Abstract
The differentiation of HSCs into myeloid lineages requires the transcription factor PU.1. Whereas PU.1-dependent induction of myeloid-specific target genes has been intensively studied, negative regulation of stem cell or alternate lineage programs remains incompletely characterized. To test for such negative regulatory events, we searched for PU.1-controlled microRNAs (miRs) by expression profiling using a PU.1-inducible myeloid progenitor cell line model. We provide evidence that PU.1 directly controls expression of at least 4 of these miRs (miR-146a, miR-342, miR-338, and miR-155) through temporally dynamic occupation of binding sites within regulatory chromatin regions adjacent to their genomic coding loci. Ectopic expression of the most robustly induced PU.1 target miR, miR-146a, directed the selective differentiation of HSCs into functional peritoneal macrophages in mouse transplantation assays. In agreement with this observation, disruption of Dicer expression or specific antagonization of miR-146a function inhibited the formation of macrophages during early zebrafish (Danio rerio) development. In the present study, we describe a PU.1-orchestrated miR program that mediates key functions of PU.1 during myeloid differentiation.
Introduction
Transcription factors play a decisive role in determining lineage fate from HSCs. PU.1 is crucial for lymphomyeloid development, and its stage-specific expression is critical in preventing leukemic transformation.1,2 Whereas the differentiating effect of PU.1 in stem cells is attributed to target gene induction, targets of negative regulation have rarely been described.3 The best-studied example of this type of negative regulation is the suppression of GATA1 activity by PU.1, which leads to a shift in cell fate toward the myeloid-lymphoid lineages.4 As a consequence of such balanced interactions, altering the levels of PU.1 expression has profound consequences for progenitor maturation.5,6 However, such direct repression mechanisms are not the only way to down-regulate alternate lineage expression programs. MicroRNAs (miRs) are a class of small, noncoding, regulatory RNAs that repress gene expression posttranscriptionally,7 which may have effects on lineage development.8 Several miRs have been implicated in the control of lineage choice in blood development; for example, miR-150 was shown to direct megakaryocyte-erythroid lineage fate9 and miR-223 was reported to direct granulocytic development,10 although this has been questioned using knockout (KO) studies in mice.11,12 Furthermore, down-regulation of the miR-17p-92 cluster can promote myeloid lineage fate.13 At a more global level, impairing mature miR expression by KO of the processing machinery in Dicer-KO experiments revealed miR dependency for HSCs14 and osteoclast development.15 We therefore investigated whether PU.1 induces the expression of miRs during myeloid differentiation to concert the myeloid lineage specification process. Profiling the early maturation stage of myelopoiesis in a PU.1-inducible cell line (PUER) yielded such miR candidates. Unbiased mapping of PU.1 binding (ChIPseq) and locus accessibility (DNase hypersensitivity) in PUER cells and primary macrophages revealed that the expression of 4 miRs (miR-146a, miR-342, miR-338, and miR-155) is PU.1 dependent during the earliest steps of myeloid progenitor maturation. One of the most prominently induced miRs (miR-146a) was analyzed further.
MiR-146a is a negative regulator of innate immune signaling and can antagonize the proliferation of developing progenitors16,17 ; it has also been reported to block megakaryopoiesis.17-19 However, this finding has recently been challenged by Opalinska et al,20 who did not detect any effects on megakaryocyte development after ectopic expression of miR-146a in a BM transplantation assay.
In the present study, we show that miR-146a is dynamically expressed during adult hematopoiesis, and reveal that PU.1 is required to control this dynamic pattern in vitro and in vivo. We demonstrate that, functionally, miR-146a fulfills a surprisingly selective role during adult myelopoiesis in that its ectopic expression specifically drives the replenishment of peritoneal macrophages from HSCs in vivo. In agreement with this, a knock-down (KD) approach in zebrafish (Danio rerio) revealed a blood-specific requirement of miR-146a for macrophage development in the embryo, which is supported by the observation of diminished macrophage development in a Dicer-KO zebrafish model. These results demonstrate that PU.1 coordinates the expression of a specific miR profile during myeloid progenitor maturation, with miR-146a inducing differentiation and specifying maturation to peritoneal macrophages from stem cells during adult hematopoiesis and initiating macrophage development during embryonic hematopoiesis.
Methods
Cell isolation and antibody staining
BM was prepared from 8- to 12-week-old C57/BL6 mice and minced through a nylon filter (40 μm), generating a single-cell suspension. HSCs, defined as Lin−Sca-1+c-Kit+ (LSK) cells,21 were isolated from the suspension as follows. First, the cells were enriched for c-Kit+ cells by immunomagnetic separation with anti-c-Kit MACS beads (Miltenyi Biotec). Antibody staining for surface markers was performed next using the following fluorescent antibodies: Sca-1 FITC (D7) and lineage PE-Cy5 (a mixture of antibodies against CD3, CD4, CD8, B220, CD19, and Gr1), c-Kit allophycocyanin (2B8). All antibodies were from Becton Dickinson. The cells were then resuspended in PBS plus 2% FCS, with propidium iodide for exclusion of dead cells, and subjected to FACS purification on a FACSAria1 high-speed sorter (Becton Dickinson) and used as indicated. Analysis of hematopoietic and nonhematopoietic tissues was performed by FACS after generating a single-cell suspension from BM, spleen, peripheral lymph nodes, peripheral blood, liver, or brain after mincing harvested organs through a nylon mesh. Lysis of erythrocytes was performed by 5 minutes of incubation in ammonium chloride-potassium chloride buffer. Peritoneal washouts were sampled by killing the animal and instilling 4-5 mL of PBS IP for 10 minutes, and then aspirating the fluid with a 20-gauge needle. Broncho-alveolar lavage fluid was collected after preparation of the lung and instilling 1-2 mL into the trachea for 5 minutes. Washouts processed for FACS were blocked with unlabeled anti-Fc-receptor antibody (Becton Dickinson) and then stained for lineage characterization with the following antibodies: F4/80, Gr-1, CD11b, CD3, CD4, CD8, CD19, B220, CD115, CD68, I-A/I-E (MHC-II), CD45.1, and CD45.2 (all Becton Dickinson). Dead cells were excluded with propidium iodide and analyzed on an LSR II flow cytometer (Becton Dickinson).
Cell culture and transplantation assays
RAW264.7 and PU.1 shRNA-transfected RAW264.7 cells were cultured and Western blot analysis was performed as described previously.22 PUER and PU.1 KO cell lines were a kind gift from H. Singh (University of Chicago). Cell lines were derived from fetal livers of PU.1 KO animals. PUER cells were generated by stable transfection of a tamoxifen-inducible PU.1 construct.6 Induction with 4-hydroxy-tamoxifen (OHT; 100nM) lead to macrophage development (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). For transplantation assays, wild-type (WT) LSK cells were isolated as described previously21 and prestimulated for 1 day in SCF (30 ng/mL), IL6 (10 ng/mL), and leukemia inhibitory factor (10 ng/mL; all from PeproTech) and infected during coincubation with virus-conditioned supernatant (see “MiR-expressing retrovirus”) in the presence of polybrene (8 μg/mL) for 24 hours. In addition, 3000-5000 infected LSK cells were transplanted into lethally irradiated (9.5 Gy) syngenic recipients (SJL/C57/Bl6 CD45.1) with 5 × 105 WT BM cells as radiation support. To analyze phagocytosis in vivo, UV-excitable, polystyrene-based latex beads (BB flouresbrite microparticles; Polysciences) were modified and used.23 In brief, for in vitro studies peritoneal washout cells were incubated in IMDM for 1 hour and then 20 μL/mL of beads were added for 90 minutes. In vivo phagocytosis was measured by instilling a 1:25 diluted bead solution IP 18 hours before collection of peritoneal washout. FACS analysis of bead uptake was performed on a FACSAria 3 sorter. Bead-positive macrophages were FACS isolated from WT treated animals and stained with Wright-Giemsa stain, and were shown to generate phagocytotic vacuoles (data not shown).
MiR-expressing retrovirus
To generate a miR-146a–expressing retroviral construct, the pre-miR sequence ± 200 bp was PCR amplified from murine WT genomic DNA (strain C57/Bl6) and subcloned into the murine MDH-1 vector8 (Addgene plasmid 11379) in exchange for the original miR as follows: forward primer (with PacI restriction site) ccttaattaatttttcacacatcaatccaacatgactc; reverse primer (with XhoI restriction site) ttcctcgagccagcatgtttaatgattgctt. The correct insertion of the pre-miR was confirmed by sequencing. The miR expression plasmids were then cotransfected using calcium-phosphate precipitates (Invitrogen) with gag/pol and eco-env plasmids into HEK293T cells, and virus-containing supernatant was harvested 3 and 4 days later, filtered, and stored at −80°C until use. The generation of the mature miR-146a was confirmed by RT-PCR on RNA from the virus-producing cells using the miRVana quantitative RT-PCR (qRT-PCR) detection kit (Ambion).
MicroRNA profiling and statistical analysis
RNA was isolated from PUER cells at 18 hours solvent control (t0) and induction with OHT at 18 hours (supplemental Figure 1) using the miRVana microRNA isolation kit (Ambion) in triplicate from independent cell samples. In brief, cells were lysed and RNA was extracted with phenol/chloroform and then enriched with micro-columns (miRVana microRNA kit; Ambion). The presence of small RNAs was confirmed on a bioanalyzer (Agilent).
MiR-expression profiling was conducted with the Applied Biosystems real-time PCR assay with 50 ng of total RNA, which allowed measurement of all 189 miRs in miRBase 8.2. Stem-loop primers were used to reverse-transcribe the respective miRs, and real-time PCR was run on an ABI 7900HT Prism (Applied Biosystems; supplemental Table 1). A miR was considered to be present during induction if the cycle threshold value was < 35 in one biologic replicate. The dataset was then normalized to the global mean. The relative expression values were calculated as 2 − δCT in relation to global mean. Differentially expressed miRs early in differentiation were identified by 2 class comparisons, mock treated (t 0) versus treated (18 hours) using the Student t test–based significance analysis of microarrays (SAM) algorithm.24 MiRs were considered to be differentially expressed if the mean measured expression changed at least 2-fold for up-regulated and at least 0.75-fold for down-regulated candidates within 18 hours with a false discovery rate (q) of < 5%. For further miR studies, cells were lysed in TRIzol (Invitrogen) and RNA was extracted with chloroform and precipitated with EtOH. RNA integrity was confirmed by analysis of the absorption spectra, for which ratios of 260/280 nm and 260/230 nm > 1.8 were achieved.
Luciferase reporter assay
A 375-bp genomic fragment located 10 kb upstream of miR-146a was PCR amplified (primer forward: 5′-ctcgaggagccggaatagaaggttcc-3′; primer reverse: 5′-aagcttaatgttaaattgaggtttttggtcg-3′) and subcloned into the pXP2 backbone (American Type Culture Collection no. 37577) via XhoI/HindIII. Mutation of the PU.1-binding motif (ccctgaggaagt to ccctgaccaagt)25 was introduced using the Quickchange IIXL kit (Stratagene). All constructs were confirmed by sequencing. RAW264.7 cells (5 × 105) were chemically (Fugene HD; Roche) transfected with 1 μg of firefly constructs and 10 ng of pRL-CMV (Promega) as a transfection control. Reporter activity was analyzed after 48 hours using the Dual-Luciferase Reporter Assay System (Promega). Alternatively, RAW264.7 cells carrying a stable PU.1 KD construct22 were used.
Zebrafish microinjections, morpholino oligonucleotides, whole-mount in situ hybridization, and FACS analysis
To knockdown miR-146a activity, we used 2 non-overlapping antisense oligonucleotide KD constructs (ASO) targeting the drosha and loopstar regions of the pre–miR-146a (drosha dre-mir-146a: cagacattaaacagattgccagaag, loopstar: dre-mir-146a gagcccatagatgaacttttcatga; Gene Tools). In addition, to control for unspecific effects of the ASOs during hematopoiesis, an anti–miR-430 KD construct (actaccccaacaaatagcacttacc) was used, as described previously.26 ASOs were injected at a working dilution of 50nM into 1- to 2-cell stage embryos, as described previously.27 Zebrafish were staged at 28.5°C, dechlorinated manually after 24-26 hours postfertilization (hpf), and fixed in 4% paraformaldehyde before further processing. All animal experiments were performed in compliance with national and institutional regulations and were approved by the animal care and use committee of the Max Delbrueck Center (Berlin).
The Dicer allele used in this study is dicerHu896.28 To generate maternal and zygotic MZdicer embryos, we performed dicer mutant germ cell transplantations into WT embryos.26 To probe for L-plastin expression, a 800-bp region from zebrafish L-plastin was amplified from WT DNA as follows: forward primer (with Bst1 restriction site): ccttaattaatttttcacacatcaatccaacatgactc, reverse primer (with BamHI restriction site): ccttaattaatttttcacacatcaatccaacatgactc, and cloned into the pGEM-T Easy Vector (Invitrogen), making use of the specified restriction sites. From the linearized vector, a DIG-labeled antisense RNA (mMESSAGE SP6 Kit Sp6; Ambion) was produced, which was used for whole-mount in situ hybridizations.29 Stained embryos were either mounted in 1% methylcellulose and imaged for oversight pictures on a binocular microscope (M205MA; Leica), and recorded using a camera (DFC420C; Leica) and Leica application suite, or for details and quantification were cleared in benzyl:benzoate (2:1) and embedded in Permount. Images were recorded on an inverted microscope (Axioplan; Zeiss) with a 10× objective using a SPOT digital camera (Diagnostic Instruments) and Meta Morph 6.2r6 software (Visitron). Z-stack image files in 8-μm steps were acquired, and the total number of positive cells was manually quantified using MetaMorph software.
To test for lineage specificity of the miR-146a KD, single-cell suspensions were generated from 24- to 26-hpf zebrafish embryos by digestion in 0.05% trypsin for 30 minutes and mechanical disruption on a 70-μm nylon mesh. Erythrocytes and macrophages were identified as described previously.30 The lineage identities of FACS-gated erythrocytes and macrophages were further confirmed by expression of DsRed or green fluorescent protein (GFP) in GATA-1-DsRed or PU.1-GFP transgenic fish, respectively (supplemental Figure 2).31,32
Results
Identification of a PU.1-dependent myeloid microRNA profile
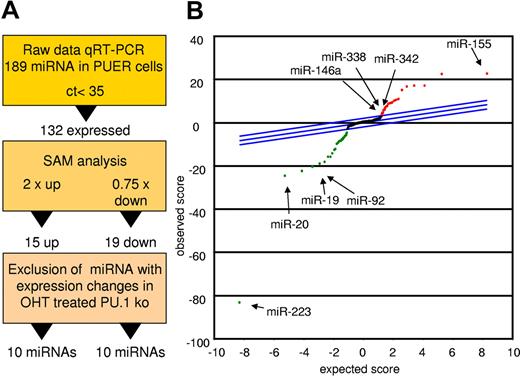
To detect miRs controlled by PU.1 during early myeloid differentiation, we compared mock-induced PUER cells with those receiving 18 hours of OHT stimulation to activate PU.1.33 Figure 1A depicts the strategy for candidate determination. Differentially expressed miRs were identified using the SAM algorithm. Data were controlled for unspecific effects by comparison with OHT-treated PU.1 KO cells lacking the inducible PU.1 construct (Figure 1B and Table 1). Using this strategy, we identified 10 miRs with increased expression and 10 miRs with decreased expression in the induced PUER cells compared with the controls. Among the increased miRs were 6 that had not been attributed to myeloid differentiation before (miR-409, miR-434-5p, miR-096, miR-151, miR-33, and miR-485-5p). However, 4 miRs (miR-146a, miR-342, miR-338, and miR-155) had previously been reported to play roles in myeloid cells, confirming the reliability of our profiling approach. In agreement with this, several of the miRs (eg, miR-223 and the miR-17p-92 cluster) that were down-regulated by PU.1 have been shown to block macrophage differentiation.10,13 Other PU.1–down-regulated miRs have been shown to be oncogenic; for example, miR-196b, miR-25, and miR-19, which was demonstrated to be the key oncogenic miR within the miR-17p-92 cluster.34-36 Our data reveal that PU.1 establishes a selective miR profile that is instructive for macrophage differentiation and inhibitory for leukemic transformation, implicating PU.1 as a master coordinator of myeloid miR expression.
PU.1 induces a specific miR profile. PUER and PU.1 KO cells were treated with solvent or with OHT (100nM) for 18 hours and profiled for miR expression. Monocytic differentiation of PUER was controlled by FACS analysis showing induction of CD11b and by phenotype (supplemental Figure 1). (A) Strategy of PU.1-regulated miR identification. Raw data of the PUER and PU.1 KO cells were normalized to global mean. PU.1-specific candidates were then identified using SAM for PUER cells (18 hours for solvent vs 18 hours for OHT) using a 2-fold change for up-regulated and a 0.75-fold change for down-regulated miRs as a cutoff. To control for unspecific OHT effects on miR expression, those miR that were also differentially expressed in PU.1 KO cells after 18 hours of OHT treatment compared with the solvent control were excluded from further analyses. (B) SAM plot of the miR expression differences between OHT and mock-treated PUER after exclusion of the miR expression changes resulting from OHT treatment of PU.1 KO cells. Selected PU.1-regulated miRs are indicated (arrow); all PU.1-regulated miRs are listed in Table 1. Arrays were performed in biologic triplicate for PUER cells and in duplicate for PU.1 KO cells.
PU.1 induces a specific miR profile. PUER and PU.1 KO cells were treated with solvent or with OHT (100nM) for 18 hours and profiled for miR expression. Monocytic differentiation of PUER was controlled by FACS analysis showing induction of CD11b and by phenotype (supplemental Figure 1). (A) Strategy of PU.1-regulated miR identification. Raw data of the PUER and PU.1 KO cells were normalized to global mean. PU.1-specific candidates were then identified using SAM for PUER cells (18 hours for solvent vs 18 hours for OHT) using a 2-fold change for up-regulated and a 0.75-fold change for down-regulated miRs as a cutoff. To control for unspecific OHT effects on miR expression, those miR that were also differentially expressed in PU.1 KO cells after 18 hours of OHT treatment compared with the solvent control were excluded from further analyses. (B) SAM plot of the miR expression differences between OHT and mock-treated PUER after exclusion of the miR expression changes resulting from OHT treatment of PU.1 KO cells. Selected PU.1-regulated miRs are indicated (arrow); all PU.1-regulated miRs are listed in Table 1. Arrays were performed in biologic triplicate for PUER cells and in duplicate for PU.1 KO cells.
List of PU.1-regulated miRs
| miRNA . | Change at 18 h, -fold . | SAM score . |
|---|---|---|
| Up-regulated | ||
| hsa-miR-146a | 29.55 | 4.40 |
| hsa-miR-342 | 23.09 | 7.01 |
| mmu-miR-409 | 6.73 | 5.84 |
| mmu-miR-434–5p | 5.17 | 7.78 |
| mmu-miR-096 | 2.50 | 16.71 |
| mmu-miR-151 | 2.44 | 9.23 |
| hsa-miR-485–5p | 2.25 | 3.64 |
| hsa-miR-338 | 2.08 | 5.66 |
| mumu-miR-155 | 2.95 | 22.56 |
| hsa-miR-033 | 2.67 | 8.86 |
| Down-regulated | ||
| mmu-miR-203 | 0.34 | −8.45 |
| hsa-miR-223 | 0.52 | −83.19 |
| hsa-miR-196b | 0.52 | −9.19 |
| hsa-miR-025 | 0.56 | −20.34 |
| hsa-miR-020a | 0.57 | −22.42 |
| hsa-miR-212 | 0.59 | −5.89 |
| hsa-miR-378 | 0.63 | −18.25 |
| hsa-miR-135a | 0.57 | −12.33 |
| hsa-miR-026a | 0.68 | −7.65 |
| hsa-miR-135b | 0.59 | −6.18 |
| miRNA . | Change at 18 h, -fold . | SAM score . |
|---|---|---|
| Up-regulated | ||
| hsa-miR-146a | 29.55 | 4.40 |
| hsa-miR-342 | 23.09 | 7.01 |
| mmu-miR-409 | 6.73 | 5.84 |
| mmu-miR-434–5p | 5.17 | 7.78 |
| mmu-miR-096 | 2.50 | 16.71 |
| mmu-miR-151 | 2.44 | 9.23 |
| hsa-miR-485–5p | 2.25 | 3.64 |
| hsa-miR-338 | 2.08 | 5.66 |
| mumu-miR-155 | 2.95 | 22.56 |
| hsa-miR-033 | 2.67 | 8.86 |
| Down-regulated | ||
| mmu-miR-203 | 0.34 | −8.45 |
| hsa-miR-223 | 0.52 | −83.19 |
| hsa-miR-196b | 0.52 | −9.19 |
| hsa-miR-025 | 0.56 | −20.34 |
| hsa-miR-020a | 0.57 | −22.42 |
| hsa-miR-212 | 0.59 | −5.89 |
| hsa-miR-378 | 0.63 | −18.25 |
| hsa-miR-135a | 0.57 | −12.33 |
| hsa-miR-026a | 0.68 | −7.65 |
| hsa-miR-135b | 0.59 | −6.18 |
PUER-induced miR (18 hours in solvent vs 18 hours in OHT) were determined by SAM analysis and controlled for OHT-induced miR expression changes in PU.1 KO cells. Presented are miRs up-regulated 2-fold or down-regulated 0.75-fold.
Dynamic PU.1 binding near genomic loci encoding microRNAs
We further investigated the role of PU.1 in driving expression of the identified miRs directly by searching for sites of PU.1 occupancy within the chromatin vicinity of the pre–miR-coding regions. For this, we used global PU.1-binding datasets (ChIP-Seq)37 of PUER and of primary macrophages and compared them with our recently generated genome-wide DNase 1 hypersensitivity (DNase HSS) datasets22 to determine the positions of regulatory elements in the chromatin. We considered a PU.1-occupied site to be associated with a particular miR or miR-host gene if it was closer to the genomic position of the pre-miR than to that of any neighboring gene and if it overlapped with an evolutionarily conserved DNase HSS. Four of the PU.1–up-regulated miRs (miR-146a, miR-342, miR-338, and miR-155) fulfilled these criteria (Figure 2A).
Validation of PU.1–up-regulated miRs. (A) UCSC browser images of ChIPseq data from PUER cells after OHT treatment for the indicated time points and of primary macrophages37 in alignment with global DNase HSS data of primary macrophages22 (second rows from the bottom) and with sequence conservation tracks (bottom rows) according to the phylop algorithm (phast package, http://compgen.bscb.cornell.edu/phast/). Genomic coordinates are indicated at the top and miR positions, host genes and PU.1 occupancy peaks are shown at the bottom. (B) Kinetics of PU.1 induction of target miR expression in PUER cells compared with PU.1 KO controls. PUER and PU.1 KO cells were incubated with OHT (100nM) for the indicated time points. MiR expression was determined from total RNA by qRT-PCR compared with U6 RNA. PUER cells were induced with OHT (100nM) in the presence of cycloheximide (2 μg/mL; CHX) for 18 hours to inhibit protein translation. (C) MiR expression was determined from total RNA by qRT-PCR compared with U6 RNA. Error bars indicate SD.
Validation of PU.1–up-regulated miRs. (A) UCSC browser images of ChIPseq data from PUER cells after OHT treatment for the indicated time points and of primary macrophages37 in alignment with global DNase HSS data of primary macrophages22 (second rows from the bottom) and with sequence conservation tracks (bottom rows) according to the phylop algorithm (phast package, http://compgen.bscb.cornell.edu/phast/). Genomic coordinates are indicated at the top and miR positions, host genes and PU.1 occupancy peaks are shown at the bottom. (B) Kinetics of PU.1 induction of target miR expression in PUER cells compared with PU.1 KO controls. PUER and PU.1 KO cells were incubated with OHT (100nM) for the indicated time points. MiR expression was determined from total RNA by qRT-PCR compared with U6 RNA. PUER cells were induced with OHT (100nM) in the presence of cycloheximide (2 μg/mL; CHX) for 18 hours to inhibit protein translation. (C) MiR expression was determined from total RNA by qRT-PCR compared with U6 RNA. Error bars indicate SD.
We identified a conserved DNAse HSS that harbored a strong PU.1-binding peak at 10 kb upstream (−10 kb) of the pre–miR-146a locus. This region has been shown to contain a functional NF-κB–binding site and also to bind PU.1.16,38 ChIP-Seq data revealed that the −10-kb region was the only major PU.1-binding site within a genomic stretch of at least 100 kb surrounding the pre–mir-146a coding region. Moreover, PU.1 binding at this site was dynamic in that PU.1 occupancy increased with differentiation of PUER cells, and was strongest in fully matured PUER cells and in primary macrophages (Figure 2A top left panel). This finding indicated that PU.1 binding to this site occurred early and increased progressively with differentiation into monocytic cells.
PU.1 occupancy at a conserved PU.1-binding motif at 15 kb upstream of the bic gene (AY096003), a noncoding RNA that includes the pre–miR-155 coding region, followed a completely different kinetic. Maximal PU.1 occupancy was reached at 1 hour after OHT induction, but declined steadily thereafter and was no longer detectable after 48 hours. In agreement with this, no PU.1 occupancy was noted in mature macrophages, in which this site was also not accessible to DNase. This result suggested that PU.1 binding is needed to initiate the expression of miR-155, but not for its maintenance.
MiR-342 is encoded in the third intron of the Evl gene (Enah/Vasp-like, NM_016337)39 and we identified a PU.1-binding site40 near the promoter region of the host gene (at 5 kb upstream of the transcription start site), which was increasingly occupied by PU.1 during PUER differentiation and was marked by a DNase HSS in mature macrophages.
MiR-338 is located in the seventh intron of the Aatk gene (serine/threonine-protein kinase LMTK1, NM_001198787). We detected PU.1 occupancy in the promoter region of the host gene (at 2 kb upstream) and more prominently within the first intron of Aatk, suggesting that PU.1 regulates miR-338 expression through these sites. Both sites are readily occupied during PUER induction and accessible to DNase in mature macrophages.
We conclude that PU.1 bound to open chromatin near 4 of its induced miR loci with 2 types of kinetics: (1) permanent (miR-146a, miR-342, and miR-338) and (2) transient (miR-155) during myeloid differentiation.
PU.1 controls microRNA expression with different kinetics
We next investigated whether the observed differential kinetics in PU.1 occupancy near the pre-miR loci were associated with differential miR expression kinetics during myeloid differentiation and, indeed, expression of miR-146a, miR-342, and miR-338 steadily increased during differentiation of PUER cells and peaked in terminally differentiated macrophages (Figure 2B). In contrast, the expression of miR-155 transiently increased at 1 and 3 days, but decreased afterward in differentiated macrophages at day 7 after induction. Therefore, the expression kinetics of all identified PU.1 target miRs followed the patterns of PU.1 occupancy at the identified regulatory loci (Figure 2A).
To examine whether PU.1 alone was sufficient to drive the expression of its target miRs or if secondary PU.1-induced factors were also required, we induced PUER cells in the presence of cycloheximide to block secondary protein synthesis. This experiment revealed that miR-146a and miR-155 up-regulation was cycloheximide insensitive, showing that PU.1 alone is sufficient to induce their expression (Figure 2C). In contrast, induction of miR-342 and miR-338 was blocked by cycloheximide treatment, indicating that cooperative factors are necessary for their induction.
PU.1 is required for miR-146a expression in vivo
Because the role of miR-146a in progenitor differentiation is still controversial,9,17-20 we first determined miR-146a expression in FACS-purified HSCs and progenitor populations, as well as in mature myeloid lineages. This experiment revealed basic transcription of miR-146a already in HSCs and myeloid progenitors, which increased markedly in mature myeloid cells (Figure 3A). However, the expression was higher in macrophages than in granulocytes, suggesting that miR-146a may have a unique role in monopoiesis.
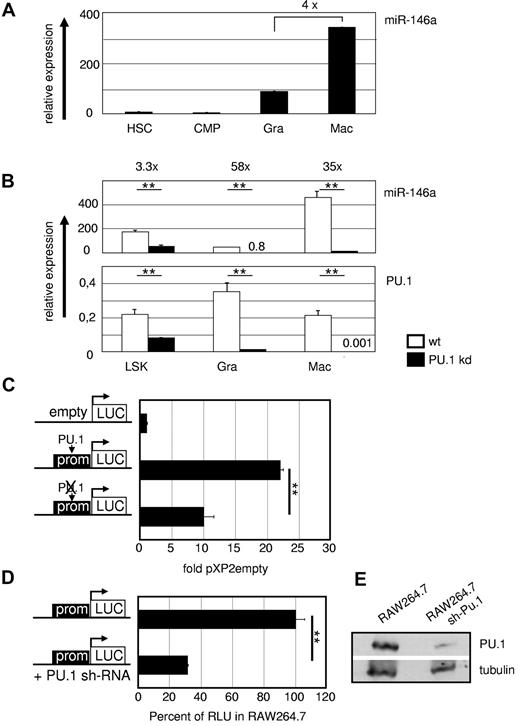
PU.1 regulates miR-146a in vivo. (A) MiR-146a expression in WT hematopoietic precursors and mature myeloid cells. miR-146a qRT-PCR was performed from FACS-purified HSCs (LSKs), common myeloid progenitors (CMPs; Lin−Sca-1−c-Kit+CD34+FcγRII/IIIlow), splenic granulocytes (Gra; Gr1+CD11b+), and splenic macrophages (Mac; Gr1−CD11b+). Data were compared with U6 RNA ± SD miR-146a and PU.1 mRNA expression in the indicated FACS-purified hematopoietic populations from WT and PU.1 KD animals. (B) MiR-146a was analyzed compared with U6 RNA, and PU.1 expression was determined relative to β-actin. **P < .001 by Student t test. (C) A 375-bp fragment containing the PU.1-binding site at 10 kb upstream of the genomic miR-146 locus was inserted in the pXP2 luciferase plasmid pXP2-(-)10kb. Mutation of the PU.1 motif (ctgaggaagt to ctgaccaagt) in the pXP2-(-)10kb construct, as indicated by the crossed-out PU.1 symbol in the pictogram, led to a marked reduction in reporter activity in transiently transfected myeloid RAW264.7 cells. Relative expression was determined 48 hours after transfection compared with pXP2 empty basal fluorescence activity (n = 3). **P < .001 by Student t test. (D) RAW264.7 cells carrying a stable PU.1 KD shRNA showed significantly lower miR146 promoter activity than plain RAW264.7 cells when transiently transfected with the WT pXP2-(-)10kb construct. Reporter activity of plain RAW264.7 cells was set to 100% (n = 3). **P < .001 by Student t test. (E) PU.1 expression in untreated and stably PU.1-shRNA–transfected RAW264.7 cells in Western blot normalized to levels of tubulin. Error bars indicate SD.
PU.1 regulates miR-146a in vivo. (A) MiR-146a expression in WT hematopoietic precursors and mature myeloid cells. miR-146a qRT-PCR was performed from FACS-purified HSCs (LSKs), common myeloid progenitors (CMPs; Lin−Sca-1−c-Kit+CD34+FcγRII/IIIlow), splenic granulocytes (Gra; Gr1+CD11b+), and splenic macrophages (Mac; Gr1−CD11b+). Data were compared with U6 RNA ± SD miR-146a and PU.1 mRNA expression in the indicated FACS-purified hematopoietic populations from WT and PU.1 KD animals. (B) MiR-146a was analyzed compared with U6 RNA, and PU.1 expression was determined relative to β-actin. **P < .001 by Student t test. (C) A 375-bp fragment containing the PU.1-binding site at 10 kb upstream of the genomic miR-146 locus was inserted in the pXP2 luciferase plasmid pXP2-(-)10kb. Mutation of the PU.1 motif (ctgaggaagt to ctgaccaagt) in the pXP2-(-)10kb construct, as indicated by the crossed-out PU.1 symbol in the pictogram, led to a marked reduction in reporter activity in transiently transfected myeloid RAW264.7 cells. Relative expression was determined 48 hours after transfection compared with pXP2 empty basal fluorescence activity (n = 3). **P < .001 by Student t test. (D) RAW264.7 cells carrying a stable PU.1 KD shRNA showed significantly lower miR146 promoter activity than plain RAW264.7 cells when transiently transfected with the WT pXP2-(-)10kb construct. Reporter activity of plain RAW264.7 cells was set to 100% (n = 3). **P < .001 by Student t test. (E) PU.1 expression in untreated and stably PU.1-shRNA–transfected RAW264.7 cells in Western blot normalized to levels of tubulin. Error bars indicate SD.
We also analyzed the PU.1 dependency of miR-146a in vivo in mice carrying a deletion of the PU.1 upstream cis-regulatory element, which leads to approximately 80% reduced PU.1 expression and results in PU.1 KD mice.41 This decrease in PU.1 causes a severe hematopoietic phenotype with a differentiation block in myelopoiesis and, as a consequence, the development of acute myeloid leukemia. Sorted fractions of stem cells and myeloid cells were analyzed, which showed that miR-146a expression was reduced in parallel with reduced PU.1 expression (Figure 3B), as well as in acute myeloid leukemia lines derived from diseased PU.1 KD mice (data not shown).
To evaluate whether PU.1 functionally controls miR-146a expression through the above described binding motif at 10 kb upstream of the pre-miR locus, we conducted luciferase reporter assays in RAW264.7 macrophages with constructs carrying a 375-bp genomic fragment containing this region with or without a mutation in the PU.1-binding motif25 (Figure 3C). This experiment indicated that the −10-kb region is a PU.1-dependent promoter. This was further confirmed by transfection of the WT reporter construct into RAW264.7 cells stably expressing a PU.1 KD hairpin (shPU.1; Figure 3D-E). These results showed that miR-146a expression requires PU.1 binding to its upstream promoter.
MiR-146a drives selective differentiation of adult HSCs into peritoneal macrophages in vivo
Because other studies failed to detect a lineage specification effect of miR-146a during adult hematopoiesis in vitro,38,42 we chose to analyze its function in vivo using a BM transplantation approach. Recently, Starczynowski et al17 found a transient increase in peripheral blood myeloid cell numbers after transplanting miR-146a–overexpressing, 5-fluoruracil–treated donor BM cells, which diminished over time. Another report of miR-146a function in lineage-negative BM progenitors failed to show a specific role in hematopoietic differentiation,20 calling into question the inhibitory effect on megakaryopoiesis that was attributed to miR-146a by previous reports.17-19 However, all of the mentioned studies analyzed miR-146a function in heterogeneous progenitor populations, not in purified stem cells, thus possibly precluding the observation of selective lineage-promoting effects. Furthermore, they omitted the analysis of peripheral nonhematopoietic tissues, a prime location at which myeloid cells reside after their terminal maturation.
To circumvent these problems, we chose to examine miR-146a function in adult hematopoiesis after transfer of a GFP-tagged miR-146a expression virus into FACS-purified C57/Bl6 CD45.2+ HSCs (defined as the LSK BM fraction). These donor cells were transplanted into SJL CD45.1+ recipient mice, which then were subjected to a comprehensive analysis of hematopoietic donor cells in multiple hematopoietic and nonhematopoietic tissues at 6-8 weeks after transplantation.
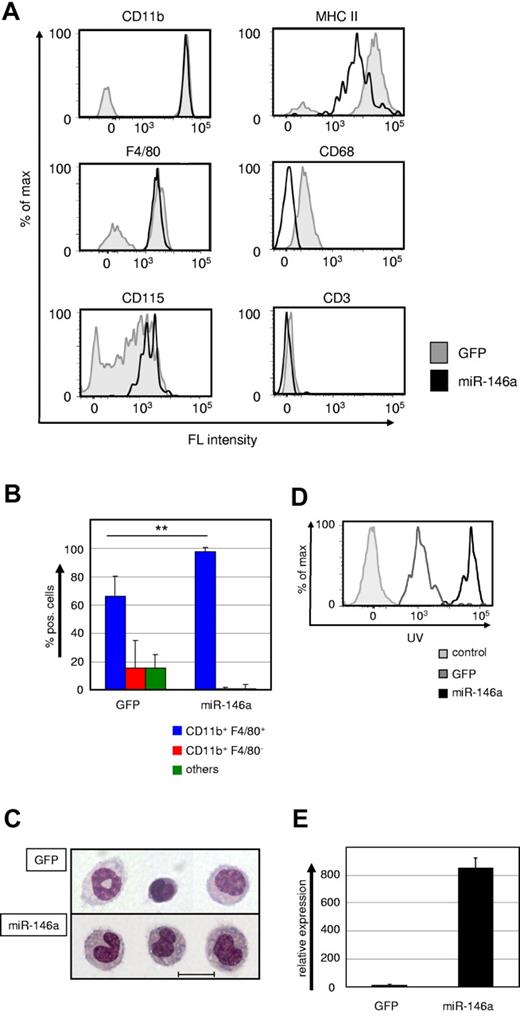
We failed to detect any GFPhigh-tagged miR-146a–expressing donor cells in the peripheral blood, spleen, BM, brain, liver, or lung (data not shown). Surprisingly, however, we readily detected GFPhigh donor cells exclusively in the peritoneum of the recipient mice. All donor cells had a typical monocyte phenotype, as indicated by morphology and surface expression of the myelomonocytic markers CD11b, F4/80, CD115, and MHC-II, but lack of expression of the granulocyte marker Gr1, the T-cell marker CD3, and the B-cell marker B220 (Figure 4A-C and data not shown). These monocytes were capable of maturing into phagocytotic macrophages when challenged with latex beads both in vivo (Figure 4D) and in vitro (data not shown). We observed a higher bead uptake in miR-146a–induced monocytes, which suggested enhanced phagocytotic activity, but this will require further investigation.
Ectopic miR-146a expression drives differentiation of HSC into peritoneal macrophages in vivo. (A) Representative FACS plots showing CD11b, F4/80, CD115, MHC-II, CD68, and CD3 expression within the GFP+ donor cell gates of peritoneal washouts from recipient mice that received transplantations with miR-146a (pre-miR-146a-GFP, solid black line) or empty vector (GFP, shaded gray line)–transduced LSK cells for 6-8 weeks. (B) Statistical analysis of FACS phenotypes of GFP+ donor-derived peritoneal cells (n = 6). **P < .001 by Mann Whitney U test. (C) Wright-Giemsa staining of cytospin preparations from FACS-purified GFP+ cells of peritoneal washouts of mice transplanted with control (GFP) or miR-146a–transduced LSK cells. The scale bar represents 10 μm. (D) In vivo phagocytosis capability was measured with FACS after IP challenge for 18 hours with UV-labeled latex beads. IP-challenged mice were transplanted with miR-146a (solid black line) or control (GFP, gray line)–transduced LSK cells for 6-8 weeks previously. Unstained control cells are depicted with a shaded gray line. (E) Quantification of ectopic miR-146a expression in GFP+ donor cells from peritoneal washouts of recipient mice transplanted with control or miR-146a–transduced LSK cells. Bars show the miR-146a qRT-PCR values in total RNA samples analyzed compared with U6 RNA. **P < .001 by Student t test. Error bars indicate SD.
Ectopic miR-146a expression drives differentiation of HSC into peritoneal macrophages in vivo. (A) Representative FACS plots showing CD11b, F4/80, CD115, MHC-II, CD68, and CD3 expression within the GFP+ donor cell gates of peritoneal washouts from recipient mice that received transplantations with miR-146a (pre-miR-146a-GFP, solid black line) or empty vector (GFP, shaded gray line)–transduced LSK cells for 6-8 weeks. (B) Statistical analysis of FACS phenotypes of GFP+ donor-derived peritoneal cells (n = 6). **P < .001 by Mann Whitney U test. (C) Wright-Giemsa staining of cytospin preparations from FACS-purified GFP+ cells of peritoneal washouts of mice transplanted with control (GFP) or miR-146a–transduced LSK cells. The scale bar represents 10 μm. (D) In vivo phagocytosis capability was measured with FACS after IP challenge for 18 hours with UV-labeled latex beads. IP-challenged mice were transplanted with miR-146a (solid black line) or control (GFP, gray line)–transduced LSK cells for 6-8 weeks previously. Unstained control cells are depicted with a shaded gray line. (E) Quantification of ectopic miR-146a expression in GFP+ donor cells from peritoneal washouts of recipient mice transplanted with control or miR-146a–transduced LSK cells. Bars show the miR-146a qRT-PCR values in total RNA samples analyzed compared with U6 RNA. **P < .001 by Student t test. Error bars indicate SD.
To confirm ectopic expression of miR-146a in these cells, we FACS-purified them and performed real-time PCR (Figure 4E). The level of miR-146a expression in the retrovirally transduced cells was comparable to that detected in induced PUER cells and furthermore comparable to the extent of induction seen in macrophages derived from LSK cells. In summary, enforced expression of miR-146a drives HSCs to selectively mature into peritoneal macrophages during adult hematopoiesis.
Macrophage development requires microRNA expression
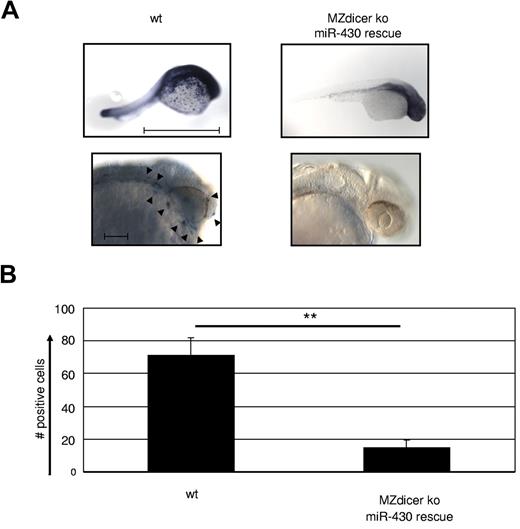
To examine the role of miR in macrophage development at a more global level, we made use of a maternal-zygote Dicer KO zebrafish (MZdicer KO).26 Embryos of this model lack miR-processing capacity from the one-cell stage on, and as a consequence display developmental lethality on day 5 because of defects in brain formation, somatogenesis, and heart development (eg, tubular hearts and pericardial edema).26 However, many of these developmental defects are overcome by supplementing the mutant embryos with mature miR-430 (MZdicer+430), which allows normal early development.26 HSCs in the zebrafish arise from the aortic endothelium43 ; macrophage development occurs from 18 hpf in the yolk sac and distributes macrophages during the first wave of primitive hematopoiesis through the blood circulation and migration into the mesenchyme of the head. Therefore, we examined macrophage development in the MZdicer+430 mutants at 24-30 hpf by whole-mount in situ hybridization against macrophage-specific L-plastin, which is strongly expressed from 18 hpf (the emergence of macrophages) on.44 Staining for L-plastin was profoundly reduced in the MZdicer+430 mutants compared with WT controls (Figure 5A-B), confirming the lack in macrophage generation upon loss of DICER activity. Consequently, we conclude that miRs possess a crucial role in the development of macrophages.
Macrophage development is dependent on microRNA expression. (A) MZdicer KO embryos were rescued in development by injection of mature miR-430 RNA.26 Comparison of L-plastin+ macrophages between WT and rescued MZdicer KO (MZdicer+430) zebrafish embryos 24-30 hpf. Macrophage development can be detected in early stages in the yolk sac and after that in the mesenchyme of the head and the blood circulation. Each embryo is shown as an overview (scale bar indicates 1 mm) and as a lateral magnified view of the head (scale bar indicates 100 μm). Nine embryos were analyzed in each experiment. (B) Quantification of L-plastin+ macrophage numbers in z-stack image files of whole-mount in situ hybridizations from controls or MZdicer+430 zebrafish as shown in panel A. **P < .001 by Student t test. Error bars indicate SD.
Macrophage development is dependent on microRNA expression. (A) MZdicer KO embryos were rescued in development by injection of mature miR-430 RNA.26 Comparison of L-plastin+ macrophages between WT and rescued MZdicer KO (MZdicer+430) zebrafish embryos 24-30 hpf. Macrophage development can be detected in early stages in the yolk sac and after that in the mesenchyme of the head and the blood circulation. Each embryo is shown as an overview (scale bar indicates 1 mm) and as a lateral magnified view of the head (scale bar indicates 100 μm). Nine embryos were analyzed in each experiment. (B) Quantification of L-plastin+ macrophage numbers in z-stack image files of whole-mount in situ hybridizations from controls or MZdicer+430 zebrafish as shown in panel A. **P < .001 by Student t test. Error bars indicate SD.
MiR-146a is required for macrophage emergence during embryogenesis
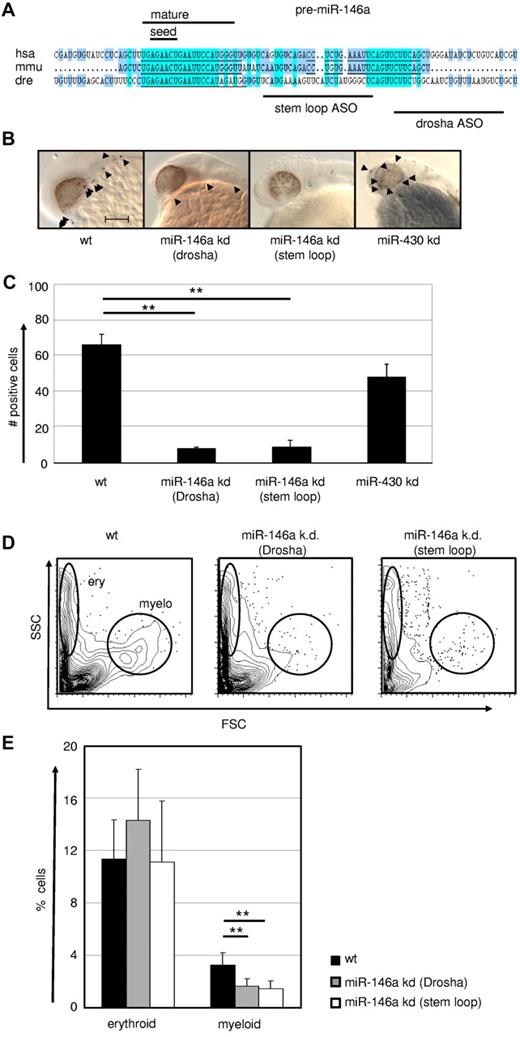
Because the genetic DICER ablation demonstrated a general requirement of miR for macrophage development, we next evaluated more specifically the role of the PU.1–regulated miR-146a in this process by applying a KD approach in early zebrafish embryos. The expression pattern and function of PU.1 is highly conserved between zebrafish and mammals.44,45 Furthermore, zebrafish miR-146a shares the same seed region with its mouse and human homologs, suggesting conserved roles in these species (Figure 6A). Microinjection of 2 different antisense oligonucleotides (one targeting the stem loop and another the drosha cleavage region) into zebrafish eggs resulted in a massive reduction in L-plastin+ macrophage numbers (Figure 6B-C). Furthermore, using FACS, we observed a selective loss of myeloid, but not erythroid cells, in miR-146a KD fish, confirming specificity of miR-146a function for the myeloid lineage in this assay (Figure 6D-E and supplemental Figure 2). No other obvious morphological or developmental abnormalities were observed in miR-146a–depleted zebrafish embryos, indicating a hematopoietic system–specific function during early embryogenesis (data not shown). In comparison, specific KD of the unrelated control miR miR-430 had no effect on macrophage development, but caused several other previously reported morphological abnormalities, including dysmorphogenic brain and heart development such as underinflated brain ventricles, slight microcephaly, pericardial edema, and string-like hearts (data not shown).
MiR-146a is required for emergence of macrophages during early zebrafish embryogenesis. (A) Conservation of pre–miR-146a sequences in the genomes of human, mouse, and zebrafish. Highlighted are the seed regions and the mature miR sequence. The target regions of the 2 independent ASOs to induce miR KD are indicated. Scale bar indicates 100 mm. Comparison of L-plastin+ macrophages (indicated by arrowheads) in WT, miR-146a KD (drosha cleavage), miR-146a KD (stem loop), and miR-430 KD in 24-28 hpf zebrafish embryos. Each control/KD embryo is shown with a lateral magnified view of the head. (C) Quantification of the L-plastin+ macrophage numbers in z-stack image files of whole-mount in situ hybridizations from controls or KD zebrafish as shown in Figure 5B (n = 5). **P < .001 by Student t test. (D-E) Generation of erythrocytes and macrophages in WT and ASO-treated zebrafish determined by FACS analysis of scatter plots of whole fish single-cell suspensions. Representative FACS plots (D) and statistical analysis (E) are displayed (n = 8). **P < .001 by Student t test. Error bars indicate SD.
MiR-146a is required for emergence of macrophages during early zebrafish embryogenesis. (A) Conservation of pre–miR-146a sequences in the genomes of human, mouse, and zebrafish. Highlighted are the seed regions and the mature miR sequence. The target regions of the 2 independent ASOs to induce miR KD are indicated. Scale bar indicates 100 mm. Comparison of L-plastin+ macrophages (indicated by arrowheads) in WT, miR-146a KD (drosha cleavage), miR-146a KD (stem loop), and miR-430 KD in 24-28 hpf zebrafish embryos. Each control/KD embryo is shown with a lateral magnified view of the head. (C) Quantification of the L-plastin+ macrophage numbers in z-stack image files of whole-mount in situ hybridizations from controls or KD zebrafish as shown in Figure 5B (n = 5). **P < .001 by Student t test. (D-E) Generation of erythrocytes and macrophages in WT and ASO-treated zebrafish determined by FACS analysis of scatter plots of whole fish single-cell suspensions. Representative FACS plots (D) and statistical analysis (E) are displayed (n = 8). **P < .001 by Student t test. Error bars indicate SD.
The loss-of-function experiment in the zebrafish model revealed that generation of the macrophage compartment in embryos requires the regulatory activity of miR-146a.
Discussion
In the present study, we identified a specific miR profile that is under the transcriptional control of PU.1 during stem/progenitor cell differentiation into monocytes/macrophages. In addition to miRs with thus-far-unknown functions in hematopoietic cells, we identified several miRs with previously reported (partly controversial) roles in myelopoiesis and/or leukemia as regulated by PU.1.10,16,46-48 Therefore, our data indicate that PU.1 orchestrates the expression of an entire miR program necessary to guide correct differentiation and counteract malignant transformation of developing myeloid progenitors (supplemental Figure 3). A recent attempt to delineate PU.1-dependent miRs showed a limited number of miR candidates, of which the miR-23 cluster was shown to block lymphopoiesis; however, that study did not clarify a specific myeloid profile.42 Therefore, our analysis of PU.1-dependent miRs reveals PU.1 as a master coordinator of myeloid miR expression.
In addition to positive regulation of several miRs (discussed further below), we observed that PU.1 suppresses the expression of several miRs. For example, we found that PU.1 down-regulates members of the miR-17p-92 cluster. Inhibition of this cluster was shown to represent an essential step in allowing physiologic myelopoiesis,13 and preventing malignant transformation.36 MiR-17p-92 is a direct target of c-MYC and can inhibit E2F to drive cellular proliferation.49 The finding that PU.1 suppresses miR-17p-92 during myeloid differentiation suggests that PU.1 stops the transient proliferation of amplifying progenitors to ensure correct terminal differentiation. Moreover, we observed that PU.1 down-regulates the expression of miR-223. This miR has been reported to stimulate granulopoiesis,10 although controversy has been raised about this particular function.11,12 Nevertheless, down-regulation of miR-223 may represent a mechanism by which PU.1 predisposes myeloid progenitors to differentiate into monocytes and not granulocytes.
Using global PU.1-binding data in combination with genome-wide DNase HSS maps, we found that 4 of the PU.1 up-regulated miRs (miR-146a, miR-342, miR-338, and miR-155) harbored PU.1-binding peaks within open chromatin regions in their direct genomic vicinity. Remarkably, this approach revealed that PU.1 occupied these regions with different kinetics, which was directly paralleled by transcription kinetics of the related miRs. Whereas PU.1 occupancy and expression of miR-146a, miR-342, and miR-338 loci increased permanently with myeloid differentiation and peaked in terminally differentiated macrophages, miR-155 expression was induced by transient PU.1 occupancy. PU.1 binding at the Bic gene, which hosts the miR-155 coding sequence, was not detectable in untreated PUER cells, but was induced within 1 hour of OHT stimulation. This effect lasted until 24 hours, but disappeared thereafter and was again undetectable at 48 hours. These binding kinetics were closely paralleled by the transient expression of miR-155 during PUER differentiation, suggesting that miR-155 is tightly regulated by PU.1 through the identified binding site. Moreover, because cycloheximide treatment did not affect PU.1-induced up-regulation of miR-155, we conclude that PU.1 is the major factor required for its expression control in early myeloid cells. MiR-155 was shown to potently induce proliferation of myeloid cells and as a consequence to cause a myeloproliferative syndrome after ectopic expression in myeloid progenitors (O'Connell et al48 and S.G. and F.R., unpublished data). It is therefore likely that PU.1 initiates the expression of miR-155 to escort quiescent HSCs into cycle and thus activates their initial differentiation into a transient amplifying myeloid progenitor stage. However, PU.1 may be removed from its binding site at the Bic gene thereafter to allow termination of miR-155 expression as a prerequisite to stopping proliferation (and thus preventing leukemia) and starting terminal myeloid maturation.
PU.1 bound the chromatin near the miR-342 and miR-338 coding loci, but PU.1-induced transcription of these miRs was blocked by cycloheximide, indicating that expression of both of them required other factors in addition to PU.1. In contrast, miR-146a was identified as another direct, cycloheximide-insensitive PU.1 target, thus extending the results of a previous study38 by showing that PU.1 is the main regulator of miR-146a expression during the development of myeloid cells. Moreover, we found that miR-146a expression is dependent on PU.1 throughout myeloid differentiation from HSCs. However, macrophages express more miR-146a as granulocytes, although both cell lineages express comparably high PU.1 levels. This discrepancy might be caused by the presence of as-yet-unknown additional regulatory factors in macrophages that cooperate with PU.1 to induce high-level miR-146a expression. Regulation of miR-146a operates through a conserved PU.1-binding motif at 10 kb upstream of the miR-146 genomic locus. Mir-146a expression has been shown to be inducible on activation in a set of mature hematopoietic cells, namely in T cells,50 dendritic cells,38 and macrophages.16 Loss of expression of miR-146a, together with miR-145 loss, was implicated in the pathology of the 5q− myelodysplastic syndrome,19 indicating its potential role as a tumor suppressor. This role is supported the very recent observation that miR-146a KO mice develop granulocytic hyperplasia and have a reduced macrophage compartment.51
In contrast to previous reports, in the present study, we made the surprising observation that ectopic expression of miR-146a in purified adult HSCs enforced their selective development into peritoneal monocytes capable of maturing into phagocytosing macrophages. Although the molecular mechanism of this function is still unclear, it appears that miR-146a may control 2 transcriptional programs during myelopoiesis, one driving terminal differentiation along a selective myeloid path and another directing the specific migration of monocyte precursors into the peritoneum.
Using a Dicer KO approach, we found that the global loss of miR activity leads to a block in the emergence of macrophages in zebrafish embryos. This effect was phenocopied by selective antagonization of miR-146a function, demonstrating that miR-146 is a critical regulator in the initiation of fetal macrophage development. The combination of gain- and loss-of-function experiments in mice and zebrafish suggest that miR-146a plays an important role not only in inhibition of proinflammatory signaling or proliferation,16,17,19 but also in directing an early, evolutionarily conserved differentiation program to promote the development of macrophages in both embryos and adults.
In summary, we describe herein a comprehensive PU.1-orchestrated miR expression profile in which individual members can mediate the key functions of PU.1 to act on proliferation, induce myeloid differentiation, and suppress leukemic transformation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank H. Singh and P. Laslo for providing them with cell lines; W.-D. Ludwig and L. Karawajew for helpful discussions and support; and H. Lammert, M. Renz, J. Richter, V. Malchin, N. Endruhn, C. Graubmann, A. Dietze, and M. Steiner for technical assistance.
F.R. was supported by a grant from the Helmholtz association of German research centers (HGF) and S.G. was a postgraduate fellow of the clinical-cooperation program (KAP) between the Charité and the Max-Delbrück Center.
Authorship
Contribution: S.G., P.R., J. Schönheit, D.L., J. Stumm, M.H., A.L., S.H., and F.R. conducted the experiments; S.G. and F.R. wrote the manuscript; S.G. generated the figures; C.B., J.B., S.A.-S., and M.H. provided ideas and essential experimental support; and F.R. supervised the project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.G. is Department of Hematology, Oncology, and Tumor Immunology, HELIOS Clinic, Berlin, Germany.
Correspondence: Frank Rosenbauer, Max Delbrück Center for Molecular Medicine, Robert-Rössle-Strasse 10, 13125 Berlin, Germany; e-mail: f.rosenbauer@mdc-berlin.de.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal