Abstract
EBV, an oncogenic human herpesvirus, can transform primary B lymphocytes into immortalized lymphoblastoid cell lines (LCLs) through multiple regulatory mechanisms. However, the involvement of protein tyrosine kinases in the infinite proliferation of B cells is not clear. In this study, we performed kinase display assays to investigate this subject and identified a specific cellular target, Recepteur d'Origine Nantais (RON) tyrosine kinase, expressed in LCLs but not in primary B cells. Furthermore, we found that latent membrane protein 1 (LMP1), an important EBV oncogenic protein, enhanced RON expression through its C-terminal activation region-1 (CTAR1) by promoting NF-κB binding to the RON promoter. RON knockdown decreased the proliferation of LCLs, and transfection with RON compensated for the growth inhibition caused by knockdown of LMP1. Immunohistochemical analysis revealed a correlation between LMP1 and RON expression in biopsies from posttransplantation lymphoproliferative disorder (PTLD), suggesting that LMP1-induced RON expression not only is essential for the growth of LCLs but also may contribute to the pathogenesis of EBV-associated PTLD. Our study is the first to reveal the impact of RON on the proliferation of transformed B cells and to suggest that RON may be a novel therapeutic target for EBV-associated lymphoproliferative diseases.
Introduction
Primary EBV infection usually is asymptomatic but persists, so that more than 90% of adults worldwide are persistently infected with EBV.1 However, as the first human tumor virus identified, EBV is associated with several disorders of lymphocytes, including infectious mononucleosis, Burkitt lymphoma, Hodgkin lymphoma, T-cell lymphoma, NK-cell lymphoma, AIDS-associated lymphoma, and posttransplantation lymphoproliferative disorder (PTLD).1 Being an oncogenic virus, EBV can immortalize primary B cells into lymphoblastoid cell lines (LCLs). EBV-immortalized LCLs are characterized by several features, including aberrant production of cytokines and chemokines, unlimited proliferation, clumping morphology, and display of an EBV latency III phenotype, characteristics similar to those of PTLD.2 Indeed, LCLs are good in vitro models to understand the pathogenesis of, and to discover novel therapeutic targets for, PTLD.
EBV encodes several proteins which mimic or associate with host cellular factors and these factors are well documented in B-cell survival, differentiation, proliferation, and immortalization.1 (1) The latent membrane protein 1 (LMP1) acts as a constitutively activated CD40 receptor, which provides signals of survival and differentiation in B cells. In LMP1 transgenic mice, constitutively activated LMP1 drives B-cell lymphoma formation, suggesting that LMP1 acts as an oncogene and plays an important role in EBV-mediated tumorigenesis.1,3 (2) The latent membrane protein 2A (LMP2A) mimics a functional B-cell receptor (BCR). It interacts with Syk and Lyn tyrosine kinases to deliver survival signals through PKC and intracellular calcium. In LMP2A transgenic mice, LMP2A can rescue BCR-lacking B cells from apoptosis, suggesting that the BCR-driven survival signals can be replaced by LMP2A.4 (3) The Epstein-Barr nuclear antigen 2 (EBNA2) mimics Notch signaling. It interacts with the DNA-binding protein RBPJκ/CBP to interfere with Notch-mediated inhibition of B-cell proliferation and differentiation.1 (4) Zta, a transactivator, mimics a functional and structural AP-1 protein. It plays an important role in regulating cytokine expression by immune cells, such as IL-13.5 (5) BCRF-1, which is an IL-10 homologue, serves as a growth factor for B cells and suppresses antiviral immune responses.6 So, we wondered whether any other specific cellular genes are involved in EBV-mediated immortalization. In this study, tyrosine kinases are our target.
Among the proteins encoded by the huge human genome, about ninety protein tyrosine kinases (PTKs) have been defined.7 PTKs are tightly regulated in cells because they play vital roles in crucial biologic functions, such as survival, proliferation, differentiation, and development.8 Dysregulation of PTK activity is one of the most common mechanisms leading to cellular transformation and cancer formation.7 Of note, the discovery of PTKs stemmed from the study of an oncogenic virus, Rous sarcoma virus encoding v-src.9 Generally, viruses can activate PTKs to transform host cells in many ways. One is when a virus genome encodes a PTK that is constitutively activated and expressed, and that PTK transforms the host cells, such as v-src in Rous sarcoma virus, v-kit in Hardy-Zuckerman-4 feline sarcoma virus, and v-abl in Abelson murine leukemia virus.10 Another mechanism involves viral products that function as ligands of PTKs and activate kinase activity. For example, v-sis of Simian sarcoma virus is a homologue of PDGF and can activate the tyrosine kinase PDGFR.10 Thus, oncogenic viruses may alter the activity of PTKs to promote their oncogenicity. However, the role of PTKs in EBV infection, which may promote the unlimited proliferation of LCLs, has not been explored yet. Taking the advantage of systematical analysis by kinase display assays, we found that Recepteur d'Origine Nantais (RON) tyrosine kinase is up-regulated in LCLs.
RON belongs to the c-Met family of receptor tyrosine kinases.11 RON is a heterodimer membrane protein composed of a transmembrane β-chain, which has tyrosine kinase activity, and a short extracellular α-chain linked by one disulfide bond.12 Expression of RON, driven by a specific tissue promoter in distal lung epithelial cells, results in the formation of multiple lung adenomas in mice.13 In clinical studies, RON overexpression has been reported in several human carcinomas, including breast cancer, head and neck tumors, colorectal carcinoma, ovarian cancer, and pancreatic cancer.14-16 Knockdown of RON leads to apoptosis of colorectal carcinoma cells and pancreatic cancer, while blockage of expression of RON reduces metastasis in colon carcinoma, suggesting a requirement for RON in tumor cell growth and its diverse roles in carcinogenesis.17-19
Our results from kinase display assays revealed that RON is overexpressed in LCLs but undetectable in primary B cells. Furthermore, the EBV oncogenic protein LMP1 was shown to induce RON through the NF-κB signaling pathway. Biologically, RON expression was found to be essential for LCL proliferation. To our knowledge, this is the first reported tyrosine kinase crucial for EBV-mediated immortalization. Impressively, we found that there is a correlation between LMP1 and RON expression in PTLD biopsies, implying that LMP1-induced RON expression is involved in the pathogenesis of EBV-associated PTLD and may be a novel therapeutic target for PTLD.
Methods
B-cell purification and EBV infection
PBMCs were obtained from anonymous donors (Taiwan Blood Service Foundation, Taipei, Taiwan) and CD19-positive B cells were purified using Dynabeads (Invitrogen), according to the manufacturer's instructions. The methods of EBV virion (B95-8 strain) production and EBV infection of primary B cells have been described previously.5 Experiments of human samples have been approved by Institutional Review Boards of National Taiwan University Hospital (Taipei, Taiwan).
Cell culture
EBV-negative Akata and BJAB cells are Burkitt lymphoma cell lines. Stable LMP1-expressing and vector control BJAB cells were established by electroporation (Neon kit; Invitrogen) of 0.3 μg of LMP1 and vector plasmids with a pulse of 1400 V for 30 ms and cells were selected with 5 μg/mL puromycin (Sigma-Aldrich) for 4 weeks. All lymphoid cell lines were cultured in complete RPMI medium (containing 10% FCS, 1mM glutamine, 100 U/mL penicillin and 100 μg/mL streptomycin). Experiments of LCL were performed from various cultured days ranged from 50-200 after EBV infection.
Kinase display assay
The tyrosine kinase display assays were described in a previous report.20 Briefly, total RNA was extracted and cDNA of PTKs was generated using degenerate primers designed for the conserved kinase domain. The PTK cDNAs were amplified with [γ-33P]–labeled primers.21 The resulting 170-bp PCR products were gel-purified and digested separately with 16 restriction enzymes. The digestion products were resolved in a denaturing polyacrylamide gel. The patterns of restriction enzyme digestion were identified for each specific PTK in the data bank.21
Plasmids
The EBNA1 (pSG5-EBNA1)–, Rta (pSG5-Rta)–, LMP2A (pSG5-LMP2A)–, and Zta (pRC-Zta)–expressing plasmids were as reported previously.5 The pPuro and LMP1 expressing plasmids are gifts from Dr Jen-Yang Chen (National Health Research Institutes, Miaoli, Taiwan).22 The pCMV-based RON expression and control plasmids are gifts from Dr Hsing-Jien Kung (UC Davis Cancer Center, Sacramento, CA). The siLMP1, siRON and siGFP plasmids were constructed by insertion of siRNA sequences into the pSuper plasmids at 5′-BglII and 3′-HindIII sites. The siRNA was targeted to the sequences 5′-GGGCGACAGAAATGAGAGT-3′ of RON and 5′-GCTCATTATTGCTCTCTAT-3′ of LMP1. The siGFP plasmids were described previously.23
RNA extraction and quantitative RT-Q-PCR
Total RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RT and quantitative PCR (RT-Q-PCR) have been described previously.24 The RON mRNAs were amplified by PCR using forward primer: 5′-ATCCACCCAGTGCCAACCTAGTT-3′ and reverse primer: 5′-GCAGCAGGATACCAAGGAGCGT-3′. The hypoxanthine phosphoribosyltransferase (HPRT) primers were described previously.20 RON and GAPDH mRNAs were detected using the TaqMan primer/probe sets Hs00234013 and Hs99999905, respectively (Applied Biosystems). β-actin mRNA was detected using Roche universal probe (no. 64) and forward primer, 5′-CCAACCGCGAGAAGATGA-3′ and reverse primer, 5′-CCAGAGGCGTACAGGGATAG-3′.
Western blotting and Abs
Cells were harvested in cell lysis buffer (20mM Tris-HCl, 150mM NaCl, 1mM Na2EDTA, 1mM EGTA, 1% Triton X-100, 2.5mM sodium pyrophosphate, 1mM β-glycerophosphate, 1mM sodium orthovanadate, and 1mM PMSF). Cell lysates (12-25 μg) were resolved by electrophoresis in 8% SDS–polyacrylamide gels and electrotransferred onto PVDF membranes (Millipore). Five-percent nonfat milk blocked blots were incubated with primary Abs at 4°C overnight and PBST-washed blots were incubated with HRP-conjugated secondary Abs (Jackson ImmunoResearch Laboratories) at room temperature for 1 hour. The target proteins were detected by using the Western Lightning chemiluminescence reagent (Perkin Elmer Life Sciences). Abs were used in this study as followed: RON-β (clone: C20; Santa Cruz Biotechnology), β-actin (clone: AC-15; Sigma-Aldrich), GAPDH (clone: 6C5; Biodesign), phospho-IκB-α Ser32/36 (clone: 9246; Cell Signaling), LMP1(clone: CS1-4; DAKO), p65 (clone: C20; Santa Cruz Biotechnology), phospho-Akt Ser473 (clone: 9271; Cell Signaling Technology), Akt (clone: H136; Santa Cruz Biotechnology), phospho-JNK Thr183/Tyr185 (clone: 9251; Cell Signaling Technology), JNK (clone: 06-748; Millipore), phospho-p38 Thr180/Tyr182 (clone: 9211; Cell Signaling Technology), p38 (clone: 9212; Cell Signaling Technology), phospho-ERK Thr202/Tyr204 (clone: 9106; Cell Signaling Technology), ERK (clone: K23; Santa Cruz Biotechnology), CD20 (clone: L26; DAKO), and EBNA1 (clone: NPC47).25
Electroporation
Cells were electroporated using a Neon kit (Invitrogen) according to the manufacturer's instructions. Briefly, cells were washed with PBS and resuspended in 10 μL of R buffer (provided in the Neon kit) and mixed with the relevant plasmids before electroporation. After pulse analysis, the samples were transferred into fresh medium and then incubated for the indicated time before being subjected to further assays. In general, 2-4 × 105 cells were electroporated with 0.3-0.8 μg of plasmids with a pulse of 1350-1400 V for 30 ms. For compensation assay, LCLs were electroporated with 0.8 μg of specific siRNA plasmids with the pulse of 1400 V for 30 ms and incubated for 3 days then transfected with 0.2 μg of RON expression or vector plasmids for another 3 days.
Preparation and infection of LMP1 and shLMP1-expressing lentiviruses
The LMP1-expressing lentivirus plasmids, pSIN-LMP1, were constructed by the insertion of a full-length LMP1 cDNA into pSIN vector at 5′-NdeI and 3′-MluI sites. The LMP1-deleted CTAR1 (ΔCTAR1) and deleted CTAR2 (ΔCTAR2) plasmids were constructed with the deletion of LMP1 amino acids (aa) 194 to 232, aa 351 to 386, by site-directed mutagenesis in pSIN-LMP1 expression plasmids with the following primers: ΔCTAR1 primer (5′-CCATGGACAACGACACAGTGATGACGGACCCCCACTCTG-3′) and ΔCTAR2 primer (5′-GCGGCGGTCATAGTCATGATTAAGGCCATGGCGGCG-3′). The LMP1-deleted CTAR1/2 (ΔCTAR1/2) plasmids were constructed by site-directed mutagenesis in pSIN-LMP1 ΔCTAR2 expression plasmids with the ΔCTAR1 primers. The shLMP1 lentivirus plasmids were constructed by the insertion of 5′-GCTCATTATTGCTCTCTAT-3′ of LMP1 into the pLKO.1 plasmids at the 5′-AgeI and 3′-EcoRI sites. The method of production and infection with lentiviruses was described previously.5 For lentivirus infection, 2-5 × 105 cells were infected with lentiviruses at the indicated multiplicity of infection (MOI) of 0.2-4.
Treatment with NF-κB inhibitor
EBV-negative Akata cells were infected with pSIN-vector or pSIN-LMP1 lentiviruses at the MOI of 2 for 3 days, then reseeded in 1 × 106 cells/mL and treated with various concentrations of NF-κB inhibitor (BAY 11-7082; Merck) for 48 hours.
ChIP assay
Briefly, BJAB vector control and LMP1-expressing cells were harvested and the complex of DNA and p65 was immunoprecipitated using anti-p65 Ab. The DNA was extracted and analyzed by PCR with the RON promoter spanning the p65-binding sites. The PCR primers and conditions were according to the report of Narasimhan et al.26 The amplification of the GAPDH promoter region was as described previously.5
[3H]-thymidine incorporation assay
LCLs were electroporated as described above and then plated at 1 × 104 cells per well in 96 well plates for 1, 2, 4, and 6 days. Before the indicated time point, 1 μCi [3H]-thymidine (Moravek Biochemicals) was added and incubated for 18 hours. The cells were harvested and the incorporated [3H]-thymidine was measured using a β-counter (LS6000 IC; Beckman).
IHC assay
PTLD specimens were obtained from National Taiwan University Hospital. Formalin fixed and paraffin-embedded sections were de-paraffinized and rehydrated. Immunohistochemistry (IHC) assays were performed by using Super Sensitive Link-Label IHC Detection System (BioGenex) according to the manufacturer's protocol. Samples were heated in 10mM citrate buffer (pH 6.0) for 10 minutes for Ag retrieval, reacted with 3% hydrogen peroxide for blocking endogenous peroxidase activities, and followed by blocked with 5% FCS for 1 hour. Sections were incubated with adequate Ab against RON, LMP1, or CD20, and then reacted with the biotinylated secondary Ab. The signal was presented with diaminobenzidine solution and counterstained with hematoxylin (BioGenex) which indicated the localization of nucleus. The photographs were observed by using a photomicroscope (Axioskop 40 FL; Zeiss).
In situ hybridization assay for EBER
An in situ hybridization assay was performed for EBER using an EBER probe (5′-CTCCTCCCTAGCAAAACCTCTAGGACGGCG-3′) labeled with a DIG-labeling Kit (Roche) according to the manufacturer's manual. Sections were deparaffinized, rehydrated, digested with proteinase K (Roche), hybridized with the EBER probe, and then incubated with alkaline phosphatase-conjugated DIG Ab (Roche). The signal was detected with nitroblue tetrazolium chloride/bromochloroindolyphosphate chromogen (Roche) and counterstained with nuclear fast red. The stained sections were observed by using a photomicroscope (Axioskop 40 FL; Zeiss).
Results
Expression of RON is induced in LCLs by EBV infection
Based on the important roles of PTKs in cellular survival and proliferation, we analyzed systematically PTK expression profiles by kinase display assays in primary B cells and EBV-immortalized LCLs. The expression of PTKs in LCLs was analyzed by amplification of PTK transcripts using degenerate primers designed for the conserved domains of PTKs.20,21 The benefits of this assay are that all the PTKs that can be detected simultaneously and each PTK has its own specific and unique pattern after restriction enzyme digestion.21 Based on the results of 3 restriction enzymes, MwoI, HaeIII, and RsaI, RON is a prominent, differentially expressed tyrosine kinase in LCLs (Figure 1A). To confirm the results of the kinase display assays, RON-specific primers were used for RT-PCR assay. Consistently, up-regulation of RON was detected in 6 LCLs but not in primary B cells (Figure 1B). The kinetics of RON expression were analyzed during EBV immortalization. Expression of RON transcripts and proteins reached peaks on day 7 and day 14, respectively, after EBV infection and then remained at a stable level until day 28 (Figure 1C). Expression of EBNA1 and LMP1 showed that EBV infection of primary B cells was successful. To confirm that up-regulation of RON was a common phenomenon during EBV immortalization, the expression of RON transcripts was detected in another 9 LCLs by RT-Q-PCR. The RON transcripts were increased about 7- to 50-fold in various LCLs, compared with primary B cells (Figure 1D). Thus, these results suggested that RON is induced in B cells on EBV infection and is constantly expressed when they become LCLs, even with some variation in the levels of expression.
RON is induced by EBV infection. (A) Total RNA was extracted from primary B cells and 3 LCLs and cDNA was synthesized using [33P]–labeled PTK degenerate primers. RON-specific cDNAs could be digested by 3 restriction enzymes, MwoI, HaeIII, and RsaI. (B) The amounts of RON transcripts were detected by RT-PCR using specific RON primers. Detection of HPRT served as an internal control. (C) Total RNA and protein were harvested at the days post-EBV infection indicated. RON transcripts were detected by RT-Q-PCR and the relative fold increase was normalized to the amounts of RON transcripts in uninfected primary B cells (top panel). The expression of RON, EBNA1, LMP1, and β-actin proteins was measured by Western blotting (bottom panel). SE indicates short exposure; and LE, long exposure. (D) The expression of RON transcripts in various EBV-immortalized LCLs was detected by RT-Q-PCR and the relative fold increase of RON transcripts was normalized to the amounts of RON transcripts in uninfected primary B cells.
RON is induced by EBV infection. (A) Total RNA was extracted from primary B cells and 3 LCLs and cDNA was synthesized using [33P]–labeled PTK degenerate primers. RON-specific cDNAs could be digested by 3 restriction enzymes, MwoI, HaeIII, and RsaI. (B) The amounts of RON transcripts were detected by RT-PCR using specific RON primers. Detection of HPRT served as an internal control. (C) Total RNA and protein were harvested at the days post-EBV infection indicated. RON transcripts were detected by RT-Q-PCR and the relative fold increase was normalized to the amounts of RON transcripts in uninfected primary B cells (top panel). The expression of RON, EBNA1, LMP1, and β-actin proteins was measured by Western blotting (bottom panel). SE indicates short exposure; and LE, long exposure. (D) The expression of RON transcripts in various EBV-immortalized LCLs was detected by RT-Q-PCR and the relative fold increase of RON transcripts was normalized to the amounts of RON transcripts in uninfected primary B cells.
EBV-induced expression of RON is triggered by LMP1
To determine which EB viral gene products contribute directly to RON expression, EBV-negative Akata cells were transfected with several important EB viral genes, including EBNA1, LMP1, LMP2A, Zta, and Rta. As shown in Figure 2A and B, RON was increased in LMP1-expressing cells, but not in other viral transfectants. Furthermore, RON expression could be induced by LMP1 in a dose-dependent manner at both the transcriptional and translational levels (Figure 2C). Thus, these results suggested that LMP1 can regulate RON expression. To verify further whether LMP1 is responsible for RON expression in EBV-immortalized LCLs, the expression of LMP1 was inhibited using an shRNA approach. The knockdown of LMP1 expression in LCLs led to significant suppression of RON expression (Figure 2D). These results supported the finding that LMP1 can trigger RON expression not only in LMP1-expressed EBV-negative Akata cells but also in EBV-immortalized LCLs.
LMP1 mediates RON expression. EBV-negative Akata cells were electroporated with plasmids expressing EBNA1, LMP1, LMP2A, Zta, or Rta, as indicated. Total RNA and protein were harvested from each transfectant at 72 hours posttransfection. (A) RON transcripts were measured by RT-Q-PCR and the relative fold increase was normalized to the amounts of RON transcripts in vector control cells. (B) Expression of RON, EBNA1, LMP2A, Rta, LMP1, Zta, and GAPDH was detected by Western blotting. (C) EBV-negative Akata cells were infected with pSIN vector or pSIN-LMP1-expressing lentiviruses at the MOI indicated for 5 days. The RON transcripts were detected by RT-Q-PCR and the relative fold increase was normalized to the amounts of RON transcripts in pSIN lentivirus-infected cells (top panel). The protein expression of RON, LMP1, and β-actin was measured by Western blotting (bottom panel). (D) LCLs (2 × 105) were infected with shLuciferase or shLMP1 lentiviruses at an MOI of 4 for 5 days. The RON transcripts were detected by RT-Q-PCR and the relative fold expression of RON was normalized to the amounts of RON transcripts in shLuciferase lentivirus-infected cells (top panel). The protein expression of RON, LMP1 and GAPDH was measured by Western blotting (bottom panel).
LMP1 mediates RON expression. EBV-negative Akata cells were electroporated with plasmids expressing EBNA1, LMP1, LMP2A, Zta, or Rta, as indicated. Total RNA and protein were harvested from each transfectant at 72 hours posttransfection. (A) RON transcripts were measured by RT-Q-PCR and the relative fold increase was normalized to the amounts of RON transcripts in vector control cells. (B) Expression of RON, EBNA1, LMP2A, Rta, LMP1, Zta, and GAPDH was detected by Western blotting. (C) EBV-negative Akata cells were infected with pSIN vector or pSIN-LMP1-expressing lentiviruses at the MOI indicated for 5 days. The RON transcripts were detected by RT-Q-PCR and the relative fold increase was normalized to the amounts of RON transcripts in pSIN lentivirus-infected cells (top panel). The protein expression of RON, LMP1, and β-actin was measured by Western blotting (bottom panel). (D) LCLs (2 × 105) were infected with shLuciferase or shLMP1 lentiviruses at an MOI of 4 for 5 days. The RON transcripts were detected by RT-Q-PCR and the relative fold expression of RON was normalized to the amounts of RON transcripts in shLuciferase lentivirus-infected cells (top panel). The protein expression of RON, LMP1 and GAPDH was measured by Western blotting (bottom panel).
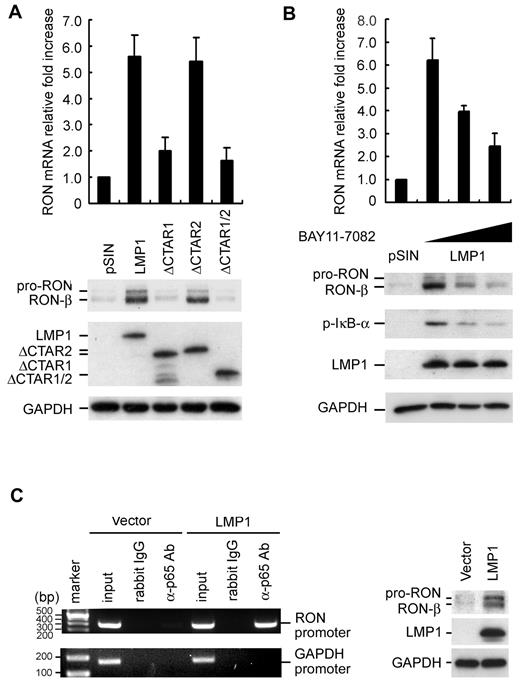
LMP1 regulates RON expression through NF-κB signaling
Structurally, LMP1 contains 6 transmembrane domains and a long C-terminal region with 2 major CTAR1 and CTAR2 domains, which associate with TNFR-associated factors, TNFR-associated death domain protein, and activate PI3K/Akt, MAPKs, IKK, and NF-κB–inducing kinase, and then constitutively activate AP-1 and NF-κB pathways.3 To further explore how LMP1 regulates RON expression, we first analyzed the contribution of 2 functional domains of LMP1 to the induction of RON. EBV-negative Akata cells were infected with lentiviruses containing pSIN vector control, LMP1 wild-type, LMP1ΔCTAR1, LMP1ΔCTAR2, or LMP1ΔCTAR1/2. Compared with the vector control, LMP1-wt and ΔCTAR2 induced RON by approximately 6-fold. However, ΔCTAR1 and ΔCTAR1/2 LMP1 significantly lost the capacity to induce the expression of RON (Figure 3A). These data suggest that the downstream signaling molecules of CTAR1 pathways are required for RON induction. It is known that CTAR1 activates the NF-κB signaling pathway and Narasimhan et al reported that p65 is the key transcription factor for RON expression.3,26 Therefore, we investigated whether LMP1 induced RON through p65. As shown in Figure 3B, LMP1 did increase RON expression; however, LMP-mediated RON expression was significantly reduced in the presence of BAY11-7082, an inhibitor of NF-κB. The same result was observed in BJAB cells constitutively expressing LMP1 (data not shown). A ChIP assay was performed to determine whether p65 binds directly to the RON promoter in vivo. As shown in Figure 3C, p65 binds the RON promoter directly in the LMP1 transfectants but not in the vector control cells. To investigate further whether NF-κB activity was essential for RON expression in LCLs, LCLs were treated with BAY11-7082. As shown in Figure 4A, inhibition of NF-κB activity significantly decreased RON transcripts and proteins in LCLs. Furthermore, knockdown of LMP1 using an shRNA approach abolished p65 binding to the RON promoter in LCLs (Figure 4B). Taken together, these data suggested that LMP1-induced RON expression is mediated by transducing p65 binding to the RON promoter in EBV-immortalized LCLs.
LMP1-induced RON expression is through its CTAR1 domain–mediated NF-κB activation. (A) EBV-negative Akata cells were infected with pSIN, LMP1, ΔCTAR1, ΔCTAR2, or ΔCTAR1/2-expressing lentiviruses at an MOI of 2 for 5 days. The RON transcripts were detected by RT-Q-PCR and the relative fold increase of RON was normalized to the amounts of RON transcripts in pSIN lentivirus-infected cells (top panel). The protein expression of RON, full-length LMP-1, CTAR1-deleted LMP1, CTAR2-deleted LMP1, CTAR1/2-deleted LMP1, and GAPDH was measured by Western blotting (bottom panel). (B) EBV-negative Akata cells were infected with pSIN or pSIN-LMP1 lentiviruses for 3 days and then cells were reseeded at a density of 1 × 106 cells/mL. Cells were treated 2.5μM or 5μM BAY11-7082 for 48 hours. Total RNA and protein were harvested and the expression of RON transcripts was measured by RT-Q-PCR and the relative fold increase of RON was normalized to the amounts of RON transcripts in pSIN lentivirus-infected cells (top panel). The protein expression of RON, p-IκB-α, LMP1, and GAPDH was measured by Western blotting (bottom panel). (C) Cell lysates were harvested from vector or LMP1-expressed BJAB cells. The complexes of DNA and p65 were immunoprecipitated using anti-p65 Ab or rabbit IgG. RON promoter DNA and control GAPDH promoter DNA were detected in the immunoprecipitates by PCR. Total DNA was harvested from vector or LMP1-expressed BJAB cells and used as the input control (left panel). Vector and LMP1-expressing BJAB cells were harvested and total protein extracted. The expression of RON, LMP1, and GAPDH was measured by Western blotting (right panel).
LMP1-induced RON expression is through its CTAR1 domain–mediated NF-κB activation. (A) EBV-negative Akata cells were infected with pSIN, LMP1, ΔCTAR1, ΔCTAR2, or ΔCTAR1/2-expressing lentiviruses at an MOI of 2 for 5 days. The RON transcripts were detected by RT-Q-PCR and the relative fold increase of RON was normalized to the amounts of RON transcripts in pSIN lentivirus-infected cells (top panel). The protein expression of RON, full-length LMP-1, CTAR1-deleted LMP1, CTAR2-deleted LMP1, CTAR1/2-deleted LMP1, and GAPDH was measured by Western blotting (bottom panel). (B) EBV-negative Akata cells were infected with pSIN or pSIN-LMP1 lentiviruses for 3 days and then cells were reseeded at a density of 1 × 106 cells/mL. Cells were treated 2.5μM or 5μM BAY11-7082 for 48 hours. Total RNA and protein were harvested and the expression of RON transcripts was measured by RT-Q-PCR and the relative fold increase of RON was normalized to the amounts of RON transcripts in pSIN lentivirus-infected cells (top panel). The protein expression of RON, p-IκB-α, LMP1, and GAPDH was measured by Western blotting (bottom panel). (C) Cell lysates were harvested from vector or LMP1-expressed BJAB cells. The complexes of DNA and p65 were immunoprecipitated using anti-p65 Ab or rabbit IgG. RON promoter DNA and control GAPDH promoter DNA were detected in the immunoprecipitates by PCR. Total DNA was harvested from vector or LMP1-expressed BJAB cells and used as the input control (left panel). Vector and LMP1-expressing BJAB cells were harvested and total protein extracted. The expression of RON, LMP1, and GAPDH was measured by Western blotting (right panel).
The LMP1-NF-κB pathway is required for RON expression in EBV-harboring LCLs. (A) LCL-1 cells were treated with 2.5μM or 5μM BAY11-7082 for 48 hours. The RON transcripts were measured by RT-Q-PCR and the relative fold expression of RON was normalized to the amounts of RON transcripts in untreated LCLs (top panel). The expression of RON, p-IκB-α, LMP1, and GAPDH proteins was measured by Western blotting (bottom panel). (B) 2 × 105 LCL-1 cells were infected with control shRNA or LMP1 shRNA-expressing lentiviruses at an MOI of 4 for 5 days. A ChIP assay of RON promoter was carried out as described in Figure 3.
The LMP1-NF-κB pathway is required for RON expression in EBV-harboring LCLs. (A) LCL-1 cells were treated with 2.5μM or 5μM BAY11-7082 for 48 hours. The RON transcripts were measured by RT-Q-PCR and the relative fold expression of RON was normalized to the amounts of RON transcripts in untreated LCLs (top panel). The expression of RON, p-IκB-α, LMP1, and GAPDH proteins was measured by Western blotting (bottom panel). (B) 2 × 105 LCL-1 cells were infected with control shRNA or LMP1 shRNA-expressing lentiviruses at an MOI of 4 for 5 days. A ChIP assay of RON promoter was carried out as described in Figure 3.
LMP1-induced RON expression is required for growth of LCLs
RON has been shown to induce cell proliferation, scatter, motility, survival, and most of these effects have been reported in an epithelial cell model.16 However, the biologic activity of RON in B cells has not been investigated. Therefore, we explored whether LMP1-induced expression of RON was required for the growth of LCLs. The proliferation of LCLs with RON knockdown was monitored using an [3H]-thymidine incorporation assay at days 1, 2, 4, and 6 posttransfection. As shown in Figure 5A, knockdown of RON inhibited LCL proliferation. To establish further the relationship between LMP1 and RON in cell proliferation, LMP1-knockdown cells were transfected with vector control or RON-expressing plasmids. Reconstitution of RON could restore growth of LMP1-knockdown cells almost completely (Figure 5B). Thus, our results suggested that LMP1-induced RON expression contributed significantly to EBV-mediated proliferation of LCLs.
LMP1-induced RON expression plays a critical role in the proliferation of LCLs. (A) LCL-1 (left panel) and LCL-2 (right panel) cells were electroporated with control siRNA or RON siRNA plasmids. Before the indicated time point, 1 μCi [3H]-thymidine was added and incubation continued for another 18 hours. The amount of incorporated [3H]-thymidine was measured using a β-counter. (B) LCL-1 cells were electroporated with control siRNA or LMP1 siRNA plasmids for 3 days and then transfected with RON expression plasmids for another 3 days. The amount of incorporated [3H]-thymidine was measured using a β-counter (left panel). The total proteins were harvested and the expression of RON, LMP1 and GAPDH was detected by Western blotting (right panel). *P < .05 by Student t test.
LMP1-induced RON expression plays a critical role in the proliferation of LCLs. (A) LCL-1 (left panel) and LCL-2 (right panel) cells were electroporated with control siRNA or RON siRNA plasmids. Before the indicated time point, 1 μCi [3H]-thymidine was added and incubation continued for another 18 hours. The amount of incorporated [3H]-thymidine was measured using a β-counter. (B) LCL-1 cells were electroporated with control siRNA or LMP1 siRNA plasmids for 3 days and then transfected with RON expression plasmids for another 3 days. The amount of incorporated [3H]-thymidine was measured using a β-counter (left panel). The total proteins were harvested and the expression of RON, LMP1 and GAPDH was detected by Western blotting (right panel). *P < .05 by Student t test.
The downstream signaling pathways are triggered by RON in LCLs
As a receptor tyrosine kinase, RON acts by triggering an array of downstream signaling, including PI3K/Akt, ERK, JNK, and p38-dependent pathways in different cell types.16 To investigate further the molecular mechanism of RON in the growth of LCLs, we determined whether depleted RON affects these downstream signals in LCLs. As shown in Figure 6, knockdown of RON decreased the phosphorylation of Akt, ERK, JNK, and p38. The data suggest that these downstream signaling pathways, which are critical for LCL growth,27,28 may be activated by RON in LCLs.
Knockdown of RON affects its downstream signaling pathways in LCLs. LCL-1 cells were electroporated with control siRNA or RON siRNA plasmids for 3 days. Cell lysates were harvested and the phosphorylation and total of Akt, JNK, ERK, and p38 were detected by Western blotting.
Knockdown of RON affects its downstream signaling pathways in LCLs. LCL-1 cells were electroporated with control siRNA or RON siRNA plasmids for 3 days. Cell lysates were harvested and the phosphorylation and total of Akt, JNK, ERK, and p38 were detected by Western blotting.
RON expression is detectable in biopsies of PTLD
Both LCLs and PTLD display similar features in many respects, so we wondered whether RON overexpression can be detected in PTLD biopsies. The expression of CD20, EBER, LMP1, and RON was examined in 9 paraffin-embedded PTLD tissues. The presence of EBV in PTLD was demonstrated by in situ hybridization of EBER, which indicates EBV transcription in the biopsies.1 All cases were positive for CD20 staining, a marker of B cells, and were also EBER positive. LMP1-positive cells are detected in 8 of 9 PTLD biopsies. Double positivity of LMP1 and RON expression was detected in 7 of 9 cases (Table 1). In positive cases, the staining signals of LMP1 and RON were both located in the cytoplasmic and membranous regions, the coexpression of LMP1 and RON in CD20-positive B cells being indicated by an arrow (Figure 7, 2 cases). The results indicated that RON expression correlated with that of LMP1, suggesting that RON may be involved in the pathogenesis of PTLD.
Patient demography, CD20, EBER, LMP1, and RON expression in PTLD biopsies
| Patient case no. . | Age/sex . | Origin of biopsy . | CD20 . | EBER . | LMP1 . | RON . |
|---|---|---|---|---|---|---|
| 1 | 35/M | Lymph node | + | + | + | + |
| 2 | 28/F | Lymph node | + | + | + | + |
| 3 | 11/M | Soft tissue, retroperitoneum | + | + | + | + |
| 4 | 27/F | Lymph node | + | + | + | + |
| 5 | 37/F | Lung | + | + | − | − |
| 6 | 1/M | Lymph node | + | + | + | + |
| 7 | 20/F | Lymph node | + | + | + | + |
| 8 | 47/M | Lymph node | + | + | + | − |
| 9 | 18/M | Lymph node | + | + | + | + |
| Patient case no. . | Age/sex . | Origin of biopsy . | CD20 . | EBER . | LMP1 . | RON . |
|---|---|---|---|---|---|---|
| 1 | 35/M | Lymph node | + | + | + | + |
| 2 | 28/F | Lymph node | + | + | + | + |
| 3 | 11/M | Soft tissue, retroperitoneum | + | + | + | + |
| 4 | 27/F | Lymph node | + | + | + | + |
| 5 | 37/F | Lung | + | + | − | − |
| 6 | 1/M | Lymph node | + | + | + | + |
| 7 | 20/F | Lymph node | + | + | + | + |
| 8 | 47/M | Lymph node | + | + | + | − |
| 9 | 18/M | Lymph node | + | + | + | + |
LMP1 indicates latent membrane protein 1; RON, Recepteur d'Origine Nantais; and PTLD, posttransplantation lymphoproliferative disorder.
LMP1 and RON are coexpressed in EBER-positive B cells in 2 PTLD biopsies. Formalin-fixed, paraffin-embedded PTLD consecutive sections of case nos. 1 and 4 were stained for the expression of CD20, EBER, LMP1, and RON. Positive CD20 staining is seen as a dark brown precipitate on the surfaces of the B cells. The EBER signal was characterized by a dense purple. Positive immunoreactivity against LMP1 and RON was seen as a light brown color. Co-localization among EBER, CD20, LMP1, and RON was observed using a Zeiss photomicrographic system (Axioskop 40 FL) equipped with Image-Pro Plus software and indicated by arrow. Magnification ×400 (scale bar, 25 μm).
LMP1 and RON are coexpressed in EBER-positive B cells in 2 PTLD biopsies. Formalin-fixed, paraffin-embedded PTLD consecutive sections of case nos. 1 and 4 were stained for the expression of CD20, EBER, LMP1, and RON. Positive CD20 staining is seen as a dark brown precipitate on the surfaces of the B cells. The EBER signal was characterized by a dense purple. Positive immunoreactivity against LMP1 and RON was seen as a light brown color. Co-localization among EBER, CD20, LMP1, and RON was observed using a Zeiss photomicrographic system (Axioskop 40 FL) equipped with Image-Pro Plus software and indicated by arrow. Magnification ×400 (scale bar, 25 μm).
Discussion
The involvement of PTKs in EBV-induced immortalization has not been investigated up to now. Here, we used kinase display assays to analyze systematically the expression profile of PTKs in LCLs and found that RON is overexpressed in LCLs but not in primary B cells (Figure 1). Furthermore, we showed that LMP1 can induce the expression of RON (Figure 2). Our results suggest that LMP1-induced cell proliferation is partially caused by RON overexpression (Figure 5B). This is the first report to define a functional role for RON in B cells.
Several EBV-regulated PTKs have been well-documented in epithelial cells. For instance, Zta, the important EBV lytic transactivator, up-regulates TKT and may contribute to the metastatic potential of nasopharyngeal carcinoma.20 In the case of LMP1, it has been reported that EGFR and c-Met are activated by LMP1 through NF-κB and Ets1, respectively.29-31 LMP2A can induce cell migration through Syk.32 Furthermore, EBV BARF0 dysregulates HER2/HER3 to promote oncogenic activity in breast cancer cells.33
Until now, only c-Mer PTK has been reported to be expressed in LCLs and expression of Lck is increased after EBV infection of primary B cells.34,35 However, the precise functions of c-Mer and Lck in EBV-induced immortalization have not been determined. Previous studies have shown that Axl, which belongs to the c-Mer family, is essential for Kaposi sarcoma cell growth and invasion.36 LMP2A mimics the structure of cellular BCR to recruit the Lyn and Syk kinases and provides the survival signals for B cells.1 Both Lyn and Syk are detectable in our LCLs and primary B cells, but do not vary at the transcriptional level.
In this study, we showed clearly that RON is overexpressed in EBV-immortalized LCLs and LMP1 is responsible for this up-regulation. In addition, we demonstrated that LMP1 induces RON expression in LCLs through its CTAR1 domain and stimulates p65 binding to NF-κB binding sites on the RON promoter (Figure 3 and 4). In an epithelial model, a series of novel studies indicated that LMP1 induces EGFR expression via CTAR1 to activate the formation and binding of p50/p50 homodimers and Bcl3 complex to the promoter of EGFR.29,30,37 According to these results, LMP1 may activate the distinct NF-κB subunits to induce the expression of a variety of cellular genes. So, it would be valuable to investigate further the detailed mechanism and signaling pathways involved in LMP1 induction of specific NF-κB complexes through CTAR1, thereby inducing RON expression in LCLs.
So far, 3 ways are known that regulation of aberrant RON activity may promote carcinogenesis, including autophosphorylation of RON because of gene overexpression or aberrant splicing, and activation of RON kinase by its ligand-macrophage stimulating protein.16,38 Here we demonstrate that LMP1-induced RON overexpression is responsible for proliferation of LCLs. Blockage of RON expression using an siRNA approach affected the proliferation of LCLs (Figure 5). Furthermore, knockdown of RON in LCLs decreased the phosphorylation of Akt, JNK, ERK, and p38, which are all known to act downstream of RON in various cell types (Figure 6). Of note, these pathways have been reported to be activated in LCLs and some are important for the growth of LCLs, even though the regulatory mechanisms are unknown.27,28 So, we assume that LMP1-induced RON can trigger downstream signaling to promote the proliferation of LCLs, which is one of the explanations why LMP1 is essential for EBV-induced immortalization.
Recently, the association between RON and various viruses has been studied. Jaagsiekte sheep virus (JSRV) causes a bronchoalveolar lung tumor in sheep that resembles bronchoalveolar carcinomas in humans. Hyaluronidase (HYAL2), a receptor for the JSRV envelope protein, acts as a negative regulator of RON activation; overexpression of JSRV envelope protein leads to binding to HYAL2, which releases RON to enhance cell transformation.39 In humans, the overexpression of RON in monocytes and macrophages decreases the binding activity of NF-κB and RNA polymerase II to the long terminal repeats of human immunodeficiency virus and inhibits proviral transcription.40
In EBV-associated diseases, Renne et al demonstrated that 6 PTKs, including RON, are expressed in Hodgkin/Reed-Sternberg cells of Hodgkin lymphoma and suggested that RON may be involved in the pathogenesis of Hodgkin lymphoma.41 Our results indicated that LMP1 induces RON expression, and that both LMP1 and RON are coexpressed in PTLD biopsies, suggesting that RON may be involved in the pathogenesis of PTLD (Figure 7, Table 1). This is the first study to show RON expression in PTLD biopsies. Furthermore, the common treatment of PTLD is to reduce immunosuppression to restore the immune responses to EBV. However, these may not recover fast enough to destroy malignant cells and the treatment also may increase the risk of graft rejection.42 Another therapy is based on the anti-CD20 Ab, rituximab. The efficacy of rituximab varies between 50% and 60% for complete regression of PTLD.43,44 Recently, a dual RON/MET inhibitor was shown to inhibit tumor growth significantly.45 Furthermore, Foretinib, a multitarget inhibitor of RON, c-Met, Axl, and VEGFR, was evaluated in a human phase 1 clinical trial.46
In conclusion, our data suggest that RON is overexpressed in patients with PTLD and may contribute to the pathogenesis of EBV-associated diseases. RON is a potential novel therapeutic target for lymphoproliferative diseases.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Tim J. Harrison of UCL Medical School (London, United Kingdom) for reviewing the manuscript critically.
This work was supported by National Science Council grants NSC 97-2320-B-002-003-MY3 and NSC100-2320-B-002-100-MY3, and National Health Research Institute grants NHRI-EX100-10031BI.
Authorship
Contribution: Y.-C.C. designed and performed experiments, analyzed the data, and co-wrote the manuscript; S.-J.L. designed experiments, analyzed the data, and co-wrote the manuscript; J.L. designed and performed experiments and analyzed the data; T.-H.Y. provided materials; C.-L.C. provided materials, performed experiments, and interpreted the data; P.-L.W. and J.-H.L. performed experiments; M.Y. provided materials; and C.-H.T. guided experimental design and co-wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Prof Ching-Hwa Tsai, PhD, Graduate Institute of Microbiology, College of Medicine, National Taiwan University, Room 719, No 1, 1st Section, Jen-Ai Road, Taipei 10051, Taiwan; e-mail: chtsai@ntu.edu.tw.

![Figure 1. RON is induced by EBV infection. (A) Total RNA was extracted from primary B cells and 3 LCLs and cDNA was synthesized using [33P]–labeled PTK degenerate primers. RON-specific cDNAs could be digested by 3 restriction enzymes, MwoI, HaeIII, and RsaI. (B) The amounts of RON transcripts were detected by RT-PCR using specific RON primers. Detection of HPRT served as an internal control. (C) Total RNA and protein were harvested at the days post-EBV infection indicated. RON transcripts were detected by RT-Q-PCR and the relative fold increase was normalized to the amounts of RON transcripts in uninfected primary B cells (top panel). The expression of RON, EBNA1, LMP1, and β-actin proteins was measured by Western blotting (bottom panel). SE indicates short exposure; and LE, long exposure. (D) The expression of RON transcripts in various EBV-immortalized LCLs was detected by RT-Q-PCR and the relative fold increase of RON transcripts was normalized to the amounts of RON transcripts in uninfected primary B cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/5/10.1182_blood-2011-02-335448/4/m_zh89991175600001.jpeg?Expires=1767716232&Signature=amgZZb3J3rB5xlH8eQ-EVtYa9EBJj7GxB3q98WZI8-6iSza2MZwDYh24PaLJi7P2uQEs-mZueD0xwFrdwioIjR27ICti750hneH3eITq72Bwh29Mv~2tA4gA5vRFh~tNWb4GibuXef79ubt9elL0Jr8zA-C9Wcrp4QHTsFxhvN8iHXjP4L3VpppFRioJbO09CFuq5djwTH77nCXCshUwpHq0MsZ~qU2u7wUhhKpNRqhxbZ3Gyi-DKkyje2DdY2SpS48u3cGgF26gKfOICx2rMY1G7UxKfCbX0FczmHSx3OG7n6e0S1JJb-p1okTWjEQjgeOLrL2xnn3~~~k2cbom7g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 5. LMP1-induced RON expression plays a critical role in the proliferation of LCLs. (A) LCL-1 (left panel) and LCL-2 (right panel) cells were electroporated with control siRNA or RON siRNA plasmids. Before the indicated time point, 1 μCi [3H]-thymidine was added and incubation continued for another 18 hours. The amount of incorporated [3H]-thymidine was measured using a β-counter. (B) LCL-1 cells were electroporated with control siRNA or LMP1 siRNA plasmids for 3 days and then transfected with RON expression plasmids for another 3 days. The amount of incorporated [3H]-thymidine was measured using a β-counter (left panel). The total proteins were harvested and the expression of RON, LMP1 and GAPDH was detected by Western blotting (right panel). *P < .05 by Student t test.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/5/10.1182_blood-2011-02-335448/4/m_zh89991175600005.jpeg?Expires=1767716232&Signature=e8EMc~uVrcwnfoO6eFgbTUU7K3vBtNvxJaaC2-FbMi3c36z3vJ-M7HbAYsw277g2e-lM5RqIxuQXS~EHTl0KI95nQbOeK7UpsEPg32Ycw1zufW8elk5hvX~0JahCptmUptIj5Mzt8CFh7jYYQgFnXqWrvgepbxpIgL4EWY8Hu4~GrUSQC5dO1ZeCAO~eL6i~j1EnUdhwEB29TGlFmEYJIngIji-O28K9Tj62loqtJ-i38IEJO3yXuL2IsN89dHAob0R1X-GExlUbacMjd4Dt-knRkL6Ugkp0E-UMY-hzsyGm2Mk6tBP6DoW0NZIQz9zyhWt0wgxnWkG7csuLDddUbw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal