Abstract
Familial hemophagocytic lymphohistiocytosis (FHL) is a potentially lethal genetic disorder of immune dysregulation that requires prompt and accurate diagnosis to initiate life-saving immunosuppressive therapy and to prepare for hematopoietic stem cell transplantation. In the present study, 85 patients with hemophagocytic lymphohistiocytosis were screened for FHL3 by Western blotting using platelets and by natural killer cell lysosomal exocytosis assay. Six of these patients were diagnosed with FHL3. In the acute disease phase requiring platelet transfusion, it was difficult to diagnose FHL3 by Western blot analysis or by lysosomal exocytosis assay. In contrast, the newly established flow cytometric analysis of intraplatelet Munc13-4 protein expression revealed bimodal populations of normal and Munc13-4–deficient platelets. These findings indicate that flow cytometric detection of intraplatelet Munc13-4 protein is a sensitive and reliable method to rapidly screen for FHL3 with a very small amount of whole blood, even in the acute phase of the disease.
Introduction
The granule-dependent cytotoxic pathway is a major immune effector mechanism used by cytotoxic T lymphocytes (CTLs) and natural killer (NK) cells.1 The pathway involves a series of steps, including cell activation, polarization of the lysosomal granules to the immunologic synapse, exocytosis of lytic proteins such as perforin and granzymes, and induction of apoptosis in the target cells.2 In addition to its central role in the defense against intracellular infections and in tumor immunity, this pathway also plays an important role in the regulation of immune homeostasis. Defects in the granule-dependent cytotoxic pathway result in a catastrophic hyperinflammatory condition known as hemophagocytic lymphohistiocytosis (HLH).1,3
HLH is a life-threatening syndrome of immune dysregulation resulting from the uncontrolled activation and proliferation of CTLs, which leads to macrophage activation and the excessive release of inflammatory cytokines.4,5 Clinical diagnosis of HLH is made on the basis of cardinal signs and symptoms including prolonged fever and hepatosplenomegaly, and by characteristic laboratory findings such as pancytopenia, hyperferritinemia, hypofibrinogenemia, increased levels of soluble IL-2 receptor, and low or absent NK cell activity.5,6 HLH can be classified into primary (genetic) or secondary (acquired) forms according to the underlying etiology, although this distinction is difficult to make in clinical practice.4,5
Familial hemophagocytic lymphohistiocytosis (FHL) encompasses major forms of primary HLH for which mutations in the genes encoding perforin (PRF1; FHL2),7 Munc13-4 (UNC13D; FHL3),8 syntaxin-11 (STX11; FHL4),9 and syntaxin-binding protein 2 (also known as Munc18-2) (STXBP2; FHL5)10,11 have been identified to date. Perforin is a cytolytic effector that forms a pore-like structure in the target cell membrane. Munc13-4, syntaxin-11, and Munc18-2 are involved in intracellular trafficking or the fusion of cytolytic granules to the plasma membrane and the subsequent delivery of their contents into target cells.1,12 Consequently, defective cytotoxic activity of CTLs and NK cells is one of the hallmark findings of FHL,7,8,13-16 although NK cell activity is also decreased in some cases of secondary HLH.15,17-20
Prompt and accurate diagnosis of FHL is mandatory to initiate life-saving immunosuppressive therapy and to prepare for hematopoietic stem cell transplantation. Detection of perforin expression in NK cells with flow cytometry is a reliable method to screen for FHL2.21 Another test analyzes the expression of CD107a on the surface of NK cells, which marks the release of cytolytic granules.22 Reduced expression of CD107a implies impaired degranulation of NK cells and predicts a likelihood of FHL3.23 However, this analysis is not available in some patients with extremely reduced NK cell numbers, such as during the acute phase of HLH.19 In addition, NK-cell degranulation is also impaired in FHL424 and FHL5,10,11 making it impossible to differentiate these disorders.
We reported previously that Munc13-4 protein is expressed in platelets and regulates the secretion of dense core granules.25 Herein we report that Munc13-4 is expressed far more abundantly in platelets than in PBMCs. We also describe the development of a new method to screen for FHL3 rapidly by detecting intraplatelet Munc13-4 expression through flow cytometry.
Methods
Patients
Between January 2008 and March 2010, whole blood samples from 85 patients were screened for FHL3. The patients had been clinically diagnosed with HLH by their referring physicians and were suspected of possible FHL. Characteristics of the enrolled patients are summarized in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). As a control, blood obtained from healthy adults at the time of patient sampling was shipped for screening along with the patient samples. Before the laboratory studies were performed, informed consent was obtained from the patients and their parents, in accordance with the institutional review board of Kyoto University Hospital and the Declaration of Helsinki.
Preparation of PBMCs and platelet samples
Whole blood samples treated with EDTA were centrifuged gently at 100g for 10 minutes, and platelets were collected from the supernatant plasma layer. Alternatively, platelets were prepared from small aliquots of blood samples by lysing red blood cells with ammonium chloride. PBMCs were obtained by Ficoll-Hypaque density gradient centrifugation from the remaining sample. CD4+, CD8+, CD14+, CD19+, and CD45+ cells were separated from PBMCs using an AutoMACS Pro (Miltenyi Biotec) and magnetic bead–conjugated mAbs according to the manufacturer's instructions. Flow cytometric analysis revealed that each cell population contained > 95% CD4+, CD8+, CD14+, CD19+, and CD45+ cells (data not shown).
Mutation analysis
Genomic DNA was isolated from the PBMCs of patients with defective Munc13-4 expression using standard procedures. Primers were designed for the amplification and direct DNA sequencing of the UNC13D-coding exons, including the adjacent intronic sequences for the identification of splice-site variants. Primer sequences are available upon request. Products were sequenced directly with an ABI3130 genetic analyzer (Applied Biosystems).
Antibodies
Rabbit polyclonal antibodies raised against the N-terminal region (residues 1-262)25 and full-length human Munc13-4 protein were used as primary antibodies for Western blot and flow cytometric analysis, respectively. Rabbit polyclonal anti-integrin αIIb (Santa Cruz Biotechnology) and mouse polyclonal anti–β-actin (Sigma-Aldrich) antibodies were used as primary antibodies for Western blotting. The mAbs used in the flow cytometric analysis were FITC-conjugated anti-CD3 (SK7; BD Pharmingen), phycoerythrin (PE)–conjugated anti-CD41a (HIP8; BD Pharmingen); allophycocyanin-conjugated anti-CD56 (N901; Beckman Coulter), and PE-conjugated anti-CD107a (H4A3; eBioscience).
Western blot analysis
Cell extracts were fractionated by SDS-PAGE, and the fractionated proteins were electrotransferred onto polyvinylidene fluoride membranes. The membranes were blocked overnight in blocking buffer (5% skim milk) and incubated for 1 hour at room temperature with the primary antibodies, followed by HRP-conjugated anti–rabbit or anti–mouse IgG polyclonal antibodies (Santa Cruz Biotechnology). Specific bands were visualized by the standard enhanced chemiluminescence method.
Flow cytometric analysis of Munc13-4 protein
After surface staining with anti-CD41a mAbs, platelets were fixed and permeabilized by Cytofix/Cytoperm (BD Biosciences) and washed 3 times with Perm/Wash buffer (BD Biosciences). After nonspecific reactions were blocked with Chrome-Pure human IgG (Jackson ImmunoResearch Laboratories), rabbit polyclonal antibody against the full-length human Munc13-4 protein was added, followed by FITC-conjugated donkey anti–rabbit IgG (Jackson ImmunoResearch Laboratories). Platelets were gated on the basis of their appearance on forward- and side-scatter plots in log/log scale and by CD41a expression. The gated platelets were analyzed for Munc13-4 expression by flow cytometry (FACSCalibur; BD Biosciences).
Lysosomal degranulation assays
To quantify lysosome exocytosis by NK cells, 2 × 105 PBMCs were mixed with 2 × 105 human erythroleukemia cell line K562 cells and incubated for 2 hours in complete medium (RPMI 1640 medium supplemented with 2mM l-glutamine and 10% FCS) at 37°C in 5% CO2. Cells were resuspended in PBS supplemented with 2% FCS and 2mM EDTA; stained with anti-CD3–FITC, anti-CD56–allophycocyanin, and anti-CD107a–PE mAbs; and analyzed by flow cytometry.
Platelet exocytosis of the lysosomal granules was analyzed as described previously26 but with a minor modification. Briefly, platelets were suspended in PBS containing 2mM EDTA and PE-conjugated anti-CD107a mAb, stimulated with 5 U/mL of thrombin (Wako Pure Chemical Industries) for 10 minutes at 25°C, and immediately analyzed by flow cytometry. The degranulation index of platelets was calculated as: (mean fluorescence value of stimulated sample − mean fluorescence value of nonstimulated sample)/mean fluorescence value of nonstimulated sample.
Statistical analysis
Statistical analyses were performed with 1-way ANOVA followed by the Tukey post hoc test to compare multiple groups, with a P < .05 level considered to be significant.
Results
Diagnosis of FHL3 by Western blot analysis using platelets
Before screening for FHL3, the Munc13-4 expression level was compared between platelets and PBMCs. Munc13-4 expression in platelets was approximately 10 times higher than that in PBMCs (Figure 1A). CD8+ cells expressed a similar level of Munc13-4 protein as other PBMC cell types (Figure 1B). Similar amounts of platelet- and PBMC-derived proteins could be obtained from a sample (data not shown). Therefore, platelets were used to perform Western blotting to screen for Munc13-4 deficiency. Of the 85 patients screened, 6 patients were diagnosed with FHL3 (Figure 1C). Munc13-4 protein was barely detected in the platelets of each FHL3 patient regardless of the gene mutation (Table 1). For each sample, no more than 1 mL of whole blood was required to perform the analysis.
Diagnosing FHL3 by Western blotting using platelet protein. The amount of Munc13-4 protein expression was compared between platelets and PBMCs (A) and among platelets, CD8+, CD4+, CD19+, and CD14+ cells (B) by Western blotting. A representative result of 5 independent experiments is shown. (C) Six FHL3 patients were diagnosed by Western blotting for Munc13-4 protein using platelets.
Diagnosing FHL3 by Western blotting using platelet protein. The amount of Munc13-4 protein expression was compared between platelets and PBMCs (A) and among platelets, CD8+, CD4+, CD19+, and CD14+ cells (B) by Western blotting. A representative result of 5 independent experiments is shown. (C) Six FHL3 patients were diagnosed by Western blotting for Munc13-4 protein using platelets.
UNC13D gene mutations of FHL3 patients
| Patient . | Age at onset . | Gender . | Mutation . | Genotype . | Predicted effect . |
|---|---|---|---|---|---|
| P1 | 14 days | Female | c.1596 + 1G → C | Homo | Splice error |
| P2 | 2 months | Male | c.322–1G → A | Hetero | Splice error |
| c.990G → C | Hetero | p.Q330H | |||
| c.3193C → T | Hetero | p.R1065X | |||
| P3 | 12 months | Female | c.754–1G → C | Hetero | Splice error |
| c.2485delC | Hetero | p.L829fs | |||
| P4 | 4 months | Female | c.754–1G → C | Hetero | Splice error |
| c.1799C → T | Hetero | p.T600M | |||
| c.1803C → A | Hetero | p.Y601X | |||
| P5 | 2 months | Female | c.754–1G → C | Hetero | Splice error |
| c.1596 + 1G → C | Hetero | Splice error | |||
| P6 | 5 months | Male | ND | ND | ND |
| Patient . | Age at onset . | Gender . | Mutation . | Genotype . | Predicted effect . |
|---|---|---|---|---|---|
| P1 | 14 days | Female | c.1596 + 1G → C | Homo | Splice error |
| P2 | 2 months | Male | c.322–1G → A | Hetero | Splice error |
| c.990G → C | Hetero | p.Q330H | |||
| c.3193C → T | Hetero | p.R1065X | |||
| P3 | 12 months | Female | c.754–1G → C | Hetero | Splice error |
| c.2485delC | Hetero | p.L829fs | |||
| P4 | 4 months | Female | c.754–1G → C | Hetero | Splice error |
| c.1799C → T | Hetero | p.T600M | |||
| c.1803C → A | Hetero | p.Y601X | |||
| P5 | 2 months | Female | c.754–1G → C | Hetero | Splice error |
| c.1596 + 1G → C | Hetero | Splice error | |||
| P6 | 5 months | Male | ND | ND | ND |
Mutations were checked for single nucleotide polymorphisms using the dbSNP Build 132 database from the National Center for Biotechnology Information.
X indicates stop; fs, frame shift; and ND, not determined.
Difficulty in diagnosing FHL3 in the acute phase of the disease
Patients in the acute phase of the disease who require screening for FHL often receive platelet transfusions because of thrombocytopenia.4-6 To study the effect of transfused platelets on screening results, FHL3 screening was attempted in a patient receiving platelet transfusions. As expected, Western blotting using platelets could not detect Munc13-4 deficiency because of the normal expression of the protein in the transfused platelets (Figure 2A left column). Surprisingly, Western blotting using PBMCs also could not clearly identify Munc13-4 deficiency because a substantial number of platelets were present in the PBMCs obtained by the standard method (Figure 2A right column). By positively selecting CD45+ cells and removing platelets, it was found that a considerable amount of the Munc13-4 protein detected in PBMC samples obtained by standard density gradient centrifugation was actually derived from the contaminating platelets (Figure 2B).
Effect of platelet transfusion on Western blot analysis. (A) Western blotting analysis for Munc13-4 expression using platelets and PBMCs from an FHL3 patient (P5) receiving platelet transfusions during the acute phase of the disease. (B) The expression of Munc13-4 was compared between PBMCs obtained by density gradient centrifugation (DGC) and CD45+ cells obtained by magnetic sorting from healthy controls. A representative result of 3 independent experiments is shown.
Effect of platelet transfusion on Western blot analysis. (A) Western blotting analysis for Munc13-4 expression using platelets and PBMCs from an FHL3 patient (P5) receiving platelet transfusions during the acute phase of the disease. (B) The expression of Munc13-4 was compared between PBMCs obtained by density gradient centrifugation (DGC) and CD45+ cells obtained by magnetic sorting from healthy controls. A representative result of 3 independent experiments is shown.
We performed a NK-cell degranulation assay for every referred sample and found the assay to be defective for every FHL3 patient identified (data not shown). All of the other patients showed a normal release of lysosomal granules by NK cells; however, the analysis could not be performed in some patients because of the extremely low NK-cell number during the acute phase of the disease (data not shown).
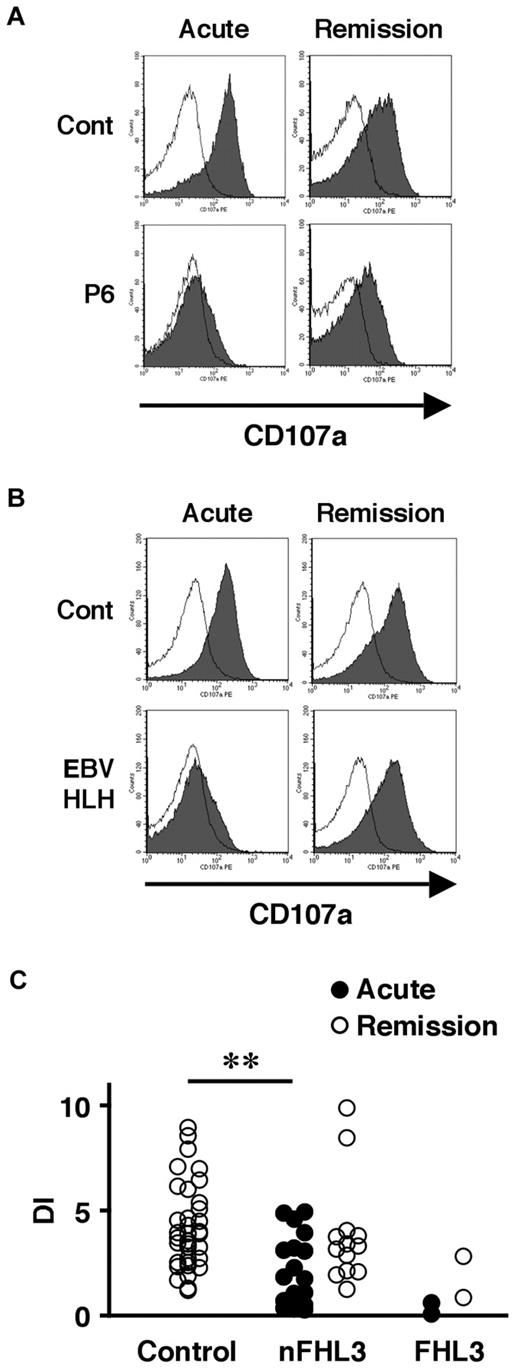
We also examined the lysosomal granule release of platelets in 31 patients to determine whether this assay could be used as a screening method for FHL3. Lysosomal exocytosis of FHL3 platelets was partially impaired at steady state, but profound impairment was observed during the acute phase of the disease (Figure 3A-C). This profound impairment was also observed in platelets obtained from some secondary HLH patients during the acute phase (Figure 3B-C). These results indicate that it is difficult to diagnose FHL3 during the acute phase of HLH either by Western blot or by lysosomal degranulation assay.
Analysis of lysosomal exocytosis using platelets from HLH patients. Platelets from an FHL3 patient (P6; A) and from a secondary (EBV-associated) HLH patient (B) along with healthy controls were left untreated (open histogram) or were stimulated with thrombin (closed histograms), and the surface expression of CD107a was analyzed by flow cytometry. Analysis was performed during the acute phase of the disease (left column) and after clinical remission (right column). (C) Degranulation index (DI) of platelets from HLH patients during the acute phase (●) and after clinical remission (○). HLH patients with normal NK-cell degranulation and Munc13-4 protein expression by Western blot analysis were defined as non-FHL3 (nFHL3). **P < .01 by the Tukey post hoc test.
Analysis of lysosomal exocytosis using platelets from HLH patients. Platelets from an FHL3 patient (P6; A) and from a secondary (EBV-associated) HLH patient (B) along with healthy controls were left untreated (open histogram) or were stimulated with thrombin (closed histograms), and the surface expression of CD107a was analyzed by flow cytometry. Analysis was performed during the acute phase of the disease (left column) and after clinical remission (right column). (C) Degranulation index (DI) of platelets from HLH patients during the acute phase (●) and after clinical remission (○). HLH patients with normal NK-cell degranulation and Munc13-4 protein expression by Western blot analysis were defined as non-FHL3 (nFHL3). **P < .01 by the Tukey post hoc test.
Rapid diagnosis of FHL3 by flow cytometric detection of intraplatelet Munc13-4
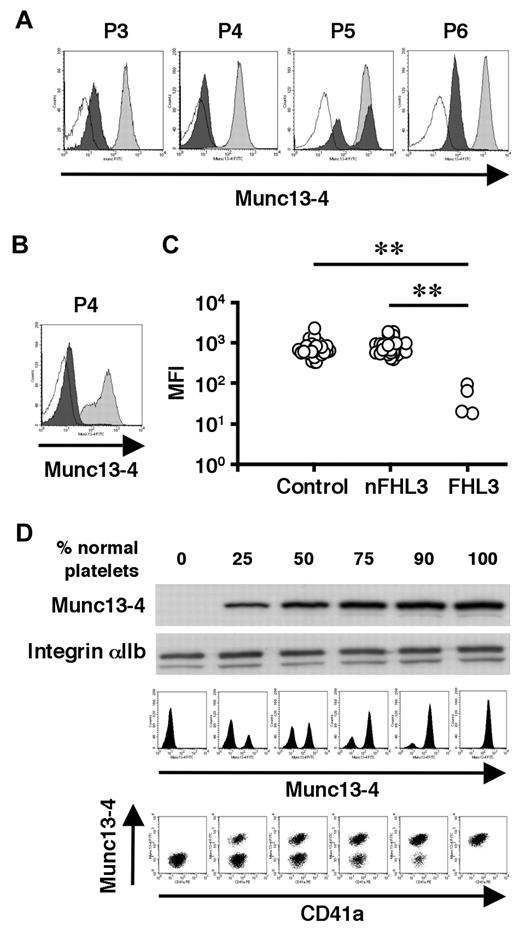
To overcome the difficulty in diagnosing FHL3 during the acute phase of HLH, antibodies were raised against the full-length human Munc13-4 protein (supplemental Figure 1) and a new method was developed to detect Munc13-4 protein in platelets by flow cytometry. A total of 35 patients, including 4 with FHL3 (P3-P6), were analyzed using this method. Munc13-4 deficiency was readily detected in all of the FHL3 patients, with a sample volume of < 100 μL of whole blood (Figure 4A-C). Munc13-4 protein was expressed at normal level in the platelets of parents and siblings of FHL3 patients carrying heterozygous UNC13D mutations (data not shown). In the FHL3 patient receiving platelet transfusions, flow cytometric analysis revealed bimodal populations of normal and Munc13-4–deficient platelets (P5 in Figure 4A). As shown in Figure 4B, the method was able to clearly identify Munc13-4–deficient platelets in whole blood samples stored at room temperature for 1 week.
Flow cytometric detection of intraplatelet Munc13-4 protein. Flow cytometric analysis of intraplatelet Munc13-4 expression in 4 FHL3 patients and healthy controls using whole blood samples shipped overnight (A) and in an FHL3 patient (P4) and a healthy control using samples stored at room temperature for a week (B). Dark closed histograms represent platelets from FHL3 patients, whereas light closed histograms represent platelets from healthy controls. Open histograms represent staining with isotype controls. (C) Mean fluorescence intensity (MFI) of intraplatelet Munc13-4 staining for HLH patients and healthy controls. All of the healthy controls (n = 35) were adults. Non-FHL3 (nFHL3) patients (n = 31), as defined in Figure 3, varied in age (2 days-39 years) and included 2 patients with FHL2. Age-related variations in the MFI of Munc13-4 staining were not observed. **P < .01 by the Tukey post hoc test. (D) The sensitivities of Western blot and flow cytometric analyses for detecting Munc13-4–deficient platelets were compared.
Flow cytometric detection of intraplatelet Munc13-4 protein. Flow cytometric analysis of intraplatelet Munc13-4 expression in 4 FHL3 patients and healthy controls using whole blood samples shipped overnight (A) and in an FHL3 patient (P4) and a healthy control using samples stored at room temperature for a week (B). Dark closed histograms represent platelets from FHL3 patients, whereas light closed histograms represent platelets from healthy controls. Open histograms represent staining with isotype controls. (C) Mean fluorescence intensity (MFI) of intraplatelet Munc13-4 staining for HLH patients and healthy controls. All of the healthy controls (n = 35) were adults. Non-FHL3 (nFHL3) patients (n = 31), as defined in Figure 3, varied in age (2 days-39 years) and included 2 patients with FHL2. Age-related variations in the MFI of Munc13-4 staining were not observed. **P < .01 by the Tukey post hoc test. (D) The sensitivities of Western blot and flow cytometric analyses for detecting Munc13-4–deficient platelets were compared.
To determine the sensitivity of the new method, Munc13-4–deficient platelets were mixed with normal platelets at varying ratios. Western blot analysis could not detect Munc13-4–deficient platelets easily, even when the proportion of normal platelets was as low as 25% (Figure 4D). In contrast, flow cytometric analysis easily identified 10% Munc13-4–deficient platelets among 90% normal platelets (Figure 4D), which proved the high sensitivity of the method in diagnosing FHL3.
Discussion
FHL is a rare but life-threatening inherited immune disorder for which mutations in 4 genes have been identified as causative factors. PRF1 encodes the cytolytic effector protein perforin that forms a pore-like structure in the target cell membrane.1,12 A mutation in PRF1 results in FHL2,7 which accounts for 20%-50% of FHL cases.4,5 UNC13D encodes the protein Munc13-4, which is crucial for the fusion of cytolytic granules to the plasma membrane and the subsequent release of perforin and granzymes.1,12 Mutations in UNC13D result in FHL3,8 which accounts for 20%-30% of FHL cases.4,12 FHL4 is caused by mutations in STX11, which encodes syntaxin-11.9 Mutations in STXBP2, which encodes Munc18-2, were recently reported to cause FHL5.10,11 Syntaxin-11 and Munc18-2 also mediate the fusion of cytolytic granules to the plasma membrane.1,5,12 The ability to screen for FHL2-5 rapidly would facilitate the initiation of life-saving immunosuppressive therapy and the preparation of FHL patients for hematopoietic stem cell transplantation.
In the present study, we found that the Munc13-4 protein is expressed abundantly in platelets (Figure 1A-B). The detection of Munc13-4 protein in platelets by Western blotting (Figure 1C) or flow cytometry (Figure 4A-B) was a reliable screening method to identify FHL3 patients. Munc13-4–deficient platelets were identified easily among normal transfused platelets by flow cytometry, which indicated that this method could be applied to patients who are receiving platelet transfusions during the acute phase of the disease (P5 in Figure 4A). Detection of intraplatelet Munc13-4 was enabled by the use of highly specific antibodies against the full-length human Munc13-4 (supplemental Figure 1).
There is a possibility that FHL3 patients with residual Munc13-4 protein expression could be overlooked by the screening methods described in this study. Most FHL3 patients have mutations that result in the absence or significant reduction of Munc13-4 protein expression,16,23 as was the case with the patients screened in this study (Figure 1C), which suggests that the mutated Munc13-4 protein is unstable. The NK-cell degranulation assay, which was performed for every referred sample with a sufficient number of NK cells, revealed defective degranulation only in the identified FHL3 patients (date not shown). These results indicate that the majority of mutations in UNC13D are likely amenable to rapid detection by the new methods described in this study. Comparative studies on the UNC13D genotype, Munc13-4 protein expression, and the lysosomal exocytosis assay must be performed to confirm the reliability of these methods.
It was also investigated whether the analysis of lysosomal release by platelets could be used as an alternative method to screen for FHL3. Profound impairment of lysosomal exocytosis by platelets during the acute phase of the disease and restoration of this impairment after clinical remission was observed in FHL3 and in some secondary HLH patients (Figure 3). It is not clear whether this transient impairment of platelet degranulation is involved in HLH pathogenesis or if it merely reflects in vivo platelet activation by diffuse endothelial damage during the acute phase of the disease that renders them unresponsive to ex vivo stimulation. The release of lysosomal granules by Munc13-4–deficient platelets was impaired only minimally at steady state (Figure 3A and 3C), which is in contrast to a recent study showing the involvement of the Munc13-4 protein in the release of lysosomal granules in mouse platelets.27 Although the precise reason for this discrepancy is unclear, platelet degranulation is likely to be regulated differentially between species; for example, Munc13-4–deficient mice have bruising and bleeding tendencies27 that are not commonly associated with human FHL3. Further studies are warranted to elucidate the exocytosis pathways of platelets and their role in the pathophysiology of HLH.
With the development of tools for rapid screening, the diagnostic approach for FHL has changed over the years. Impaired NK cytotoxicity was the first reported signature clinical finding of FHL patients.13,14 Defective CTL activity was subsequently reported as another hallmark of FHL.7,8,16,28 However, NK-cell activity is also decreased in some cases of secondary HLH,15,17-20 and the CTL cytotoxicity assay is not readily accessible to most clinicians. The NK-cell lysosomal exocytosis assay is a comprehensive method to identify patients with a degranulation defect.10,11,22-24 However, this analysis is not available in some patients with extremely reduced NK-cell numbers, which are often observed during the acute phase of HLH.19 Although CTLs can be an alternative tool to perform the lysosomal exocytosis assay,24,28,29 it remains impossible to differentiate FHL3-FHL5.10,11,23,24 Impairment in these assays warrants the genetic confirmation of FHL, but sequencing all of the candidate genes is not a suitable approach for rapid diagnosis. Flow cytometric detection of perforin expression in NK cells is a reliable and rapid way of identifying patients with FHL2,21 and the new method described in this study for the detection of Munc13-4 expression in platelets would add to the rapid diagnosis of FHL3.
Platelets could also be used for the screening of FHL4 and FHL5 because they share some granule-transport mechanisms with other types of hematopoietic cells, including CTLs and NK cells.2,30,31 Indeed, in the present study, both syntaxin-11 and Munc18-2 were expressed abundantly in platelets (data not shown). We are currently using platelet proteins to screen for FHL4-FHL5 by Western blot analysis, although no cases have been found so far because of the extreme rarity of these disorders.
In summary, platelets abundantly express Munc13-4 protein and are a useful tool to screen for FHL3. By detecting intraplatelet Munc13-4 expression by flow cytometry, it is now possible to rapidly screen for FHL3 with a very small sample of whole blood, even in the acute disease phase requiring platelet transfusion. Because platelets share some of their granule transport systems with other types of hematopoietic cells, they could also be used to screen for other types of immune disorders, including FHL4 and FHL5.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to all of the participating patients, their families, and the referring physicians for their generous cooperation in this study.
This study was supported by grants from The Morinaga Foundation for Health and Nutrition; from the Japanese Ministry of Education, Culture, Sports, Science, and Technology; and from the Japanese Ministry of Health, Labor, and Welfare.
Authorship
Contribution: T.Y., R.N., T.N., H.H., and H.T. designed the research; Y.M., K.I., and M.S. performed the Western blot and flow cytometric analyses; K.O. and O.O. performed the genetic analyses; R.S. and H.H. prepared the anti–Munc13-4 antibodies and started the FHL3 screening; Y.M., T.Y., R.S., K.I., H.S., J.A., N.T., T.K., R.N., E.I., T.N., H.H., and T.H. analyzed and discussed the results; and Y.M., T.Y., and T.H. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Takahiro Yasumi, Department of Pediatrics, Kyoto University Graduate School of Medicine, 54 Kawahara-cho, Shogoin, Sakyo-ku, Kyoto, 606-8507 Japan; e-mail: yasumi@kuhp.kyoto-u.ac.jp or Hisanori Horiuchi, Department of Molecular and Cellular Biology, Institute of Development, Aging and Cancer, Tohoku University, 4-1 Seiryo-machi, Aoba-ku, Sendai 980-8575 Japan; e-mail: horiuchi@idac.tohoku.ac.jp.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal