Abstract
Up to 15% of acute promyelocytic leukemia (APL) patients fail to achieve or maintain remission. We investigated a common G > A polymorphism at position −1377 (rs2234767) in the core promoter of the CD95 cell death receptor gene in 708 subjects with acute myeloid leukemia, including 231 patients with APL. Compared with the GG genotype, carrier status for the −1377A variant was associated with a significantly worse prognosis in APL patients. Carriers were more likely to fail remission induction (odds ratio = 4.22; 95% confidence interval, 1.41-12.6, P = .01), were more likely to die during the first 8 weeks of remission induction therapy (hazard ratio = 7.26; 95% confidence interval, 2.39-22.9, P = .0005), and had a significantly worse 5-year overall survival (odds ratio = 2.14; 95% confidence interval, 1.10-4.15, P = .03). The −1377A variant destroys a binding site for the SP1 transcriptional regulator and is associated with lower transcriptional activity of the CD95 promoter. Identifying patients at high risk of life-threatening events, such as remission induction failure, is a high priority in APL, especially because such events represent a major cause of death despite the introduction of differentiation therapy.
Introduction
Prognosis in patients with acute myeloid leukemia (AML) is heterogeneous, and dependent on both disease and host characteristics. This is exemplified by acute promyelocytic leukemia (APL), where treatment has been revolutionized by the introduction of differentiation chemotherapy targeted against the promyelocytic leukemia/retinoic acid receptor-α fusion oncoprotein (PML-RARA). However, up to 15% of patients do not achieve complete remission (CR) and some die during induction therapy, primarily because of hemorrhage, differentiation syndrome, or infection.1 Patients achieving CR have a relatively good prognosis, although up to 10% will relapse or experience an adverse event.2 Understanding the mechanisms that determine failure to achieve or maintain CR and affect risk of developing complications in APL could allow for patients at risk of poor outcome to be prospectively identified. Understanding host and leukemic features that increase the risk of chemoresistance, infection, or coagulopathy is an important clinical goal in APL where adverse events are a significant cause of mortality.3
CD95 (FAS, APO-1, TNFRSF6, and APT1) is a potent cell death receptor which, when bound by its ligand (CD95L), initiates the assembly of a signaling complex that includes the FAS-associated death domain and ultimately leads to cell death.4 The CD95 pathway is essential in eliminating self-reactive B-lymphocytes,5 mediating neutrophil6 and megakaryocyte cell death,7 and regulating T-lymphocyte homeostasis.8 As such, CD95 plays a role in controlling both myeloid and lymphoid cell functions. Furthermore, apoptosis induced by cytotoxic chemotherapeutic agents used to treat AML also occurs via CD95 signaling, and an intact CD95-dependent DNA damage-induced cell death response underlies remission induction in AML.9
We have previously shown that a genetic polymorphism (rs2234767; G > A, position −1377) in the core promoter of CD95 is associated with risk of developing AML.10 As such, we hypothesized that attenuation of CD95 death signaling via constitutional genetic variation in CD95 could also affect disease course in patients diagnosed with AML, including APL.
Methods
Study patients
DNA was available from 654 patients entered into the United Kingdom Medical Research Council (MRC) AML 12 trial (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), which included 177 patients diagnosed with APL. Cytogenetic status was based on the original MRC hierarchical prognostic classification.11 AML 12 patients were randomized to receive chemotherapy protocols, including DAT (daunorubicin, cytarabine, and 6-thioguanine), ADE (cytarabine, daunorubicin, and etoposide), and MAE (mitoxantrone, cytarabine, and etoposide).12 Seventy-eighty APL patients were entered into the MRC all-trans retinoic acid (ATRA) trial,2 in which patients were randomized to receive a 5-day course of ATRA before induction chemotherapy (short ATRA) or an extended course of ATRA begun at the same time as induction therapy and continued until morphologic CR was achieved or to a maximum of 60 days (extended ATRA). In the remaining patients, ATRA was given according to current clinical practice where patients received an extended course as per the MRC ATRA trial.
DNA was available from an additional 54 APL patients, the majority of whom were entered into the MRC AML 15 trial.13 All APL patients were treated with ATRA but were randomly assigned to receive 4 courses of intensive MRC combination chemotherapy or 4 courses of less intensive modified AIDA anthracyline-based chemotherapy (PETHEMA schedule).14
In summary, 38 and 185 APL patients were treated on the short and extended ATRA protocols, respectively. Specific ATRA protocol details were not available for 8 patients. In addition, 100, 64, 48, and 19 APL patients were treated with ADE-, MAE-, DAT-, and AIDA-based protocols, respectively. None of the patients in this study was treated with arsenic trioxide.
CR was defined as a bone marrow aspirate containing less than 5% blasts, displaying normal trilineage maturation. Treating clinicians classified remission failure as induction death (death induced by treatment or hypoplasia), resistant disease (> 15% blasts in the bone marrow), or partial remission (5%-15% blasts in the bone marrow, or < 5% blasts but a hypocellular bone marrow). The MRC AML 12 and 15 trials received multicenter research ethics committee approval, and this study received local research committee approval. Patient consent was obtained in accordance with the Declaration of Helsinki.
Statistical analysis
The Mantel-Haenszel test for trend and the χ2 test were used to examine the distribution of clinical/demographic/treatment variables as a function of CD95 genotype. Univariate analyses of remission were calculated using the Mantel-Haenszel test. Comparisons of overall survival (OS) and relapse free survival (RFS) between genotype groups were performed using the Kaplan-Meier method and the log-rank test. Multivariate analysis using logistic and Cox regression was used to test for independency of polymorphic status and clinical/demographic variables. Point estimates of effect size (odds ratio [OR]; hazard ratio [HR]) are given together with 95% confidence intervals. Statistical significance was set at P < .05. Given the low number of AA homozygotes at the −1377 position, these were grouped with heterozygotes for the majority of analyses.
CD95 allelic discrimination and gene expression
DNA and RNA were extracted from peripheral blood and bone marrow smears using the QIAamp DNA Blood Mini Kit and the RNeasy Mini kit (QIAGEN), respectively. Genotype for rs2234767 (−1377 G > A) was determined using a TaqMan allelic discrimination assay (assay identification C_12123966_10; Applied Biosystems) and validated by DNA sequencing. HL60 and NB4 cells were confirmed as wild-type homozygote (GG) for the CD95 −1377 promoter variant. CD95 gene expression was determined by quantitative real-time PCR, using a TaqMan gene expression assay (assay identification Hs00531110_m1; Applied Biosystems).
Cell lines and cell culture
HL60, NB4, AML-2, AML-3, Jurkat, and LoVo cell lines were maintained in RPMI 1640 medium supplemented with 10% FCS, 2nM l-glutamine, 50 U/mL penicillin, and 50 μg/mL streptomycin (Invitrogen) in a humidified atmosphere with 5% CO2 (37°C).
EMSAs
HL60 and NB4 cell pellets were lysed by resuspension in 10mM HEPES, 1.5mM MgCl2, 10mM KCl, 0.5mM DTT, 10mM PMSF, 4°C for 10 minutes followed by centrifugation (10 seconds, 14 000g). The nuclear pellet was resuspended in 100 μL 15mM HEPES, 1.5mM MgCl2, 0.42M NaCl, 0.5mM DTT, 10mM PMSF, 25% volume/volume glycerol at 4°C for 20 minutes, followed by centrifugation at 14 000g for 2 minutes. The supernatant containing nuclear extract was quantified using the BioRad Protein Assay reagent (Bio-Rad). Electrophoretic mobility shift assays (EMSAs) were performed as previously described.10
SP1 siRNA
HL60 and NB4 cells were electroporated with siRNA specific to SP1 (Santa Cruz Biotechnology) or an siRNA specifically directed against the fusion breakpoint of the MLL-AF4 fusion gene transcript (Invitrogen). Exponentially growing cells (500 μL at 107 cells/mL) were incubated with SP1 or MLL-AF4 siRNAs at a final concentration of 100nM, and electroporation was carried out using a rectangle pulse of 350 V for 10 ms in a 4-mm cuvette (Fischer). Degree and specificity of SP1 knockdown were assessed in each experiment by immunoblot. Immunoblots were scanned using a Fuji LAS-3000 Image Reader (Fuji Photo Film), and densitometric analysis was performed using AIDA Image Analyser Version 3.28 (LI-COR Biosciences). Statistical significance was tested using the Student t tests, with P values < .05 considered significant.
ATRA treatment and NBT reduction assay
HL60 and NB4 cells were treated with 10μM ATRA or vehicle only (DMSO), and granulocytic differentiation of cells was confirmed 48 hours after treatment by nitroblue tetrazolium (NBT) reduction assay. Briefly, ATRA and vehicle only treated cells were resuspended in 0.3μM phorbol 12-myristate 13-acetate (Sigma-Aldrich) diluted in 1 mg/mL NBT chloride solution (Sigma-Aldrich) and incubated for 40 minutes at 37°C. Cytospins were prepared from each cell suspension before microscopic examination. ATRA inhibited cell growth by approximately 10% and 20% at 48 and 72 hours, respectively, compared with DMSO-treated controls. Cells were examined using a 40×/0.6× objective on a Zeiss Axio Observer Z1 microscope (Carl Zeiss Ltd). Images were captured using a Hitachi CCD Color HV-D30P camera (Hitachi), with PALM Robo Version 4.0.0.10 software (P.A.L.M. Microlaser Technologies).
Caspase-8 and caspase-3 cleavage assay
Cell suspensions at a density of 5 × 105/mL were cultured for 24 hours in media supplemented with 1 μg/mL Enhancer for Ligands (Enzo Life Sciences) and either 50 or 100 ng/mL recombinant human FAS ligand (Enzo Life Sciences). Cytosolic extracts were prepared from 106 washed cells, and caspase activation was assessed by immunoblot.
Plasmid constructs and Luciferase reporter gene assays
CD95 promoter sequences were cloned into the pGL4.10[luc2] reporter vector. LoVo cells were seeded at a density of 4 × 105 cells/mL and transfected using Lipofectamine LTX and PLUS Reagent (Invitrogen); 500 ng of the pGL4.10[luc2] vector containing the appropriate CD95 promoter region construct and 100 ng of the pRL-TK[hRluc] control vector were combined with 0.5 μL of PLUS Reagent and 3 μL of Lipofectamine LTX, in Opti-MEM I reduced-serum medium (Invitrogen), to a final volume of 500 μL. The ratio of firefly and Renilla luciferase activities was determined 48 hours after transfection using the Dual Luciferase Reporter Assay (Promega) and a 96-well plate luminometer (EG&G Berthold).
Results
CD95 genotype and outcome in AML
DNA was available from 654 patients recruited to the MRC AML12 clinical trial. G > A genotype (rs2234767) distribution at position −1377 of CD95 was not significantly associated with demographic or clinical characteristics (supplemental Table 1). Carrier status for the rare allele (GA + AA) was not significantly associated with failure to achieve CR (carriers, 18%; noncarriers, 15%; OR = 1.26; 95% CI, 0.75-2.12), resistant disease (carriers, 8%; noncarriers, 7%; OR = 1.22; 95% CI, 0.58-2.56) or induction death (carriers, 10%; noncarriers, 8%; OR = 1.27; 95% CI, 0.65-2.49). However, carriers had a moderately worse OS (P = .09) and RFS (P = .06) compared with noncarriers (supplemental Figure 1).
CD95-1377 genotype and outcome in APL
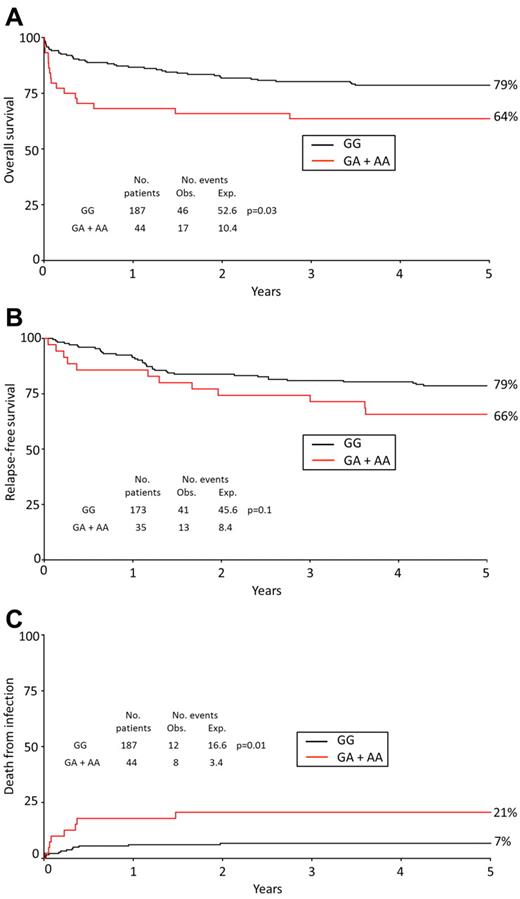
The PML gene encodes a tumor suppressor that is crucial for CD95-mediated cell death signaling.15 Because most cases of APL are characterized by a fusion gene involving PML and APL is treated differently from other forms of AML, we stratified the analysis by APL status, which revealed a strong association between CD95 genotype and prognosis in APL specifically. Within the 231 APL patients recruited to this study, CD95 genotype was not significantly associated with clinical and demographic features (Table 1). Carriers of the variant allele at position −1377 (GA + AA) had a significantly worse OS (Table 2; Figure 1A) and were significantly more likely to die within the first 8 weeks after diagnosis compared with noncarriers (23% vs 6%; HR = 7.26; 95% CI, 2.39-22.9; P = .0005; Table 2). There was also a significant association between CD95 genotype and risk of death in the first 30 days and a trend toward an association within the first 7 days (Table 2). The high risk of early death is probably the result of a poor response to induction therapy, as indicated by an association between the variant allele and failure to achieve CR (OR = 4.22; 95% CI, 1.41-12.6; P = .01, Table 2). In multivariate Cox regression analysis that included performance status, white blood cell (WBC) count, age, sex, and secondary status in the model, carrier status for the CD95 variant allele remained a significant predictor of OS in APL (CD95 rare variant, HR = 2.21; 95% CI, 1.22-4.00, P = .007) and was the strongest predictor of death within 8 weeks (CD95 variant, HR = 5.53; 95% CI, 2.19-13.97, P < .0001). The variant allele at position −1377 was also associated with a moderately worse RFS, but this was not statistically significant (Table 2; Figure 1B). There was no evidence of any association between the −1377 variant and outcome in non-APL AML (supplemental Table 2).
Demographic and clinical characteristics of APL cases stratified by CD95 −1377 genotype
| . | Total, N (%) . | CD95 −1377 status . | P . | |
|---|---|---|---|---|
| GG, N (%) . | GA + AA, N (%) . | |||
| Total | 231 | 187 | 44 | |
| Sex | ||||
| Female | 124 (54) | 102 (55) | 22 (50) | .6 |
| Male | 107 (46) | 85 (45) | 22 (50) | |
| Age, y | ||||
| 0-14 | 2 (1) | 1 (0) | 1 (2) | .2* |
| 15-29 | 65 (28) | 56 (30) | 9 (20) | |
| 30-39 | 57 (25) | 46 (25) | 11 (25) | |
| 40-49 | 51 (22) | 41 (22) | 10 (23) | |
| 50-59 | 48 (21) | 38 (20) | 10 (23) | |
| ≥ 60 | 8 (3) | 5 (3) | 3 (7) | |
| Median (range) | 38 (1-63) | 38 (1-62) | 40 (13-61) | |
| Diagnosis | ||||
| De novo | 223 (97) | 182 (97) | 41 (93) | .18 |
| Secondary | 8 (3) | 5 (3) | 3 (7) | |
| WHO performance status | ||||
| 0 | 149 (65) | 122 (65) | 27 (61) | .8* |
| 1 | 63 (27) | 49 (26) | 14 (32) | |
| 2 | 10 (4) | 9 (5) | 1 (2) | |
| 3 | 6 (3) | 5 (3) | 1 (2) | |
| 4 | 3 (1) | 2 (1) | 1 (2) | |
| WBC, × 109 L−1 | ||||
| < 10 | 165 (72) | 131 (70) | 34 (79) | .15* |
| 10-19.9 | 22 (10) | 15 (8) | 6 (14) | |
| 20-49.9 | 21 (9) | 20 (11) | 1 (2) | |
| 50-99.9 | 16 (7) | 17 (9) | 0 (0) | |
| ≥ 100 | 5 (2) | 3 (2) | 2 (5) | |
| Median (range) | 3 (0.2-195) | 3.3 (0.2-140) | 2.1 (0.6-195) | |
| Platelets, × 109 L−1 | ||||
| < 10 | 14 (6) | 12 (6) | 2 (5) | .15* |
| 10-19.9 | 75 (34) | 62 (33) | 13 (31) | |
| 20-49.9 | 102 (44) | 83 (45) | 19 (45) | |
| 50-99.9 | 23 (10) | 19 (10) | 4 (10) | |
| ≥ 100 | 13 (6) | 9 (5) | 4 (10) | |
| Median (range) | 23 (5-232) | 22 (6-232) | 26 (5-146) | |
| . | Total, N (%) . | CD95 −1377 status . | P . | |
|---|---|---|---|---|
| GG, N (%) . | GA + AA, N (%) . | |||
| Total | 231 | 187 | 44 | |
| Sex | ||||
| Female | 124 (54) | 102 (55) | 22 (50) | .6 |
| Male | 107 (46) | 85 (45) | 22 (50) | |
| Age, y | ||||
| 0-14 | 2 (1) | 1 (0) | 1 (2) | .2* |
| 15-29 | 65 (28) | 56 (30) | 9 (20) | |
| 30-39 | 57 (25) | 46 (25) | 11 (25) | |
| 40-49 | 51 (22) | 41 (22) | 10 (23) | |
| 50-59 | 48 (21) | 38 (20) | 10 (23) | |
| ≥ 60 | 8 (3) | 5 (3) | 3 (7) | |
| Median (range) | 38 (1-63) | 38 (1-62) | 40 (13-61) | |
| Diagnosis | ||||
| De novo | 223 (97) | 182 (97) | 41 (93) | .18 |
| Secondary | 8 (3) | 5 (3) | 3 (7) | |
| WHO performance status | ||||
| 0 | 149 (65) | 122 (65) | 27 (61) | .8* |
| 1 | 63 (27) | 49 (26) | 14 (32) | |
| 2 | 10 (4) | 9 (5) | 1 (2) | |
| 3 | 6 (3) | 5 (3) | 1 (2) | |
| 4 | 3 (1) | 2 (1) | 1 (2) | |
| WBC, × 109 L−1 | ||||
| < 10 | 165 (72) | 131 (70) | 34 (79) | .15* |
| 10-19.9 | 22 (10) | 15 (8) | 6 (14) | |
| 20-49.9 | 21 (9) | 20 (11) | 1 (2) | |
| 50-99.9 | 16 (7) | 17 (9) | 0 (0) | |
| ≥ 100 | 5 (2) | 3 (2) | 2 (5) | |
| Median (range) | 3 (0.2-195) | 3.3 (0.2-140) | 2.1 (0.6-195) | |
| Platelets, × 109 L−1 | ||||
| < 10 | 14 (6) | 12 (6) | 2 (5) | .15* |
| 10-19.9 | 75 (34) | 62 (33) | 13 (31) | |
| 20-49.9 | 102 (44) | 83 (45) | 19 (45) | |
| 50-99.9 | 23 (10) | 19 (10) | 4 (10) | |
| ≥ 100 | 13 (6) | 9 (5) | 4 (10) | |
| Median (range) | 23 (5-232) | 22 (6-232) | 26 (5-146) | |
WBC (n = 229) and platelet (n = 227) data not available on all patients.
Test for trend.
Prognosis of APL cases stratified by CD95 −1377 genotype
| Outcome . | CD95 −1377 status, events/patients* (%) . | Unadjusted OR/HR (95% CI); P . | Adjusted OR/HR (95% CI); P . | |
|---|---|---|---|---|
| GG . | GA + AA . | |||
| Failure to achieve complete remission | 14/187 (7) | 9/44 (20) | 4.22 (1.41-12.6); .01 | 4.31 (1.40-13.26); .007 |
| Death within 7 d | 5/187 (3) | 3/44 (7) | 3.39 (0.58-20.0); .18 | 2.00 (0.34-11.68); .4 |
| Death within 30 d | 11/187 (6) | 9/44 (20) | 6.02 (1.93-18.7); .002 | 4.82 (1.85-12.55); .0004 |
| Death within 8 wk | 11/187 (6) | 10/44 (23) | 7.26 (2.39-22.9); .0005 | 5.53 (2.19-13.97); .0001 |
| 5-year OS | 46/187 (79) | 17/44 (64) | 2.14 (1.10-4.15); .03 | 2.21 (1.22-4.00); .007 |
| 5-year RFS | 41/173 (79) | 13/35 (66) | 1.92 (0.92-4.02); .10 | 1.92 (1.27-2.90); .03 |
| Death from infection | 12/187 (7) | 8/44 (21) | 5.26 (1.63-16.99); .01 | 4.63 (1.79-11.99); .0006 |
| Outcome . | CD95 −1377 status, events/patients* (%) . | Unadjusted OR/HR (95% CI); P . | Adjusted OR/HR (95% CI); P . | |
|---|---|---|---|---|
| GG . | GA + AA . | |||
| Failure to achieve complete remission | 14/187 (7) | 9/44 (20) | 4.22 (1.41-12.6); .01 | 4.31 (1.40-13.26); .007 |
| Death within 7 d | 5/187 (3) | 3/44 (7) | 3.39 (0.58-20.0); .18 | 2.00 (0.34-11.68); .4 |
| Death within 30 d | 11/187 (6) | 9/44 (20) | 6.02 (1.93-18.7); .002 | 4.82 (1.85-12.55); .0004 |
| Death within 8 wk | 11/187 (6) | 10/44 (23) | 7.26 (2.39-22.9); .0005 | 5.53 (2.19-13.97); .0001 |
| 5-year OS | 46/187 (79) | 17/44 (64) | 2.14 (1.10-4.15); .03 | 2.21 (1.22-4.00); .007 |
| 5-year RFS | 41/173 (79) | 13/35 (66) | 1.92 (0.92-4.02); .10 | 1.92 (1.27-2.90); .03 |
| Death from infection | 12/187 (7) | 8/44 (21) | 5.26 (1.63-16.99); .01 | 4.63 (1.79-11.99); .0006 |
Events refers to failure to achieve CR, deaths, relapse, or infection-related death, depending on the analysis.
Prognosis of APL patients stratified by CD95 genotype. OS (A), RFS (B), and infection-related death (C) in APL cases by CD95 −1377 genotype (GG vs GA + AA). Percentages shown on Kaplan-Meier plot are at 5 years. The analysis for death from infection censors at deaths from other or unknown causes. Obs indicates observed; and Exp, expected.
Prognosis of APL patients stratified by CD95 genotype. OS (A), RFS (B), and infection-related death (C) in APL cases by CD95 −1377 genotype (GG vs GA + AA). Percentages shown on Kaplan-Meier plot are at 5 years. The analysis for death from infection censors at deaths from other or unknown causes. Obs indicates observed; and Exp, expected.
CD95-1377 genotype and infection-related death in APL
There were 62 (27%) APL deaths, and the specific cause was recorded for 54 patients, which included infection (N = 20), cerebral vascular accident (N = 9), recurrent leukemia (N = 9), cardiac failure (N = 5), respiratory failure (N = 3), graft-versus-host disease (N = 3), other cancers (N = 2), resistant disease (N = 1), renal failure (N = 1), and hemorrhage (N = 1). Six patients died of unspecified multiple causes, and the cause of death was not recorded for 2 patients. There were no deaths because of retinoic acid syndrome. However, carriers of the −1377A variant (GA + AA) were significantly more likely to die from infection compared with GG homozygotes (21% vs 7%, HR = 5.26; 95% CI, 1.63-16.99, P = .01; Table 2; Figure 1C).
CD95-1377 genotype and outcome in APL stratified by WBC count
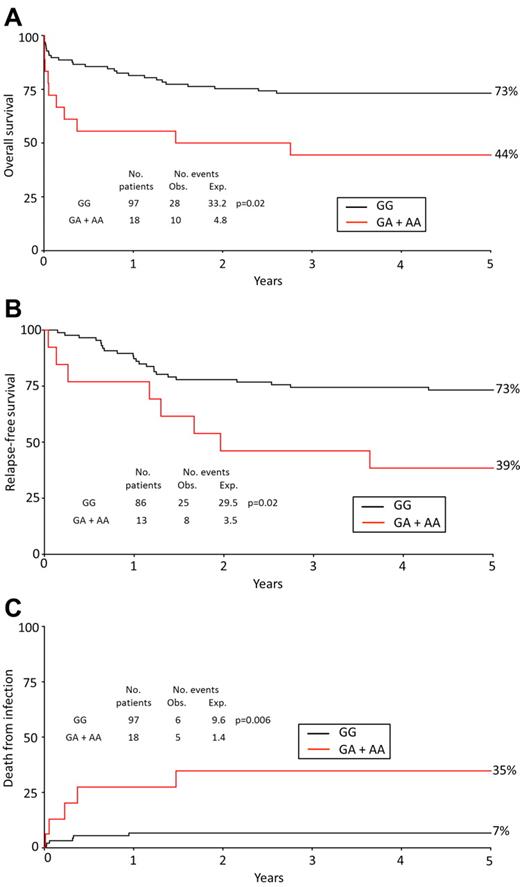
APL patients presented with a median WBC count of 3 × 109 L−1 at diagnosis (data were available for 229 of 231 [99%] patients). In patients with a WBC count ≥ 3 × 109 L−1, carriers of the rare allele had a significantly worse OS (HR = 3.54; 95% CI, 1.35-9.27) and RFS (OR = 4.22; 95% CI, 1.39-12.77) compared with noncarriers (Table 3; Figure 2A-B). Likewise, carriers of the rare allele with a WBC count ≥ 3 × 109 L−1 were significantly more likely to experience a fatal infection (HR = 20.07; 95% CI, 3.37-119.5; Table 3; Figure 2C). By contrast, in patients with a WBC count < 3 × 109 L−1, CD95 −1377 status was not significantly associated with OS, RFS, or infection-related death (Table 3).
Prognosis of APL cases stratified by WBC count at presentation and CD95 genotype
| Stratified at 3 × 109 L−1 . | CD95 −1377 status (*Events/Patients [%]) . | H.R. (95% CI) . | |
|---|---|---|---|
| GG . | GA + AA . | ||
| Overall Survival (5-y OS) | |||
| WBC count < 3 × 109 L−1 | 18/89 (84) | 6/25 (80) | 1.36 (0.50-3.70) |
| WBC count ≥ 3 × 109 L−1 | 28/97 (73) | 10/18 (44) | 3.54 (1.35-9.27) |
| Test for heterogeneity between subgroups: χ21 = 1.8; P = 0.18 | |||
| Relapse-free survival (5-y RFS) | |||
| WBC count < 3 × 109 L−1 | 16/86 (84) | 5/22 (82) | 1.31 (0.45-3.86) |
| WBC count ≥ 3 × 109 L−1 | 25/86 (73) | 8/13 (39) | 4.22 (1.39-12.77) |
| Test for heterogeneity between subgroups: χ21 = 2.2; P = 0.14 | |||
| Death from infection (5-y incidence) | |||
| WBC count < 3 × 109 L−1 | 6/89 (7) | 3/25 (12) | 2.18 (0.43-1.10) |
| WBC count ≥ 3 × 109 L−1 | 6/97 (7) | 5/18 (35) | 20.07 (3.37-119.5) |
| Test for heterogeneity between subgroups: χ21 = 3.3; P = 0.07 | |||
| Stratified at 10 × 109 L−1 | |||
| Overall Survival (5-y OS) | |||
| WBC count < 10 × 109 L−1 | 27/131 (17) | 9/34 (23) | 1.49 (0.65-3.43) |
| WBC count ≥ 10 × 109 L−1 | 19/55 (67) | 7/9 (22) | 7.08 (1.93-25.59) |
| Test for heterogeneity between subgroups: χ21 = 3.9; P = 0.05 | |||
| Disease-free survival (5-y RFS) | |||
| WBC count < 10 × 109 L−1 | 26/125 (82) | 10/30 (30) | 1.99 (0.85-4.69) |
| WBC count ≥ 10 × 109 L−1 | 15/47 (68) | 3/5 (40) | 2.96 (0.56-15.79) |
| Test for heterogeneity between subgroups: χ21 = 0.2; P = 0.7 | |||
| Death from infection (5-y incidence) | |||
| WBC count < 10 × 109 L−1 | 7/131 (5) | 4/34 (13) | 3.08 (0.68-13.88) |
| WBC count ≥ 10 × 109 L−1 | 5/55 (9) | 4/9 (44) | 7.23 (2.10-24.87) |
| Test for heterogeneity between subgroups: χ21 = 3.8; P = 0.05 | |||
| Stratified at 3 × 109 L−1 . | CD95 −1377 status (*Events/Patients [%]) . | H.R. (95% CI) . | |
|---|---|---|---|
| GG . | GA + AA . | ||
| Overall Survival (5-y OS) | |||
| WBC count < 3 × 109 L−1 | 18/89 (84) | 6/25 (80) | 1.36 (0.50-3.70) |
| WBC count ≥ 3 × 109 L−1 | 28/97 (73) | 10/18 (44) | 3.54 (1.35-9.27) |
| Test for heterogeneity between subgroups: χ21 = 1.8; P = 0.18 | |||
| Relapse-free survival (5-y RFS) | |||
| WBC count < 3 × 109 L−1 | 16/86 (84) | 5/22 (82) | 1.31 (0.45-3.86) |
| WBC count ≥ 3 × 109 L−1 | 25/86 (73) | 8/13 (39) | 4.22 (1.39-12.77) |
| Test for heterogeneity between subgroups: χ21 = 2.2; P = 0.14 | |||
| Death from infection (5-y incidence) | |||
| WBC count < 3 × 109 L−1 | 6/89 (7) | 3/25 (12) | 2.18 (0.43-1.10) |
| WBC count ≥ 3 × 109 L−1 | 6/97 (7) | 5/18 (35) | 20.07 (3.37-119.5) |
| Test for heterogeneity between subgroups: χ21 = 3.3; P = 0.07 | |||
| Stratified at 10 × 109 L−1 | |||
| Overall Survival (5-y OS) | |||
| WBC count < 10 × 109 L−1 | 27/131 (17) | 9/34 (23) | 1.49 (0.65-3.43) |
| WBC count ≥ 10 × 109 L−1 | 19/55 (67) | 7/9 (22) | 7.08 (1.93-25.59) |
| Test for heterogeneity between subgroups: χ21 = 3.9; P = 0.05 | |||
| Disease-free survival (5-y RFS) | |||
| WBC count < 10 × 109 L−1 | 26/125 (82) | 10/30 (30) | 1.99 (0.85-4.69) |
| WBC count ≥ 10 × 109 L−1 | 15/47 (68) | 3/5 (40) | 2.96 (0.56-15.79) |
| Test for heterogeneity between subgroups: χ21 = 0.2; P = 0.7 | |||
| Death from infection (5-y incidence) | |||
| WBC count < 10 × 109 L−1 | 7/131 (5) | 4/34 (13) | 3.08 (0.68-13.88) |
| WBC count ≥ 10 × 109 L−1 | 5/55 (9) | 4/9 (44) | 7.23 (2.10-24.87) |
| Test for heterogeneity between subgroups: χ21 = 3.8; P = 0.05 | |||
Events refers to failure to achieve deaths, relapses/deaths in CR or infection-related death, depending on the analysis.
H.R. indicates hazard ratio; 95% CI, 95% confidence interval; APL, acute promyelocytic leukemia; and WBC, white blood cell.
Prognosis of APL patients with WBC ≥ 3 × 109/L stratified by CD95 genotype. OS (A), RFS (B), and death from infection (C) in APL cases with a WBC count ≥ 3 × 109 L−1 by CD95 −1377 genotype (GG vs GA + AA). Percentages shown on Kaplan-Meier plot are at 5 years. The analysis for death from infection censors at deaths from other or unknown causes. Obs indicates observed; and Exp, expected.
Prognosis of APL patients with WBC ≥ 3 × 109/L stratified by CD95 genotype. OS (A), RFS (B), and death from infection (C) in APL cases with a WBC count ≥ 3 × 109 L−1 by CD95 −1377 genotype (GG vs GA + AA). Percentages shown on Kaplan-Meier plot are at 5 years. The analysis for death from infection censors at deaths from other or unknown causes. Obs indicates observed; and Exp, expected.
Sixty-four patients (28%) had a median WBC count ≥ 10 × 109 L−1 at diagnosis. In this group, carriers had a poorer OS (HR = 7.08; 95% CI, 1.93-25.59) and were significantly more likely to experience a fatal infection compared with noncarriers (4 of 9 [44%] carriers; 5 of 55 [9%] noncarriers; HR = 7.23; 95% CI, 2.10-24.87; Table 3).
Of the total 231 APL patients recruited to this study, only 3 patients were AA homozygotes, consistent with the low frequency of AA homozygosity (2%) reported in European Caucasians.10 All 3 AA homozygotes had a low WBC count at presentation (< 3 × 109 L−1), and all achieved CR and were alive at last follow-up (mean follow-up time 3608 days). Because all 3 patients had a low WBC count, these data do not inform on whether the risk of poor outcome is affected by allele dosage.
SP1 binding to the CD95 promoter consensus sequence containing −1377G/A
Nucleotide position −1377 of the CD95 promoter lies within a SP1 transcription factor consensus binding site (5′GGCACGCCCA3′; position −1377 is underlined). SP1 from HL60 AML and NB4 APL cells has a significantly greater binding affinity for the −1377G allele compared with the −1377A allele (P = .019 for HL60 and P = .04 for NB4; Figure 3A-B). The signal produced by SP1 can be supershifted by the addition of anti–SP1 antibody (Figure 3A), confirming the identity of this protein.
The −1377 G > A variant affects SP1 binding and transcriptional activity of the CD95 promoter. (A-B) EMSA illustrating binding of nuclear proteins to the polymorphic site in the CD95 core promoter. (A) 32P-labeled CD95 −1377 DNA probe (G allele, lanes 1, 2, 5, 6, 9 to 11; A allele, lanes 3, 4, 7, and 8) was incubated with 15 μg HL60 or NB4 nuclear extract (lanes 2, 4, 6, 8, and 11). Incubation with anti–human SP1 antibody (lane 10) generated a supershifted complex confirming the identity of the DNA binding protein as SP1. SP1 binding was quantified using densitometry and is shown in panel B. Error bars represent the mean ± SEM from 3 independent experiments, and statistical analysis was performed using the unpaired Student t test for SP1 binding. (C-D) siRNA-mediated knockdown of SP1 gives rise to a concomitant reduction in CD95 protein expression. RNA interference of SP1 in HL60 or NB4 cells was performed using an siRNA specific to SP1 (sc-29487, Santa Cruz Biotechnology). Cells were electroporated with SP1 or MLL-AF4 siRNA (negative control), and the degree and specificity of SP1 knockdown and effect on CD95 protein expression were assessed by immunoblot analysis 8 hours after transfection (C). (D) The results of quantification using densitometry. Results were normalized against β-actin levels. Error bars represent the mean ± SEM from 3 independent experiments, and statistical analysis was performed using the unpaired Student t test. (E) Reporter gene assay illustrating the effect of the −1377 variant on CD95 promoter activity. Promoter activity was analyzed using the Dual-Luciferase Reporter assay system, with reporter vector luminescence (firefly [FF]) normalized against control luminescence (TK-renilla [Rh]). Luciferase activity was assessed from 6 independent clones for each −1377 variant. Error bars represent the mean ± SEM from 6 independent experiments. Statistical analysis was performed using the unpaired Student t test.
The −1377 G > A variant affects SP1 binding and transcriptional activity of the CD95 promoter. (A-B) EMSA illustrating binding of nuclear proteins to the polymorphic site in the CD95 core promoter. (A) 32P-labeled CD95 −1377 DNA probe (G allele, lanes 1, 2, 5, 6, 9 to 11; A allele, lanes 3, 4, 7, and 8) was incubated with 15 μg HL60 or NB4 nuclear extract (lanes 2, 4, 6, 8, and 11). Incubation with anti–human SP1 antibody (lane 10) generated a supershifted complex confirming the identity of the DNA binding protein as SP1. SP1 binding was quantified using densitometry and is shown in panel B. Error bars represent the mean ± SEM from 3 independent experiments, and statistical analysis was performed using the unpaired Student t test for SP1 binding. (C-D) siRNA-mediated knockdown of SP1 gives rise to a concomitant reduction in CD95 protein expression. RNA interference of SP1 in HL60 or NB4 cells was performed using an siRNA specific to SP1 (sc-29487, Santa Cruz Biotechnology). Cells were electroporated with SP1 or MLL-AF4 siRNA (negative control), and the degree and specificity of SP1 knockdown and effect on CD95 protein expression were assessed by immunoblot analysis 8 hours after transfection (C). (D) The results of quantification using densitometry. Results were normalized against β-actin levels. Error bars represent the mean ± SEM from 3 independent experiments, and statistical analysis was performed using the unpaired Student t test. (E) Reporter gene assay illustrating the effect of the −1377 variant on CD95 promoter activity. Promoter activity was analyzed using the Dual-Luciferase Reporter assay system, with reporter vector luminescence (firefly [FF]) normalized against control luminescence (TK-renilla [Rh]). Luciferase activity was assessed from 6 independent clones for each −1377 variant. Error bars represent the mean ± SEM from 6 independent experiments. Statistical analysis was performed using the unpaired Student t test.
We next demonstrated that siRNA-mediated knockdown of SP1 message in HL60 and NB4 cells gives rise to a concomitant decrease in both SP1 (HL60, P < .001; NB4, P < .001), and CD95 protein levels assessed by Western analysis (HL60, P < .001; NB4, P < .01; Figure 3C-D). These data provide further evidence that SP1 is a critical factor for CD95 expression and identify the mechanism by which the −1377 promoter polymorphism affects transcriptional activity and CD95 expression levels.
Functional activity of CD95-1377 promoter variant
We next determined whether the −1377 variant affected CD95 expression. Leukemic blast cells from APL patients homozygous for the G allele had higher mean expression of CD95 compared with carriers of the A variant at position −1377 measured by quantitative PCR, but the difference was not statistically significant (P = .424; n = 89; data not shown). However, because this approach is significantly compromised by genetic heterogeneity between patients, we tested transcriptional activity of the variant CD95 promoter in a fully isogenic reporter gene system. Consistent with the data from primary leukemic cells, the −1377G variant exhibited significantly higher basal transcriptional activity compared with the −1377A variant (P = .047, Figure 3E), confirming the functionality of the −1377 CD95 variant.
Retinoic acid-mediated differentiation down-regulates SP1 and CD95
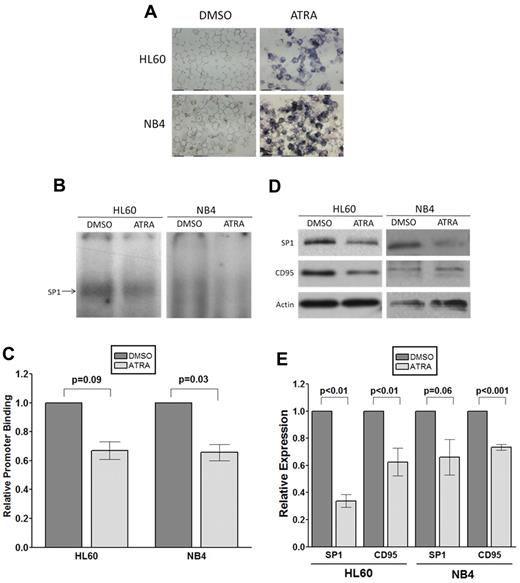
We next investigated possible reasons for the specific association between the CD95 variant and poor outcome in APL. Because ATRA is used as specific therapy for APL, we treated HL60 and NB4 cells with this agent to determine the effects on SP1 and CD95 expression. Exposure to 10μM ATRA resulted in differentiation of both HL60 and NB4, confirmed by the NBT assay (Figure 4A) and induced a reduction in SP1 binding to the CD95 promoter sequence (Figure 4B-C) and SP1 protein levels (Figure 4D-E). Consistent with a role for SP1 as a transcriptional activator of CD95, there was also a concomitant down-regulation of CD95 expression in both HL60 and NB4, although this was relatively modest (Figure 4D-E). Although these data demonstrate that ATRA-mediated differentiation affects both SP1 and CD95 expression, there is no discernible difference in the SP1/CD95 response of NB4 APL cells and HL60 non-APL cells.
ATRA-induced differentiation of AML cells downregulates SP1 promoter binding and CD95 expression. (A) Assessment of HL60 and NB4 ATRA-induced differentiation by NBT reduction assay. Cells were treated with 10μM ATRA or vehicle only (DMSO) for 48 hours. After treatment, cells were resuspended in 3 × 10−7M phorbol 12-myristate 13-acetate diluted in 1 mg/mL NBT solution. Cytospins were prepared from 100 μL of each cell suspension, with blue staining in ATRA-treated cells indicative of phagocytic capability and therefore confirming differentiation. (B-C) EMSA illustrating down-regulation of SP1 binding to the polymorphic site in the CD95 core promoter after ATRA treatment. (B) 32P-labeled CD95 −1377 DNA probe (G allele, all lanes) was incubated with 15 μg HL60 or NB4 nuclear extract from cells treated with 10μM ATRA or vehicle alone (DMSO). (C) SP1 binding was quantified using densitometry. Error bars represent the mean ± SEM from 3 independent experiments, and statistical analysis was performed using the unpaired Student t test. (D-E) ATRA-induced differentiation of HL60 and NB4 down-regulates SP1 and CD95 expression. HL60 or NB4 cells were incubated with 10μM ATRA or vehicle-alone (DMSO). Down-regulation of SP1 and CD95 protein expression was confirmed by immunoblot analysis (D) and quantified using densitometry (E). Error bars represent the mean ± SEM from at least 3 independent experiments, and statistical analysis was performed using the unpaired Student t test (E).
ATRA-induced differentiation of AML cells downregulates SP1 promoter binding and CD95 expression. (A) Assessment of HL60 and NB4 ATRA-induced differentiation by NBT reduction assay. Cells were treated with 10μM ATRA or vehicle only (DMSO) for 48 hours. After treatment, cells were resuspended in 3 × 10−7M phorbol 12-myristate 13-acetate diluted in 1 mg/mL NBT solution. Cytospins were prepared from 100 μL of each cell suspension, with blue staining in ATRA-treated cells indicative of phagocytic capability and therefore confirming differentiation. (B-C) EMSA illustrating down-regulation of SP1 binding to the polymorphic site in the CD95 core promoter after ATRA treatment. (B) 32P-labeled CD95 −1377 DNA probe (G allele, all lanes) was incubated with 15 μg HL60 or NB4 nuclear extract from cells treated with 10μM ATRA or vehicle alone (DMSO). (C) SP1 binding was quantified using densitometry. Error bars represent the mean ± SEM from 3 independent experiments, and statistical analysis was performed using the unpaired Student t test. (D-E) ATRA-induced differentiation of HL60 and NB4 down-regulates SP1 and CD95 expression. HL60 or NB4 cells were incubated with 10μM ATRA or vehicle-alone (DMSO). Down-regulation of SP1 and CD95 protein expression was confirmed by immunoblot analysis (D) and quantified using densitometry (E). Error bars represent the mean ± SEM from at least 3 independent experiments, and statistical analysis was performed using the unpaired Student t test (E).
We next tested whether penetrance of the CD95 −1377 variant for poor outcome was related to specific treatment protocol and performed survival analysis stratified by remission induction regimen (ADE, N = 100; MAE, N = 64; DAT, N = 48; or AIDA, N = 19) and also by length of ATRA schedule: short (N = 38) or extended (N = 185). There was no evidence that anthracycline-based induction regimen affected CR rate (test for heterogeneity between treatment subgroups, P = 1.0, supplemental Table 3). However, penetrance of the variant allele for remission induction failure was marginally higher in patients who received the short ATRA protocol compared with those who received the extended ATRA protocol, but the difference was not statistically significant (test for heterogeneity between ATRA treatment subgroups, P = .06, supplemental Table 3). These data should be interpreted with caution because of the low numbers in this subgroup analysis.
CD95-mediated cell death signaling is compromised in PML/RARA-positive NB4 cells
We next looked for evidence that the association between the CD95 promoter variant and outcome in APL specifically could be related to the PML-RARA fusion gene. PML is essential for cell death signaling via the CD95 pathway.15 However, the PML-RARA fusion protein significantly attenuates CD95-mediated cell death signaling when ectopically expressed in myeloid bone marrow cells of transgenic mice.15 To investigate whether human cells expressing the PML-RARA fusion gene also have limited capacity for executing cell death via the CD95 pathway, we treated AML cells with CD95 ligand (CD95L) and monitored caspase-8 and caspase-3 cleavage as markers of cell death activation. Three cell lines negative for the PML-RARA fusion (AML-2, AML-3, and Jurkat) all showed increased caspase-8 cleavage in response to CD95L (Figure 5). Caspase-8 processing was accompanied by downstream caspase-3 cleavage (Figure 5) in all 3 cell lines. In contrast, there was only very limited caspase-8 cleavage or caspase-3 cleavage in NB4 APL cells in response to CD95L (Figure 5), showing that NB4 APL cells have compromised capacity for death signaling via this pathway. These observations are consistent with data recently published by Tao et al, who demonstrated using isogenic systems that PML-RARA has a dominant negative effect on wild-type PML function and inhibits CD95-mediated apoptosis in primary human APL cells, cell lines and mice.16 HL60 myelomonocytic cells were also relatively unresponsive to CD95L (Figure 5), consistent with previous reports documenting this phenotype in HL60, despite surface expression of CD95.17
CD95 ligand-induced caspase-8 and caspase-3 cleavage. PML-RARA-positive cells (NB4, lanes 1-3), and PML-RARA-negative cells (AML-2, AML-3, HL60, and Jurkat, lanes 4-15) were cultured for 24 hours in media supplemented with 0, 50, or 100 ng/mL FAS ligand. Caspase-8 activation (top panels) and capase-3 activation (middle panels) were assessed by monitoring the formation of cleaved product (p41/43 and p18 for caspase-8 and p19/17 for caspase-3) using immunblot analysis.
CD95 ligand-induced caspase-8 and caspase-3 cleavage. PML-RARA-positive cells (NB4, lanes 1-3), and PML-RARA-negative cells (AML-2, AML-3, HL60, and Jurkat, lanes 4-15) were cultured for 24 hours in media supplemented with 0, 50, or 100 ng/mL FAS ligand. Caspase-8 activation (top panels) and capase-3 activation (middle panels) were assessed by monitoring the formation of cleaved product (p41/43 and p18 for caspase-8 and p19/17 for caspase-3) using immunblot analysis.
Given that PML-RARA inhibits CD95-mediated apoptosis, we next investigated whether penetrance of the CD95 −1377 variant for poor outcome was affected by PML-RARA isoform type. PML breakpoint data were available for 150 APL patients; and for analytical purposes, patients with a PML intron 6 breakpoint (BCR-1, long isoform) and exon 6 breakpoint (BCR-2, variable isoform) were grouped (n = 98) and compared against those with an intron 3 (BCR-3, short isoform) breakpoint (n = 52). Carriers of the CD95 variant allele were less likely to achieve CR than noncarriers, but this difference was greater in those patients with the BCR-3 breakpoint where 3 of 7 carriers failed to achieve CR compared with only 2 of 45 noncarriers (test for heterogeneity between BCR subgroups P = .03; supplemental Table 3). These data should be treated with caution given the low numbers in the subgroups but nevertheless suggest that penetrance of the CD95 variant for poor outcome is greater in APL patients with a PML BCR-3 breakpoint.
Discussion
We identified a common genetic variant in the core promoter of the CD95 cell death receptor gene as a strong predictor of OS in APL. Particularly striking was the association with risk of death during the first 8 weeks after diagnosis, which in our study occurred predominantly because of infection. Evidence suggests that poor outcome in carriers was the result of poor response to induction therapy, as indicated by a strong association between the variant allele and failure to achieve CR. We have also shown that the CD95 core promoter is bound by the SP1 transcription factor at the site of the polymorphism, and that SP1 from both APL and non-APL cells binds with lower affinity to the poor prognosis promoter variant. We have also demonstrated that targeted knockdown of SP1 down-regulates expression of CD95 protein. These data provide evidence that the poor risk variant allele gives rise to lower CD95 expression relative to the common allelic variant, a conclusion that is supported by reporter gene analysis.
CD95 is a potent mediator of cell death, which functions as part of the extrinsic apoptosis pathway in response to genotoxic stress. Apoptosis induced by cytotoxic anthracyclines used to induce remission in AML occurs via CD95 signaling.9 Defective CD95 signaling attenuates apoptosis after treatment with chemotherapy and radiotherapy and is an important cause of cancer cell resistance to therapy.18,19 Moreover, CD95 expression is inversely correlated with chemosensitivity in leukemia cell lines,20 as is FAS-associated death domain (downstream of CD95) expression in primary leukemic blast cells from AML patients.21 As such, compromised CD95 expression, such as that associated with the variant CD95 allele, confers an antiapoptotic phenotype on leukemic blast cells and is predicted to increase risk of remission failure and poor clinical response.
Although consistent with our data, this simple model is not sufficient to explain the association between CD95 genotype and poor outcome in APL specifically, and the lack of a prognostic effect in non-APL AML, both in adults (this study) and children.22 However, evidence demonstrating that the PML-RARA fusion oncoprotein modulates CD95 signaling suggests a possible explanation for the specific association with APL. Using isogenic cells engineered to express PML-RARA, Tao et al demonstrated that the fusion protein inhibits CD95-mediated death signaling in APL cell lines, primary human APL cells, and mice.16 PML-RARA binds to CD95 and recruits the caspase-8 inhibitor c-FLIP, preventing downstream signaling via caspase-8 activation. In addition, PML-RARA also interferes with binding of CD95L by interacting with the death domain of CD95.16 We have extended these observations by showing that caspase-3 (downstream of caspase-8) is not activated in PML-RARA–positive NB4 cells after exposure to CD95L. Taken together, the molecular and cellular data suggest a model where low CD95 expression conferred by the −1377 promoter variant further compromises chemotherapy-induced death signaling via this pathway in APL blast cells already significantly compromised by virtue of the inhibitory effect of the PML-RARA fusion protein on death signaling. This model is predicted to confer a high risk of remission induction failure and poor outcome on carriers of the variant CD95 allele. Poor response to induction therapy is associated with a high risk of life-threatening complications, such as infection, and is consistent with the data reported in our study.
It remains to be determined whether the dominant negative effect of PML-RARA on CD95 signaling is affected by PML-RARA isoform type. Our data provide some evidence that penetrance of the CD95 variant allele for remission induction failure is strongest in APL expressing the short isoform associated with BCR-3 PML breakpoints. However, the weak association seen in our data could be related to a high median WBC count associated with this isoform,23-25 which our analysis suggests also increases penetrance of the variant CD95 allele for poor outcome.
We have shown that ATRA treatment of HL60 and NB4 gives rise to a modest reduction in CD95 expression, which is consistent with published data demonstrating a similar reduction in CD95 expression in HL60 and U937 myeloid cells after retinoid therapy.26 ATRA-induced differentiation also down-regulates CD95 expression in primary blasts from APL patients.26 Concomitant down-regulation of SP1 expression and reduced SP1 binding to the site of the −1377 polymorphism in the CD95 promoter, as demonstrated in our study, identify one mechanism by which CD95 expression is regulated in response to retinoid therapy. Loss of surface CD95 expression induced by differentiation therapy is predicted to render AML cells less sensitive to chemotherapy induced death signaling, and this phenotype has been demonstrated in HL60 and U937.26,27 In contrast, however, differentiation of NB4 PML-RARA–positive APL cells sensitizes to CD95-mediated death signaling.16 As an explanation for these apparently contradictory observations, Tao et al have recently demonstrated that destruction of the PML-RARA fusion protein after arsenic trioxide-driven differentiation releases the block on CD95 function and reinstates functional death signaling in PML-RARA-expressing mouse cells.16 Presumably, ATRA-mediated destruction of PML-RARA also restores CD95 death signaling via the same mechanism, rendering leukemic cells more sensitive to cytotoxic chemotherapy, despite a modest reduction in CD95 expression. Consistent with this model, Burnett et al reported a significantly superior outcome in APL patients when ATRA was administered concurrently with cytotoxic chemotherapy until CR was achieved (extended schedule) compared with a short course of ATRA before commencement of cytotoxic therapy.2 If high penetrance of the variant CD95 allele for poor outcome is dependent on a background of compromised death signaling, then restoration of death signaling in APL cells via extended differentiation therapy is predicted to attenuate penetrance of the CD95 variant allele for adverse events. Consistent with this model, our data show that penetrance of the risk allele was lower in patients treated with extended ATRA given concurrently with cytotoxic chemotherapy compared with patients who received a curtailed course of ATRA before cytotoxic chemotherapy. However, the limited statistical power of these subgroup analyses warrants caution, and investigation in larger prospective studies is suggested.
There was a high proportion of fatal infections in our study (37% of all deaths), and CD95 genotype was significantly associated with risk of infection-related death. The low number of deaths from hemorrhage or retinoic acid syndrome does not allow us to determine whether CD95 genotype associates with risk of death via these mechanisms. However, there was some evidence of interaction between CD95 genotype and WBC count. High WBC count is an independent poor prognostic factor in APL2,28 ; and although advances in supportive care have led to significant improvements in outcome, the risk of induction failure and relapse remains a particular problem in this group of patients.29 Elderly patients diagnosed with APL also have a relatively poor outcome. In a multicenter study of newly diagnosed cases, Ades et al reported a significantly higher frequency of death during maintenance therapy in patients older than 60 years (18.6%) compared with younger patients (5.7%), with infection the major cause of death.30 The median age at diagnosis of APL in our study was 38 years, with only 8 patients older than 60 years. As such, we were unable to investigate the prognostic value of CD95 genotype in this particularly vulnerable age group. Nevertheless, the −1377 CD95 promoter variant might prove a valuable prognostic marker in elderly and other APL subgroups where the risk of a potentially fatal infection or other life-threatening events is elevated. Understanding the role of CD95 in mediating risk of infection is particularly important in those patients given intensive chemotherapy and who are more likely to experience bone marrow hypocellularity and concomitant neutropenia. Designing therapeutic strategies to reduce chemotherapy-induced mortality and morbidity in susceptible patient groups is a high priority. To this end, nonmyeloablative molecularly targeted therapeutic regimens have been developed for APL, and evidence suggests that remission can be achieved without the need for genotoxic chemotherapy.31,32 As such, it will be interesting to determine whether CD95 genotype retains its prognostic significance in APL patients treated without cytotoxic chemotherapy.
The available evidence suggests a model where low CD95 expression conferred by the promoter variant further attenuates CD95 death signaling in APL leukemic blast cells already compromised because of the dominant negative effect of the PML-RARA fusion protein. However, we cannot exclude other mechanisms from also contributing to the poor outcome in carriers of the rare allele, particularly because our data identify a strong association with infective death and that CD95 functions as regulator of T- and B-cell homeostasis as part of the adaptive immune response,5,8,33 and also plays a role in innate immunity.6
CD95 genotype might also be prognostically relevant in other cancers. Although the PML-RARA fusion gene is specific to t(15;17) APL, loss or reduction of wild-type PML function has a similar inhibitory effect of CD95 death signaling.16 PML expression is ubiquitous in normal tissues, but reduced or complete loss of expression occurs with high frequency in numerous hematologic and epithelial cancers, such as prostate adenocarcinoma, breast carcinoma, and lymphoma.34 The observed lack of caspase cleavage in HL60 cells after CD95L exposure warrants investigation of PML function in these cells. Low CD95 expression associated with the −1377 promoter variant could further compromise death signaling in PML-deficient cancer cells, conferring a chemoresistant phenotype.
In conclusion, our data identify the −1377 promoter variant in the CD95 cell death receptor as a major determinant of poor outcome in APL. The risk allele is carried by 20% of European Caucasians and could be used to highlight vulnerable patients at high risk of infection or who might particularly benefit from chemotherapeutic regimens designed to maximally promote CD95 death signaling in APL blast cells. Identifying and protecting patients at high risk of life-threatening events, such as remission induction failure and infection, is a high priority in APL,35 especially because such events still represent a major cause of death in APL despite the introduction of differentiation therapy.3
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank contributing patients and the MRC/NCRI Adult Leukaemia Working Party for their support of this study and the clinical investigators who entered and managed patients in clinical trials.
This work was supported by specialist programme funding from Leukaemia & Lymphoma Research (United Kingdom; grants 06002 and 11006, J.M.A.). D.G. was supported by Leukaemia & Lymphoma Research and the Guy's and St Thomas' Hospital Charity.
Authorship
Contribution: N.J.S., K.S., S.T., S.E.F., V.J.F., and L.J.W. performed research; R.H. analyzed data and interpreted results; D.G., G. Jackson, G. Jones, and S.B. contributed reagents and interpreted data; J.M.A. designed the study, performed research, interpreted data, and wrote the paper; and all authors contributed to the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: James M. Allan, Northern Institute for Cancer Research, Medical School, Framlington Place, Newcastle University, Newcastle upon Tyne, NE2 4HH, United Kingdom; e-mail: james.allan@newcastle.ac.uk.
References
Author notes
N.J.S. and K.S. contributed equally to this study.

![Figure 3. The −1377 G > A variant affects SP1 binding and transcriptional activity of the CD95 promoter. (A-B) EMSA illustrating binding of nuclear proteins to the polymorphic site in the CD95 core promoter. (A) 32P-labeled CD95 −1377 DNA probe (G allele, lanes 1, 2, 5, 6, 9 to 11; A allele, lanes 3, 4, 7, and 8) was incubated with 15 μg HL60 or NB4 nuclear extract (lanes 2, 4, 6, 8, and 11). Incubation with anti–human SP1 antibody (lane 10) generated a supershifted complex confirming the identity of the DNA binding protein as SP1. SP1 binding was quantified using densitometry and is shown in panel B. Error bars represent the mean ± SEM from 3 independent experiments, and statistical analysis was performed using the unpaired Student t test for SP1 binding. (C-D) siRNA-mediated knockdown of SP1 gives rise to a concomitant reduction in CD95 protein expression. RNA interference of SP1 in HL60 or NB4 cells was performed using an siRNA specific to SP1 (sc-29487, Santa Cruz Biotechnology). Cells were electroporated with SP1 or MLL-AF4 siRNA (negative control), and the degree and specificity of SP1 knockdown and effect on CD95 protein expression were assessed by immunoblot analysis 8 hours after transfection (C). (D) The results of quantification using densitometry. Results were normalized against β-actin levels. Error bars represent the mean ± SEM from 3 independent experiments, and statistical analysis was performed using the unpaired Student t test. (E) Reporter gene assay illustrating the effect of the −1377 variant on CD95 promoter activity. Promoter activity was analyzed using the Dual-Luciferase Reporter assay system, with reporter vector luminescence (firefly [FF]) normalized against control luminescence (TK-renilla [Rh]). Luciferase activity was assessed from 6 independent clones for each −1377 variant. Error bars represent the mean ± SEM from 6 independent experiments. Statistical analysis was performed using the unpaired Student t test.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/119/1/10.1182_blood-2011-04-349803/4/m_zh89991184210003.jpeg?Expires=1765908454&Signature=SdE3LAYnYHhjxbAKXllLcktRstb-XGO4yXl6C~jAb8~JRRdy7gW~vPobZPm4s0r433SXKuXQWFZ5OfbGJzUbtJPta~9NXisRI9-LDswKp2jw-brdLK84VdWTXQOccYNU0wuJuLVpGlIga4qcrp0R8di1v8fucT6UM7T92yfoq3mi0sQa8TfvrNJKWoTKpEcf6ahtvUm0vQHfCpzX8m6WS8RfL210H3F7v04slPf~ZP4-H5iUwtv1WHWlS-eWW1PfyfK1RGu12ObJKnVbHU5vOMbX0g07smKsRxjP3k41HVgx8ApmMApfLkGYrT9ovQYiRhve8jst-riHew6J0FetYg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal