Abstract
Imatinib-resistant tyrosine kinase (TK) fusions involving FGFR1, JAK2, or FLT3 are rare but recurrent in patients with eosinophilia-associated neoplasms. We report here 2 male patients with ETV6-FLT3+ myeloid/lymphoid neoplasms with eosinophilia who were treated with the multitargeted TK inhibitors sunitinib and sorafenib. Patient 1 achieved rapid complete hematologic response and complete cytogenetic response after 3 months of taking sunitinib. A secondary blast phase caused by clonal evolution was diagnosed after 6 months. He achieved a second complete hematologic response after taking sorafenib but relapsed 2 months later. An N841K point mutation within the TK domain of FLT3, previously reported in acute myeloid leukemia and potentially conferring resistance to sorafenib, was subsequently identified. Patient 2 was heavily pretreated according to the initial diagnosis of T-lymphoblastic lymphoma and died in sunitinib-induced pancytopenia. This report highlights the importance of a careful diagnostic workup for eosinophilia-associated neoplasms to evaluate the possibility of TK inhibitor therapy.
Introduction
Eosinophilia-associated myeloid neoplasms are frequently associated with constitutive activation of disparate tyrosine kinase (TK) fusion genes. The World Health Organization's classification includes a subcategory termed “myeloid/lymphoid neoplasms with eosinophilia and rearrangement of PDGFRA, PDGFRB or FGFR1.” These cases are often characterized by the concurrent diagnosis of a myeloid neoplasm, a high-grade lymphoma (most frequently of T-cell phenotype), and eosinophilia. In the present report, we refer to this entity as MLN-eo. Other TK fusion genes, for example, involving ABL or JAK2, have also been reported in diverse eosinophilia-associated myeloid neoplasms. Excellent response rates and long-term clinical outcomes have been reported for fusions involving PDGFRA and PDGFRB after treatment with imatinib.1-3 In contrast, disorders with FGFR1 and JAK2 fusion genes are resistant to imatinib and other clinically available TK inhibitors (TKIs).4
The FMS-like tyrosine kinase 3 (FLT3) is a class III receptor TK involved in the pathogenesis of de novo acute myeloid leukemia through activation of mutations in the juxtamembrane or TK domains. These mutations are potential targets for multitargeted TKIs such as midostaurin (PKC412), sunitinib, or sorafenib, which have selective activity against FLT3 and other TKs. The efficacy of FLT3 inhibitors in acute myeloid leukemia with FLT3 mutations is currently being explored in large phase III trials. Fusion genes involving FLT3 are uncommon. Vu et al5 characterized an ETV6-FLT3 fusion in a female patient with eosinophilia-associated myeloproliferative neoplasm (MPN-eo) who died rapidly because of progression to blast phase. Additional experiments confirmed the transforming properties of the ETV6-FLT3 fusion protein in Ba/F3 cells and mice.5,6 Grand et al7 reported a female patient with atypical chronic myeloid leukemia (CML) and an SPTBN1-FLT3 fusion who achieved long-term remission after receiving an allograft. SPTBN1-FLT3–transformed Ba/F3 cells were sensitive to several FLT3 inhibitors. We here report for the first time on the efficacy of an FLT3 TKI in ETV6-FLT3+ MLN-eo.
Results and discussion
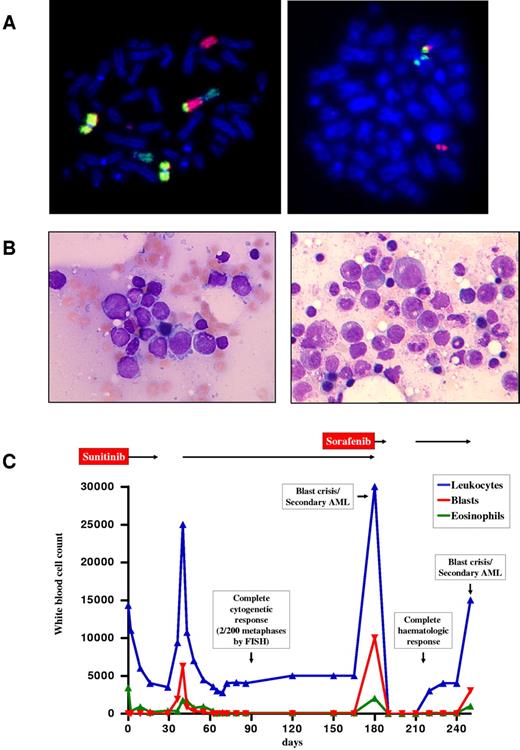
Patient 1 was a 60-year-old man who presented with widespread lymphadenopathy and splenomegaly. Leukocytes were elevated at 14.3 × 109/L, with 24% eosinophils and 8% blasts. Hemoglobin was 9.7 g/dL, and platelets were 177 × 109/L. Bone marrow morphology was consistent with an MPN-eo in an accelerated phase. FIP1L1-PDGFRA was negative, and conventional cytogenetic analysis revealed a complex karyotype: 46,XY,del(9)(q22),der(12)t(12;13)(p13;q14)t(9;13)(q34;q22),der(13)t(12;13)(p13;q14),[4] 46,XY[2]. Lymph node histology and immunohistochemistry suggested a peripheral T-cell lymphoma. FISH analysis indicated a rearrangement of ETV68 (Figure 1), and 3′ rapid amplification of cDNA ends PCR revealed a fusion between ETV6 exon 4, a 16-bp insert derived from ETV6 intron 4, and a truncated FLT3 exon 14 (Figure 2A). After approval was obtained from the patient, sunitinib was initiated at a dose of 50 mg/d (off-label use). The patient rapidly achieved complete normalization of peripheral blood counts, spleen size, and enlarged lymph nodes (Figure 1C). No dose-limiting toxicities were observed. The patient achieved a complete cytogenetic response after 3 months. Unexpectedly, a rapid increase of myeloid blasts was seen after 6 months (Figure 1B left). The karyotype was hyperdiploid, with 51 chromosomes in addition to the known complex translocation. No FLT3 mutations were detected. Ten days after initiation of sorafenib (400 mg BID), pancytopenia occurred, which lasted for 3 weeks after cessation of treatment. After a slow recovery, sorafenib was restarted, but normal blood counts were maintained for only 5 weeks (Figure 1B right). At this point, a C > A (codon AAC > AAA) sequence variant (www.ensembl.org, ENST00000241453) was identified that predicted an FLT3N841K mutation in the activation loop of the TK domain. Intensive chemotherapy with cytarabine and daunorubicin was initiated, but the patient died shortly afterward because of pancytopenia and infection.
Diagnosis and clinical course of patient 1. (A) FISH with whole-chromosome painting probes for chromosomes 9 (yellow), 12 (green), and 13 (red) demonstrating the 12;13 translocation and involvement of the derivative chromosome 12 in an additional translocation with chromosome 9 (left). FISH analysis confirmed involvement of ETV6, showing the 5′-ETV6 probe labeled in red on the derivative chromosome 13 and the 3′-ETV6 probe labeled in green on the derivative chromosome 12 in patient 1 at diagnosis (right). Cytogenetic and FISH analyses with break-apart probes for the detection of ETV6 rearrangements (Abbott) were performed according to standard procedures.14,15 (B) Bone marrow morphology of patient 1 at diagnosis of sunitinib-resistant blast phase/secondary acute myeloid leukemia 6 months after start of sunitinib (left) and 5 weeks after initiation of sorafenib (right). Micrographs were taken by an Axiolmager M1 (Zeiss) using an EC Plan-Neofluar, 100×/1.3 oil, 0.17 objective lens (Zeiss). (C) Hematologic response to treatment with sunitinib and sorafenib in patient 1 over a 250-day period. AML indicates acute myeloid leukemia.
Diagnosis and clinical course of patient 1. (A) FISH with whole-chromosome painting probes for chromosomes 9 (yellow), 12 (green), and 13 (red) demonstrating the 12;13 translocation and involvement of the derivative chromosome 12 in an additional translocation with chromosome 9 (left). FISH analysis confirmed involvement of ETV6, showing the 5′-ETV6 probe labeled in red on the derivative chromosome 13 and the 3′-ETV6 probe labeled in green on the derivative chromosome 12 in patient 1 at diagnosis (right). Cytogenetic and FISH analyses with break-apart probes for the detection of ETV6 rearrangements (Abbott) were performed according to standard procedures.14,15 (B) Bone marrow morphology of patient 1 at diagnosis of sunitinib-resistant blast phase/secondary acute myeloid leukemia 6 months after start of sunitinib (left) and 5 weeks after initiation of sorafenib (right). Micrographs were taken by an Axiolmager M1 (Zeiss) using an EC Plan-Neofluar, 100×/1.3 oil, 0.17 objective lens (Zeiss). (C) Hematologic response to treatment with sunitinib and sorafenib in patient 1 over a 250-day period. AML indicates acute myeloid leukemia.
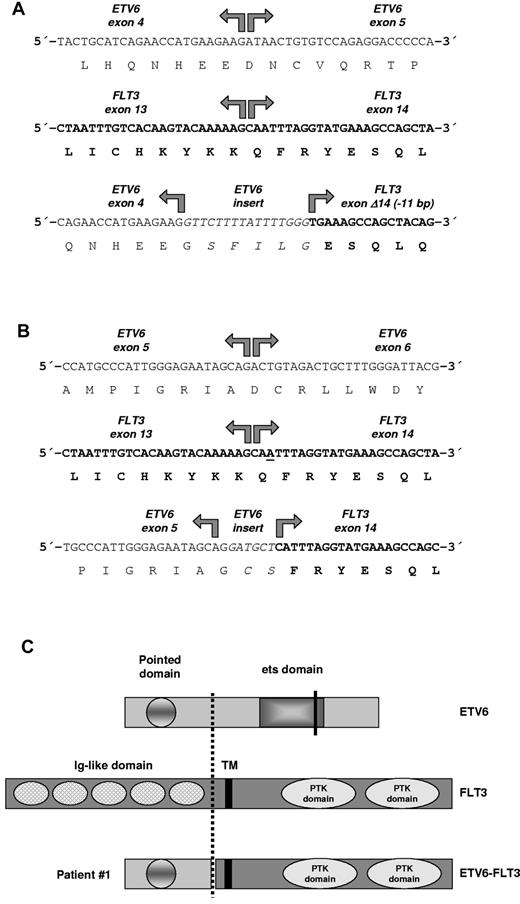
ETV6-FLT3 fusion genes and predicted protein structure. (A) RACE-PCR (3′-rapid amplification of cDNA ends) for ETV6 was performed with mRNA derived from patient 1 as described previously.8 Sequencing of RACE-PCR products revealed an in-frame fusion between ETV6 exon 4 and a truncated FLT3 exon 14 in patient 1 with insertion of a 16-bp sequence derived from ETV6 intron 4. The ETV6-FLT3 fusion gene was confirmed by RT-PCR. (B) Sequencing of RACE-PCR products revealed an in-frame fusion between ETV6 exon 5 and FLT3 exon 14 in patient 2 with insertion of a 6-bp sequence, probably derived from ETV6 intron 5. Interestingly, base pair number 2 (or 3) of FLT3 exon 14 was deleted in the fusion gene to retain the open-reading frame (underscored). The ETV6-FLT3 fusion gene was confirmed by RT-PCR. (C) The predicted ETV6-FLT3 protein of patient 1 retained the TK domain of FLT3. TM indicates transmembrane domain; PTK domain, protein-TK domain. Break point of patient 1 is indicated by a dashed line; the ETV6 break point of patient 2 as determined by RT-PCR and sequencing with primers TELFLT/f1&2 and TELFLT/r1&2 (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) is shown by a solid line, whereas its complete fusion protein is not shown.
ETV6-FLT3 fusion genes and predicted protein structure. (A) RACE-PCR (3′-rapid amplification of cDNA ends) for ETV6 was performed with mRNA derived from patient 1 as described previously.8 Sequencing of RACE-PCR products revealed an in-frame fusion between ETV6 exon 4 and a truncated FLT3 exon 14 in patient 1 with insertion of a 16-bp sequence derived from ETV6 intron 4. The ETV6-FLT3 fusion gene was confirmed by RT-PCR. (B) Sequencing of RACE-PCR products revealed an in-frame fusion between ETV6 exon 5 and FLT3 exon 14 in patient 2 with insertion of a 6-bp sequence, probably derived from ETV6 intron 5. Interestingly, base pair number 2 (or 3) of FLT3 exon 14 was deleted in the fusion gene to retain the open-reading frame (underscored). The ETV6-FLT3 fusion gene was confirmed by RT-PCR. (C) The predicted ETV6-FLT3 protein of patient 1 retained the TK domain of FLT3. TM indicates transmembrane domain; PTK domain, protein-TK domain. Break point of patient 1 is indicated by a dashed line; the ETV6 break point of patient 2 as determined by RT-PCR and sequencing with primers TELFLT/f1&2 and TELFLT/r1&2 (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) is shown by a solid line, whereas its complete fusion protein is not shown.
Patient 2 was a 29-year-old man who was diagnosed with a peripheral T-cell lymphoma. Leukocytes were elevated to 23.8 × 109/L, with 0.8 × 109/L eosinophils, whereas hemoglobin and platelets were normal. He achieved a complete response after 6 cycles of CHOP chemotherapy (cyclophosphamide, doxorubicin [Adriamycin], vincrishine, and prednisone) but rapidly developed progressive lymphadenopathy within a few days of completion of therapy. A repeat lymph node biopsy led to revised diagnosis of T-lymphoblastic lymphoma. Leukocytes were elevated to 51.6 × 109/L, with 11.2 × 109/L (22%) eosinophils. Bone marrow histology was focally hypercellular. FIP1L1-PDGFRA was negative, and conventional cytogenetics revealed a t(12;13)(p13;q12)[10] translocation. He achieved a complete remission after 3 cycles of the HyperCVAD regimen (cyclophosphamide, vincristine, doxorubicin [Adriamycin], and dexamethasone) and was consolidated with autologous stem cell transplantation with total body irradiation/cyclophosphamide. Four weeks after transplantation, the patient presented with progressive leukocytosis, thrombocytopenia, and splenomegaly. The marrow was consistent with an MPN-eo. Cytogenetics again showed a t(12;13)(p13;q12)[10] translocation with first evidence of an ETV6 rearrangement by FISH. RT-PCR identified an ETV6-FLT3 fusion between ETV6 exon 5 and FLT3 exon 14 that included a 6-bp insert, possibly derived from ETV6 intron 5. The combination of immunostaining with antibodies against CD3 and FISH with an LSI ETV6 dual-color break-apart probe (Abbott/Vysis) showed that a subset of T cells (41/100 cells) in the lymph node biopsy carried the ETV6 rearrangement (data not shown). Because of the lack of alternative treatment options, sunitinib (37.5 mg/d) was initiated after the patient gave his approval (off-label use). The patient achieved a rapid response, with complete disappearance of elevated eosinophils from 2.5 × 109/L to zero within 96 hours. Prolonged pancytopenia with markedly hypocellular bone marrow led to discontinuation of treatment after 3 weeks. FISH analysis showed a persistence of the ETV6 rearrangement (present in 44 of 100 interphase nuclei), and cytogenetic analysis revealed clonal progression (46,XY,t(12;13)(p13;q12) [4]/49,idem,+8,+15,+21[5]/46,XY[3]). The patient died 5 weeks after sunitinib was begun because of pneumonia.
Including case 2 reported here, 3 TK fusion genes (ZNF198-FGFR1, FIP1L1-PDGFRA, and ETV6-FLT3) have been reported in myeloid bone marrow cells and in CD3+ T cells in the lymph nodes of patients with both MPN-eo and lymphadenopathy.4,9,10 This clearly indicates that these diseases are stem cell disorders with predominant retention of myeloid cells in the marrow and T cells in the lymph node. The term “T-cell lymphoma” may therefore be correct for what is seen in lymph node biopsy samples through histology and immunohistochemistry but not for the disease as a whole. The diagnosis of a T-cell lymphoma in patients with significant eosinophilia should therefore only be made when clonality of myeloid cells is excluded, for example, through PCR/FISH for FIP1L1-PDGFRA and/or through cytogenetic analysis.
In addition to BCR, ETV6 is one of the very few genes that are recurrently found as fusion partner of TKs in MPN-eo or MLN-eo. The influence of partner genes on phenotype is not known precisely, but the response and duration of response because of secondary resistance might, for example, be inferior in ETV6-ABL1+ disease compared with BCR-ABL1+ CML.11 However, sustained molecular remission has been described for a case subject with ETV6-PDGFRA.12
The pattern of response and resistance because of a point mutation in the TK domain of FLT3 in 1 patient clearly resembles the clinical course during treatment with imatinib in BCR-ABL1+ CML and FIP1L1-PDGFRA+ chronic eosinophilic leukemia. FLT3 TKIs are currently also used for the treatment of acute myeloid leukemia with FLT3 mutations; however, responses are only modest or transient, and the achievement of a complete molecular remission is a rare finding.13 Although the response in case 2 reported here was also transient, on the basis of the evidence in CML, it seems reasonable to speculate that TKI treatment earlier during the disease course might have resulted in a better outcome. In summary, the present study provides the first evidence that an FLT3 TKI can induce complete hematologic and cytogenetic remissions in patients with MLN-eo with FLT3 fusion genes. These findings emphasize the importance of evaluating cases with 13q12 rearrangements for possible involvement of FLT3.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the German branch of the José Carreras Leukemia Foundation (José Carreras Leukämie-Stiftung e.V.; DJCLS R06/02, R09/29f, and H03/01; C.W., A.R., and A.H.); Leukaemia and Lymphoma Research, London, United Kingdom; the Competence Network “Acute and Chronic Leukemias” sponsored by the German Federal Ministry for Education and Research (Bundesministerium für Bildung und Forschung; Projektträger Gesundheitsforschung; DLR e.V.-01GI9980/6); and the European LeukemiaNet within the 6th European Community Framework Program for Research and Technological Development.
Authorship
Contribution: C.W., A.H., N.C.P.C., and A.R. designed the research, collected and interpreted data, and wrote the paper; P.E., C.H., T.H., S.G., and J.S. collected and interpreted data and performed experiments; and M.R., A.B., G.M., N.T., and W.-K.H. collected and interpreted data and approved the manuscript.
Conflict-of-interest statement: The authors declare no competing financial interests.
Correspondence: Professor Andreas Reiter, MD, III Medizinische Klinik, Universitätsmedizin Mannheim, Theodor-Kutzer Ufer 1-3, 68167 Mannheim, Germany; e-mail: andreas.reiter@umm.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal