It is now well-established that the anucleate platelet contains a full and functional repertoire of mRNAs. Yet how these transcripts are delivered from the megakaryocyte to the nascent platelet is unknown. In this issue of Blood, Cecchetti et al demonstrate that megakaryocytes actively sort and deliver mRNAs to platelets.1
The capacity of the small, anucleate platelet to synthesize new proteins had been questioned for some time. Warshaw et al first presented proof that platelets are capable of de novo protein synthesis nearly 50 years ago.2 A cell-free translation system developed from platelet homogenates demonstrated ribosomes in platelets and showed that this synthetic machinery was functional.3 The incorporation of radiolabeled amino acids into nascent platelet proteins also implies synthesis of new proteins by platelets. However, progress in understanding the mechanism and significance of platelet protein synthesis was relatively slow, as investigators questioned the magnitude of protein synthesis and its importance in platelet function.
Over the past decade, Weyrich and colleagues at the University of Utah have brought this discussion to an entirely new level. They demonstrated activation-dependent synthesis of several proteins including Bcl-3, IL1β, and tissue factor.4-6 The investigators not only demonstrated activation-dependent, de novo synthesis, but also described the mechanism by which platelet activation elicits new protein synthesis. They showed that signal-dependent translation can be controlled by pre-mRNA splicing machinery7 or via an mTOR-mediated mechanism.4 By addressing the fundamental questions of how new proteins are synthesized by the platelet and what the significance of this activity is, these data have broadened the landscape of possible platelet functions. Yet fundamental questions such as, “How does mRNA get from megakaryocyte to platelet?” remain unanswered.
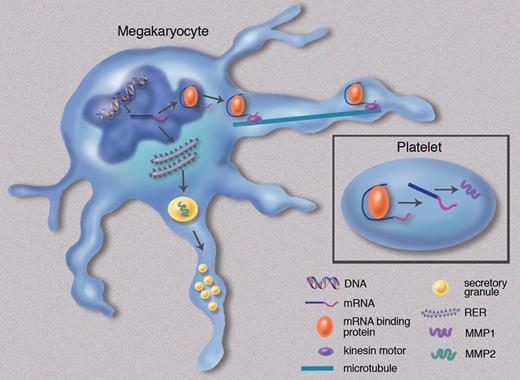
Hypothetical model of mRNA sorting in megakaryocytes. The mechanism by which mRNAs are sorted in megakaryocytes is not known. This model is based on an analogy to the more well-characterized neuronal model. DNA is transcribed into mRNA in the nucleus of the megakaryocyte. Some transcripts (eg, MMP1) bind an mRNA binding protein, forming a complex that translocates to the cytoplasm. The complex associates with a kinesin motor, is transported through the proplatelet by microtubules, and is ultimately delivered to the nascent platelet. Some transcripts that are delivered to the platelet are transcribed constitutively. Others are transcribed in an activation-dependent manner. Transcripts that fail to bind an mRNA binding protein are translated into proteins in the rough endoplasmic reticulum (RER) of the megakaryocyte and subsequently packaged in secretory granules (eg, MMP2). These secretory granules are then transported through the proplatelet and delivered to platelets. Professional illustration by Marie Dauenheimer.
Hypothetical model of mRNA sorting in megakaryocytes. The mechanism by which mRNAs are sorted in megakaryocytes is not known. This model is based on an analogy to the more well-characterized neuronal model. DNA is transcribed into mRNA in the nucleus of the megakaryocyte. Some transcripts (eg, MMP1) bind an mRNA binding protein, forming a complex that translocates to the cytoplasm. The complex associates with a kinesin motor, is transported through the proplatelet by microtubules, and is ultimately delivered to the nascent platelet. Some transcripts that are delivered to the platelet are transcribed constitutively. Others are transcribed in an activation-dependent manner. Transcripts that fail to bind an mRNA binding protein are translated into proteins in the rough endoplasmic reticulum (RER) of the megakaryocyte and subsequently packaged in secretory granules (eg, MMP2). These secretory granules are then transported through the proplatelet and delivered to platelets. Professional illustration by Marie Dauenheimer.
Now, in a collaboration between the Weyrich group and Gresele and colleagues at the University of Perugia, the question of how these mRNAs are delivered to platelets during megakaryopoiesis is addressed. The simplest mechanism would be bulk delivery, which assumes a relatively uniform distribution of transcripts. By this mechanism, mRNAs would be proportioned in platelets according to their relative abundance in the megakaryocyte. The alternative possibility is that megakaryocytes possess a mechanism for sorting mRNAs and selectively delivering them to platelets.
Cecchetti et al distinguished between these 2 mechanisms by careful analysis of matrix metalloprotease (MMP) and tissue inhibitor of metalloprotease (TIMP) mRNAs using next-generation sequencing (RNA seq).1 They showed that cord blood–derived CD34+ stem cells differentiated into megakaryocytes express 10 of 24 human MMP transcripts and 3 of 4 TIMP transcripts. However, the expression profile in platelets differed. Some megakaryocyte transcripts were absent in platelets, while others were similarly expressed or perhaps relatively increased in platelets. Although mRNA concentrations are substantially lower in platelets than in megakaryocytes, the differences in transcript profile did not result from global differences in transcript levels because equal amounts of template mRNA were used. The authors also carefully ruled out mRNA degradation or instability as the cause of differences in transcript profiles. Based on these observations, they conclude that megakaryocytes sort mRNA.
What could account for the differences in mRNA profiles between megakaryocytes and platelets? Other cell types possess an array of mRNA binding proteins that function in the delivery of specific mRNAs to different localizations in the cell. The most notable example is the neuron, which delivers mRNA from the cell body to synapses. To accomplish this task, the neuron must transport mRNAs through elongated axons and dendrites.8 The megakaryocyte has an analogous problem to solve in delivering mRNAs through proplatelet extensions. One could envision that it uses a similar machinery to transfer transcripts to platelets during megakaryopoiesis (see figure). In fact, Cecchetti et al demonstrate that megakaryocytes and platelets express several genes such as CASC3, STAU1, STAU2, and EIF4A3 that encode for proteins implicated in mRNA transport in neurons.1 Additional studies will be required to determine whether the delivery of transcripts through elongated cytoplasmic structures such as axons or proplatelet extensions is solved by an analogous mechanism using similar proteins.
The implications of mRNA sorting by megakaryocytes are substantial. If megakaryocytes actively determine which mRNAs are transferred to platelets, then platelets can be “programmed” by megakaryocytes in response to physiologic stressors. Reprogramming could occur in the setting of inflammation, sepsis, malignancy, or vascular disease. This possibility is substantiated by studies demonstrating differences in the platelet transcriptomes from patients with lupus, sickle cell, or cardiovascular disease.9-11 Modifications of the platelet transcriptome may have diagnostic significance or could potentially be engineered for therapeutic benefit. Much will need to be sorted out before we can interpret and manipulate this reprogramming function. Yet the message is clear that the platelet is not the synthetically inert cell fragment once envisioned by many platelet biologists.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■
REFERENCES
National Institutes of Health

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal