the peripheral blood of Scl/p210;Bmi1 mice. (E) Frequency of EGFP+ B-lymphoid progenitors present in the peripheral blood of Scl/p210;Bmi1 mice (n = 6 mice in each time point). (F) Frequency of annexin V+ B-lymphoid progenitors present in the peripheral blood of Scl/p210;Bmi1 mice (n = 6 mice in each time point). (G) Representative FACS contour diagram showing EGFP+ B-lymphoid progenitors present in the BM of Scl/p210;Bmi1 mice.
Discussion
In the present study, we have demonstrated that the polycomb group protein Bmi1 collaborates with BCR-ABL and transforms chronic-phase Scl/p210 B-lymphoid progenitors, giving them a proliferative advantage in inducing B-ALL in vivo and an ability to initiate leukemia in vivo over HSCs or MPPs. We used an inducer of stem cell–initiated CML with the ability to induce a myeloproliferative disorder that progress to lymphoblastic crisis,21,33,41,42 which is an advantageous system with which to determine the predominant nature of the BC leukemia initiating and propagating cells. In our specific model, FVB/N × B6 F2 mice were used. Although the pool of leukemia-initiating cells in the primary recipient mice appeared to be heterogeneous, the B-ALL was maintained and serially propagated with a predominant self-renewing tumor-initiating clone. In agreement with our previous findings,21 the mice transplanted with Scl/p210;MOCK cells also did not develop blastic transformation, and therefore survived for a long time.
Bmi1 induced the expression of HSC-specific genes, such as Scl and Runx1m in Scl/p210 B-lymphoid progenitors, supporting the idea that lineage-committed progenitors can also reacquire a stem cell–associated self-renewal program to function as cancer stem cells and be selected in vivo.8,43,44 Bmi1 overexpression collaborates with BCR-ABL to increase the serial replating efficiency of myeloid and B-lymphoid progenitors. This effect is correlated with in vitro up-regulation of the expression of Scl, but not Runx1, in expanded B-lymphoid progenitors, suggesting that Runx1 up-regulation requires an additional process of in vivo selection. Finally, the functional synergism between Bmi1 and BCR-ABL also induced blockage in B-lymphoid differentiation by repressing p16, Ebf1, Pax5, and Ikzf1 in vivo.
It is well documented that the p16Ink4a/p19Arf locus, which is required to prevent cell senescence, is repressed by Bmi1, and genetic deletion of Bmi1 increases expression of p16Ink4a and p19Arf.45,46 Deletion of p16Ink4a/p19Arf in Bmi1-deficient HSCs only partly restored self-renewal, and overexpression of Bmi1 could still increase progenitor levels in the absence of p16Ink4a/p19Arf, indicating that other Bmi1 targets are present and that p16Ink4a/p19Arf are not uniquely responsible for the self-renewal–inducing ability of Bmi1.46 Deletion of both the p16Ink4a/p19Arf locus and deletion of Ikzf1, Ebf1, and Pax5 have been implicated in the acute lymphoid, but not myeloid, leukemias initiated by BCR-ABL.35,40,41 Our data demonstrate that Bmi1 collaborates with BCR-ABL to substantially recapitulate the biologic features of human B-lymphoid BC, which is similar to the origin of tumor initiation by committed progenitors in myeloid BC.8 These biologic features include: (1) down-regulation of the expression of p16, Ikzf1, Ebf1, and Pax5, which is associated with B-lymphoid differentiation arrest such as commonly observed in human B-ALL (this is drastically different from the published models of B-lymphoid differentiation derived from cord blood-derived CD34+ cells, in which leukemias lack down-regulation of the expression of Ikzf1 and/or Pax529 ; (2) significant up-regulation of transcription factors associated with B-lymphoid expansion, including Hmgb347,48 ; (3) up-regulation of Cbx5, which along with Bmi1, forms part of the polycomb repression complex 1 (PRC1), with specific functions in chromatin remodeling and expression in MLL rearranged leukemic stem cells49 ; and (4) expression of a self-renewal–associated transcriptional program in B-lymphoid progenitors that includes expression of Scl in vitro and in vivo, and Runx1 in vivo. Whether Runx1 expression requires signals from the hematopoietic microenvironment, or is triggered by indirect mechanisms that require initial expansion of the progenitor pool is uncertain.
The advantage of an inducible murine model is that it allows the analysis of tumor initiation by immunophenotypically well-characterized HSCs and MPPs in which BCR-ABL expression is induced in vivo in adult BM, without the specific bias associated with fetal transcriptional profiles of cord blood–derived cells transduced in vitro. Our data demonstrate that Bmi1 synergistically induces in vivo transformation that occurs at the level of lymphoid progenitor, rather than at the HSC level. Although we cannot rule out that an HSC or a MPP could have initiated lymphoid leukemias 20 weeks after transplantation in the primary mice that did not develop leukemia (Figure 1C), we were unable to observe HSC/MPP–initiated leukemias at the dose administered. Our data could be interpreted such that an induction of a self-renewal program in B-lymphoid progenitors outcompetes the putative transformation effect on HSCs through a rapid expansion of a reduced number of B-lymphoid progenitor clones, which are further selected during serial transplantation. Although Bmi1 and BCR-ABL are synergistic in inducing B-cell progenitor transformation and leukemogenesis, Bmi1-driven Ph+ B-ALL–initiating cells depend entirely on BCR-ABL for leukemia initiation and maintenance, because the genetic deletion of BCR-ABL was sufficient to eradicate the tumor-initiating population in vivo. These data are in contrast to recent reports indicating that the ABL tyrosine kinase domain of BCR-ABL is dispensable for CML leukemic stem cell activity.25 However, BCR-ABL contains multiple signaling domains that may or may not depend on active ABL tyrosine kinase. Therefore, our data underscore that multidomain targeting of BCR-ABL signaling remains crucial in the eradication of B-ALL–initiating progenitors, and that, at least in this model, B-ALL tumor-propagating cells are oncogene addicted.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Claudia Huettner for providing the initial double-transgenic animals; Drs Sally Temple (Albany, NY) and H. Leighton Grimes (Cincinnati Children's Hospital, Cincinnati, OH) for providing the viral vectors; Jorden Arnett, Jeff Bailey, and Victoria Summey for technical assistance (Research Flow Cytometry Core Facility, which is supported by the National Institutes of Health/CEMH grant 1P30DK090971-01); and Drs Elke Grassman and ShivKumar Viswanathan at the Cincinnati Children's Research Foundation Translational Trial (TTDSL) Core for LAM-PCR analysis.
This study was supported by funding from the Institute of Cancer Research, United Kingdom/The Lady Tata Memorial Trust postdoctoral fellowship (to A.S.); the National Institutes of Health (grants R01-HL087159 and R01-HL087159S1); and the Department of Defense (grant CM064050 to J.A.C.). A.S. is a Ramalingaswami Fellow-Scientist awardee of the Department of Biotechnology, Government of India.
National Institutes of Health
Authorship
Contribution: A.S. performed the experiments; A.S. and J.A.C. conceived and designed the research, interpreted the data, and wrote the manuscript; A.F. and S.D. assisted in caring for the mouse colony and in genotyping; and M.M. helped with lentivirus production.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jose A. Cancelas, Division of Experimental Hematology & Cancer Biology and Hoxworth Blood Center, University of Cincinnati, 3333 Burnet Ave, Cincinnati, OH 45229; e-mail: jose.cancelas@cchmc.org or jose.cancelas@uc.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal