Abstract
IL-12 exerts several regulatory effects on natural killer (NK) cells by activating IL-12 signaling. IL-12 signaling is tightly auto-regulated to control its onset and termination, with prolonged IL-12 treatment resulting in IL-12 hyporesponsiveness. However, the mechanisms underlying IL-12 auto-regulation are still unclear. In this study we report that prolonged IL-12 treatment significantly up-regulates microRNAs (miRNAs), including miR-132, -212, and -200a in primary human NK cells. This up-regulation correlates temporally with gradually decreasing STAT4 levels and decreasing IFN-γ expression, after an initial increase within the first 16 hours of IL-12 treatment. The IL-12 hyporesponsiveness is dependent on IL-12 concentration, and associated up-regulation of miR-132, -212, and -200a. Furthermore, IL-12–hyporesponsive cells regain responsiveness of IFN-γ production 24 hours after IL-12 removal, which correlates with decreases in miR-132, -212, and -200a levels. Overexpression of miR-132, -212, and -200a by transfection into NK cells mimics IL-12 priming, inducing IL-12 hyporesponsiveness, whereas transfection of miR-132, -212, and -200a inhibitors largely abolishes IL-12 induction of IL-12 hyporesponsiveness. These data suggest that miR-132, -212, and -200a up-regulation during prolonged IL-12 treatment, negatively regulates the IL-12 signaling pathway by reducing STAT4 expression in primary human NK cells.
Introduction
IL-12 is a multifunctional cytokine that exerts several regulatory effects on T lymphocytes and natural killer (NK) cells.1,2 These include facilitating specific cytolytic T-cell responses; promoting the development of Th1-type helper T cells, thereby contributing to the development of cell-mediated immune responses; enhancing the lytic activity of NK cells; and inducing the secretion of IFN-γ by both T and NK cells.2-4 IL-12 mediates its biologic effects by signaling through specific cell surface receptors, which activate the JAK-signal transducer and activator of transcription (STAT) pathways.5 Among the STATs that are involved in IL-12 signaling, STAT4 plays a critical role. STAT4 knockout mice exhibit abrogation of all major IL-12–induced functions in both T cells and NK cells, including loss of IFN-γ production and impaired Th1 response.6,7
IL-12 and IFN-γ coordinate the link between pathogen recognition by innate immune cells and the induction of specific immunity, by mediating a positive feedback loop to amplify the Th1 response.8 Lipopolysaccharide and other pathogen-associated molecules directly trigger IL-12 production on recognition by macrophages, Dendritic cells (DCs), and neutrophils. This in turn induces IFN-γ secretion in antigen-stimulated, naive CD4+ T cells and NK cells.2,9,10 IL-12–induced IFN-γ participates in positive feedback by further promoting IL-12 production in macrophages.11 This amplification may be important in initiation or stabilization of the Th1 response (reviewed in Trinchieri et al2 and Boehm et al8 ). However, excessive amplification of this positive feedback loop and excessive production of IFN-γ may promote the development of immunopathologic conditions or immune disorders. Therefore, there need to exist mechanisms to control the production of IL-12 and the extent of ligand stimulation of IL-12 receptor signaling in vivo. Furthermore, such mechanisms might act at multiple steps in the IL-12 pathway. A variety of other cytokines, such as IL-10, IL-4, IL-l3, and TGF-β, have been implicated in down-regulating the production of IL-12 and the ability of T and NK cells to respond to IL-12.12,13 Alternatively, prolonged and repeated IL-12 stimulation may also activate an additional negative feedback mechanism to control and terminate the IL-12–induced proinflammatory immune response, representing a unique example of cytokine signaling auto-regulation.14,15 Continued IL-12 treatment induces IL-12 hyporesponsiveness in T cells in vitro through specific down-regulation of STAT4, makes T cells refractory to subsequent IL-12 stimulation and reduces their ability to produce IFN-γ in response to IL-12.14 NK cells, like T cells, are central to the immunologic effects of IL-12, particularly with respect to IFN-γ induction. In previous studies we have demonstrated that continued IL-12 treatment induces NK cells to lose responsiveness to further IL-12 stimulation through 2 mechanisms. One is down-regulation of IL-12 signaling by specific inhibition of STAT4 expression; the second is associated with IL-12–induced ROS accumulation, resulting in NK cell apoptosis.15 However, up to now, there have been no reports describing the mechanism of IL-12 regulation of the expression of STAT4.
MicroRNA (miRNA), an abundant class of highly conserved, small (18-25 nt long) noncoding RNAs, represents an entirely new paradigm in post-transcriptional regulation of gene expression. MiRNAs operate in a broad range of pathways and induce translational repression or degradation of target mRNAs by binding to the 3′-untranslated region (UTR) of target mRNAs.16,17 Recently, miRNAs have been shown to play a critical role in controlling NK cell homeostasis, activation, effector function, and in regulating diverse aspects of NK cell biology,18 but there have been no reports of IL-12 signaling pathway regulation by miRNAs in NK cells.
Starting with the hypothesis that miRNAs regulate STAT4, we analyzed the expression profile of miRNAs in human NK cells during IL-12 treatment to identify candidate miRNAs for STAT4 regulation. We found that several miRNAs, including miR-132, miR-212, and miR-200a were significantly up-regulated in NK cells by prolonged or repeated IL-12 treatment. Focusing on the regulation of IL-12 signaling by these IL-12–inducible miRNAs, we demonstrated that miR-132, -212, and -200a negatively regulate IL-12–triggered IFN-γ production by targeting STAT4, resulting in an impairment of the IL-12 pathway and the consequent development of IL-12 tolerance in NK cells. Moreover, we demonstrated that up-regulation of miR-132, -212, and -200a can mimic IL-12 induction of IL-12 tolerance in NK cells. We conclude that miR-132, -212, and -200a are negative feedback regulators of the IL-12 signaling pathways.
Methods
miRNA target predictions
Sequences of known and predicted mature human miRNAs were obtained from the miRNA Registry at miRBase (http://www.mirbase.org). STAT4 sequences were from the National Center for Biotechnology Information Nucleotide database (http://www.ncbi.nlm.nih.gov/). The prediction algorithms Targetscan human,17 miRanda,19 and RNAhybrid20 were used to search for miRNA target sites in the human STAT4 3′UTRs.
Human NK cell isolation, culture, and transfection
PBMCs were prepared from the blood by centrifugation over Ficoll-Histopaque (Sigma-Aldrich) as described.21,22 NK cells were enriched from PBMCs by immunomagnetic-negative selection using a Dynabeads Untouched Human NK cells kit (Invitrogen). The purity (% of CD56+CD3−) of NK cells measured by flow cytometer analysis was > 95%. Enriched NK cells were cultured at a density of 1 × 106 cells/mL in RPMI 1640 plus 10% FBS (Gibco), supplemented with 100 IU/mL of recombinant human IL-2 (Roche) alone or together with recombinant human IL-12 (Peprotech). For transfection, NK cells (2 × 105) were prepare and transfected with 50nM miRNA or miRNA inhibitor using Amaxa nucleofection technology with the U-001 program for high efficiency, according to the manufacturer's instructions (Human NK cell Nucleofector Kit, Lonza; catalog number VPA-1005). After electroporation, cells were immediately transferred to the culture plates. The medium was changed after 6 hours. At the indicated times after transfection, STAT4 and IFN-γ protein expression were analyzed by flow cytometry.
Flow cytometry and cytokine secretion assays
Cell surface staining and flow cytometric analysis of CD3 and CD56 (BD Biosciences) expression were performed as described elsewhere.21 Intracellular staining of STAT4, p-STAT4 and IFN-γ (all BD Biosciences) were performed after fixation and permeabilization according to BD cytofix/cytoperm manufacturer's instructions. Commercial IFN-γ ELISA kits from R&D Systems were used to measure the levels of IFN-γ in cell-free culture supernatants.
3′-UTR luciferase reporter assay
The full-length 3′ untranslated region (UTR) of the human STAT4 gene (GenBank accession no. NM_003151) was amplified from human NK cDNA and cloned into the SpeI / PmeI sites of pMIR-REPORT Luciferase vector (Ambion). The primers for STAT4 were forward, 5′-GACGTCGGACTAGTCAGGATAAACTCTGACGC-3′, and reverse, 5′-AGCTTTGTTTAA ACCTGTTAATATTGTTATTAAC-3′. The segment (base pairs 2329-2588) of the STAT4 3′UTR containing the mutated miR-200a target sequence (CAGTGTT to GTCACAA) or/and the mutated miR-132/212 target sequence (GACTGTT to TTGACAA) were also cloned into the pMIR-REPORT Luciferase vector (Ambion). The primers for mutated miR-200a target sequence in STAT4 gene (STAT4 mt200a) were forward, same as the primer for the STAT4 gene, and reverse, 5′-AGCTTTGTTTAAACCTGTTAATATTGTTATTTTGTGACGTTTCTTAAAGTTGTC-3′. The primers for mutated miR-132 and -212 target sequence in STAT4 gene (STAT4 mt132/212) were forward, 5′-CAGGATAAACTCTGACGCACCAAGAAAGGAAGCAAATG AAAAAGTTTAAATTGACAACTTTGCCCAATAACCACA-3′, and reverse, same as the primer for the STAT4 gene. HEK 293T cells (50% confluent in 24-well plates) were cotransfected with 500 ng of the indicated luciferase reporter plasmid, 100 ng of pRL-TK-Renilla-luciferase plasmid (Promega, for normalization) and the indicated miRNA mimics (final concentration, 10 or 50nM) by Lipofectamine 2000 (Invitrogen). Cell extracts were prepared 48 hours after transfection, and luciferase activities were measured by the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Data were normalized for transfection efficiency by dividing firefly luciferase activity by that of renilla luciferase.
Western blotting
Cell extracts were prepared as described previously.15,21 Protein concentration was determined using the BCA kit (Pierce). Protein was separated on 4%-12% SDS-PAGE, then electrotransferred to nitrocellulose membanes (Millipore). After the blocking step, the blots were probed with specific anti-human STAT4 antibodies (Santa Cruz Biotechnology), and then visualized with appropriate HRP-conjugated secondary antibody (Pierce) and an ECL detection system (Pierce).
Quantification of miRNA and mRNA expression levels by qRT-PCR
Total RNA was isolated from human NK cells and purified using the miR vana isolation kit (Ambion) according to the manufacturer's protocol. RNA concentration and purity were measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technology Inc), and equal amounts of each RNA (10 ng for miRNA and 8 ng for mRNA) were used for real-time RT-PCR (qRT-PCR) analysis. miRNA analyses were performed using the TaqMan MicroRNA Reverse Transcription Kit, TaqMan Universal PCR Master Mix, and TaqMan microRNA Assay primers for human miRNAs (Applied Biosystems). miRNA expression values were calculated using RNU6B as endogenous control (Applied Biosystems) following the 2-ΔΔCt method. STAT4 and IFN-γ mRNA levels were quantified by qRT-PCR using the SYBR Green Master Mix (Applied Biosystems). The following optimized forward and reverse primers for these genes and β-actin (housekeeping reference) gene were used at a final concentration of 100 nM: primers 5′-AGGGACTGTGAGGGGCGCTT-3′ and 5′-CCATTGGGCCAACAGATGCCGA-3′ for stat4; primers 5′-GCATCCAAAAGAGTGTGGAGACCA-3′ and 5′-AGCTGCTGGCGACAGTTCAGC-3′ for IFN-γ; primers 5′-GCGCGGCTACAGCTTCACCA-3′ and 5′-GGGCAGCGGAACCGCTCATT-3′ for β-actin.
Statistical analysis
Data were expressed as means ± SEM values of 3 independent experiments with different donors' cells. Each experiment was carried out in duplicate or triplicate. Statistical comparisons of the results were made using ANOVA. Statistical significance was defined as P < .05.
Results
Predicted miRNA recognition elements in the STAT4 3′UTR
To investigate the potential for miRNA regulation of STAT4, we sought miRNAs that could interact with miRNA recognition elements (MREs) in the STAT4 3′UTR. We used multiple prediction algorithms, including Targetscan human,17,23 miRanda,19 and RNAhybrid,20 to search for target sites in the human STAT4 3′UTRs. In total, 30 miRNA-mRNA target duplexes were predicted by these algorithms (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). To determine which miRNAs might be regulated by IL-12 stimulation in NK cells, we examined the expression profile of these miRNAs and found that 12 microRNAs could not be detected in human NK cells. QRT-PCR profiling of the remaining 18 microRNAs revealed that 3 microRNAs were expressed at > 2-fold (P < .01) higher levels in IL-12–treated versus control NK cells (IL-2 alone), whereas only 1 miRNA (miR-150) displayed the opposite profile after IL-12 treatment for 24 hours (supplemental Figure 2). The up-regulated miRNAs included miR-132, miR-212, and miR-200a. miR-132 is implicated in neuronal differentiation and function and the regulation of antiviral immunity.24,25 miR-132 and miR-212 are derived from the same primary microRNA transcript and these 2 miRNAs share the same seed sequence. miR-200a is implicated in the regulation of Wnt signaling.26
miR-212, -132, and -200a target STAT4
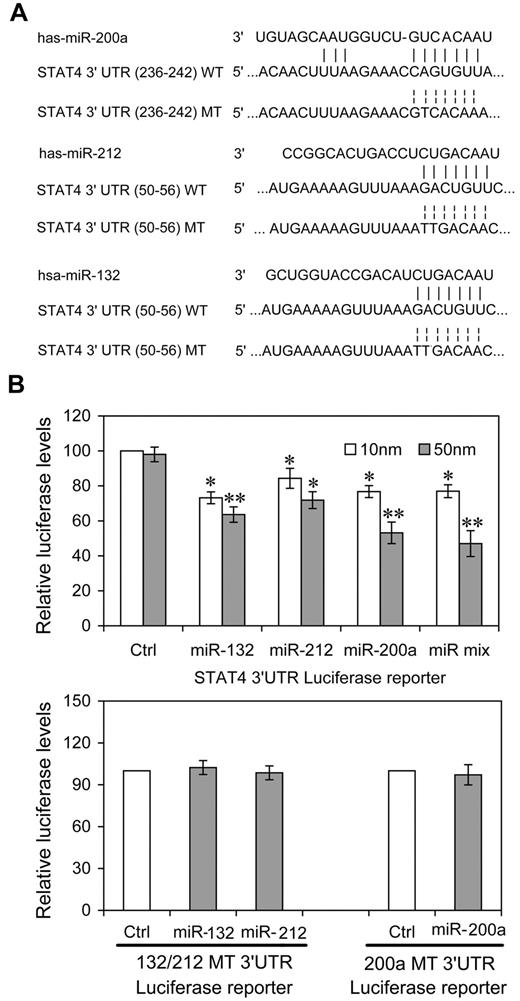
To investigate the possibility that STAT4 was regulated post-transcriptionally by these IL-12–inducible miRNAs, we constructed reporter plasmids encoding the complete wild-type 3′UTR of the human STAT4 mRNA downstream of the firefly luciferase gene (WT-3′UTR). We also constructed parallel plasmids containing mismatches in the predicted miR-212, -132 or -200a binding sites (132 or 200a MT-3′UTR) of the 3′UTR region (Figure 1A). By cotransfecting the reporter plasmids together with the indicated miRNA mimics and an internal control, pRL-TK-Renilla-luciferase, we observed that 3 of the candidate miRNAs (miR-200a, -132, -212) reduced the expression of luciferase from WT-3′UTR in HEK-293T cells compared with either control miRNA having a nucleotide composition similar to that of the mimics or to “no miRNA” control transfections (Figure 1B). At 50nM, all 3 miRNAs reduced expression significantly. For miR-200a and the equimolar mixture of all 3 miRNAs, the response was clearly dose-dependent (P < .05, respectively). That the dose response for the other miRNAs is less apparent may derive from their saturating binding sites and achieving maximal effects at lower concentrations. The equimolar mixtures (10nM = 3.33nM each; 50nM = 16.6nM each) effected a greater control than any single mimic, suggesting a cooperative effect of multiple miRNA recognition elements occupancy, as has been reported previously.27 miR-200a, -132, -212 did not down-regulate expression from MT-3′UTR plasmids containing specific mutations in their cognate binding sites (Figure 1B). These experiments validated the regulatory potential of miR-132, miR-212 and miR-200a via binding with the STAT4 3′UTR and suggested that these miRNAs might mediate the modulation of IL-12 in endogenous STAT4 in NK cells.
miRNAs directly target sequences in the STAT4 3′UTR. (A) Human STAT4 might be the molecular target of miR-132, miR-212, and miR-200a. This diagram represents a sequence alignment of miR-132, -212, and -200a and their target sites in STAT4 3′UTR, and relative mutated versions. (B) miRNAs reduced luciferase activity in cells transfected with wild-type reporter (WT-3′UTR), but not in cells transfected with mutated-type reporter (132/212 or 200a MT-3′UTR). HEK293T cells were cotransfected with wild (or mutated) type STAT4 3′UTR firefly luciferase reporter plasmids, pTK-Renilla-luciferase plasmids, together with control (ctrl), miR-132, miR-212, miR-200a mimics, or equimolar mixtures of the 3 indicated miRNAs (final concentration as indicated). After 48 hours, firefly luciferase activity was measured and normalized by renilla luciferase activity. Representative data from 3 independent experiments are shown, and each bar indicates the mean value ± SEM (n = 3). *P < .05, **P < .01 versus cells transfected control mimics.
miRNAs directly target sequences in the STAT4 3′UTR. (A) Human STAT4 might be the molecular target of miR-132, miR-212, and miR-200a. This diagram represents a sequence alignment of miR-132, -212, and -200a and their target sites in STAT4 3′UTR, and relative mutated versions. (B) miRNAs reduced luciferase activity in cells transfected with wild-type reporter (WT-3′UTR), but not in cells transfected with mutated-type reporter (132/212 or 200a MT-3′UTR). HEK293T cells were cotransfected with wild (or mutated) type STAT4 3′UTR firefly luciferase reporter plasmids, pTK-Renilla-luciferase plasmids, together with control (ctrl), miR-132, miR-212, miR-200a mimics, or equimolar mixtures of the 3 indicated miRNAs (final concentration as indicated). After 48 hours, firefly luciferase activity was measured and normalized by renilla luciferase activity. Representative data from 3 independent experiments are shown, and each bar indicates the mean value ± SEM (n = 3). *P < .05, **P < .01 versus cells transfected control mimics.
Kinetics of IL-12–induced expression of IFN-γ, STAT4, and miRNAs
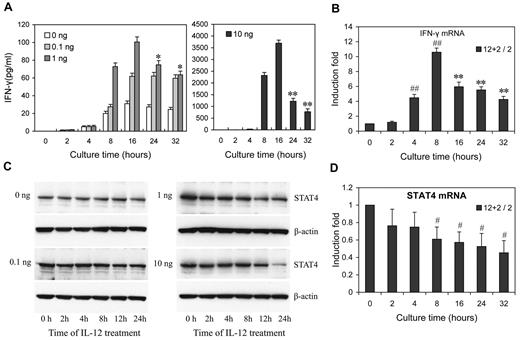
To investigate how IL-12 regulates IFN-γ and STAT4 in NK cells, we first examined the kinetics and dose response of IL-12 regulation of IFN-γ protein and STAT4 mRNA and protein expression in NK cells. A detailed time-course showed that, after the addition of IL-12, at all IL-12 concentrations tested, IFN-γ levels peaked by 16 hours after treatment, followed by either a steady decline (IL-12 at 1 and 10 ng/mL) or a plateau (IL-12 at 0.1 ng/mL; Figure 2A). (Note that because the media was changed every 8 hours, every time point after 8 hours represents IFN-γ accumulation during the previous 8 hours.) At 10 ng/mL of IL-12, the levels of IFN-γ mRNA expression showed kinetics similar to secreted IFN-γ protein, except the peak level of IFN-γ mRNA was slightly earlier, at 8 hours (Figure 2B). However, the kinetics of STAT4 expression appeared somewhat different from those of IFN-γ. Consistent with our previous study, STAT4 protein levels remained constant over 24 hours of culture in the absence of IL-12 or the presence of a low concentration of IL-12 (0.1 ng/mL), while they significantly decreased after treatment with higher concentrations of IL-12 (1 or 10 ng/mL) for 12 hours, and progressively declined at subsequent time points (Figure 2C and supplemental Figure 3). Consistent with these changes in the levels of STAT4 protein, STAT4 mRNA steadily decreased at higher concentrations of IL-12 treatment (Figure 2D). These results suggested that IL-12 initially activates NK cells to produce IFN-γ, but IL-12 treatment simultaneously down-regulates IL-12 signaling in part by reducing STAT4 mRNA levels steadily. That the STAT4 mRNA changes were less dramatic than the STAT4 protein changes, suggested that STAT4 expression might be additionally regulated at the post-transcriptional level in IL-12–treated NK cells. Similarly, although the kinetics of STAT4 protein decrease correlated with the second phase of IFN-γ (the decrease), the imperfection of the correlation suggested factors other than STAT4 also contribute to IFN-γ down-regulation by prolonged IL-12 treatment.
Dose- and time-dependent expression of IFN-γ and STAT4 in IL-12-treated NK cells. (A) NK cells were treated with 0, 0.1, 1, or 10 ng/mL IL-12 and incubated for the times indicated in the presence of IL-2. Culture supernatants were collected at the times indicated points from 2 to 32 hours. The IFN-γ levels in the medium were quantified by ELISA. Bars at 2, 4, and 8 hours indicate cumulative IFN-γ levels in culture supernatants; Bars at 16, 24 and 32 hours indicate IFN-γ production in each preceding 8 hours in culture supernatants. (B) NK cells were treated with 0 or 10 ng/mL IL-12 and incubated for indicated times in presence of IL-2. Total RNA was purified from the respective cell pellets and analyzed by qRT-PCR for the expression of IFN-γ mRNA. (C) NK cells were cultured as in panel A. Cells were collected at the times indicated and whole-cell extracts were prepared. Western blots were performed with anti-STAT4. (D) NK cells were treated with 0 or 10 ng/mL IL-12 and incubated for indicated times in presence of IL-2. Total RNA was purified from the respective cell pellets and analyzed by qRT-PCR for the expression of STAT4 mRNA. “2 + 12 / 2” represents the fold change in mRNA levels calculated by comparing the value of IL-2 + IL-12–cultured cells (2 + 12) to that of IL-2-cultured samples2 in parallel. The results are mean ± SEM of representative experiments with NK cells from 3 different donors. *P < .05, **P < .01 vs 16 hours (A) or 8 hours (B) same concentration of IL-12 treated cells. #P < .05, ##P < .01 versus 0 hours untreated cells.
Dose- and time-dependent expression of IFN-γ and STAT4 in IL-12-treated NK cells. (A) NK cells were treated with 0, 0.1, 1, or 10 ng/mL IL-12 and incubated for the times indicated in the presence of IL-2. Culture supernatants were collected at the times indicated points from 2 to 32 hours. The IFN-γ levels in the medium were quantified by ELISA. Bars at 2, 4, and 8 hours indicate cumulative IFN-γ levels in culture supernatants; Bars at 16, 24 and 32 hours indicate IFN-γ production in each preceding 8 hours in culture supernatants. (B) NK cells were treated with 0 or 10 ng/mL IL-12 and incubated for indicated times in presence of IL-2. Total RNA was purified from the respective cell pellets and analyzed by qRT-PCR for the expression of IFN-γ mRNA. (C) NK cells were cultured as in panel A. Cells were collected at the times indicated and whole-cell extracts were prepared. Western blots were performed with anti-STAT4. (D) NK cells were treated with 0 or 10 ng/mL IL-12 and incubated for indicated times in presence of IL-2. Total RNA was purified from the respective cell pellets and analyzed by qRT-PCR for the expression of STAT4 mRNA. “2 + 12 / 2” represents the fold change in mRNA levels calculated by comparing the value of IL-2 + IL-12–cultured cells (2 + 12) to that of IL-2-cultured samples2 in parallel. The results are mean ± SEM of representative experiments with NK cells from 3 different donors. *P < .05, **P < .01 vs 16 hours (A) or 8 hours (B) same concentration of IL-12 treated cells. #P < .05, ##P < .01 versus 0 hours untreated cells.
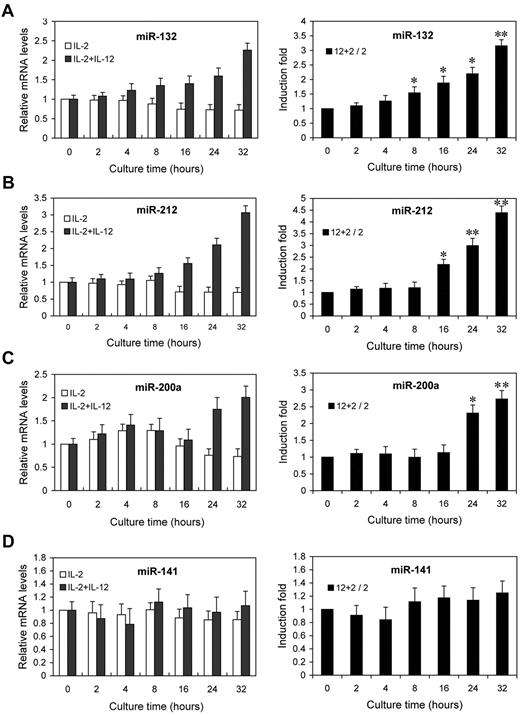
Next, we examined the kinetics of miR-132, miR-212, and miR-200a by qRT-PCR analysis on the same RNA samples. The fold changes in miRNA expression were calculated by comparing the value of IL-12–treated cells to that of untreated samples cultured in parallel. miR-132 showed an average increase of 1.6-fold after 8 hours, rising to 3.2-fold by 32 hours (Figure 3A). miR-212 and miR-200a expression showed increases of 2.2- and 2.3-fold by 16 hours and 24 hours of IL-12 treatment, respectively, and further increased at subsequent time points (Figure 3B-C). No significant change in the expression of miR-141 (which shares the same seed sequence with miR-200a) was observed (Figure 3D). Indeed, the expressions of miR-132, miR-212, and miR-200a were progressively reduced with time in NK cells cultured with IL-2 alone, whereas they progressively increased with times in NK cells cultured with IL-2 plus IL-12 (Figure 3A-C left panel), despite no similar change in the expression of miR-141 (Figure 3D left panel). These results confirmed that the expressions of miR-132, miR-212, and miR-200a are induced by IL-12 signaling, consistent with the original screen of supplemental Figure 2.
IL-12 treatment induces the expression of miR-132, miR-212, and miR-200a in NK cells. NK cells were treated with 0 or 10 ng/mL IL-12 and incubated for indicated times in presence of IL-2. Total small RNA were purified from the respective cell pellets and analyzed by qRT-PCR for the expression of miR-132 (A), miR-212 (B), miR-200a (C), and miR-141 (D). miRNA expression was normalized with control RNU6B. 2 + 12 / 2 represents that the fold changes in mRNA levels were calculated by comparing the value of IL-2+IL-12–cultured cells (2 + 12) to that of IL-2 cultured samples2 in parallel. The results are mean ± SEM of representative experiment with NK cells from 3 different donors. *P < .05, **P < .01 vs 0 hours untreated cells.
IL-12 treatment induces the expression of miR-132, miR-212, and miR-200a in NK cells. NK cells were treated with 0 or 10 ng/mL IL-12 and incubated for indicated times in presence of IL-2. Total small RNA were purified from the respective cell pellets and analyzed by qRT-PCR for the expression of miR-132 (A), miR-212 (B), miR-200a (C), and miR-141 (D). miRNA expression was normalized with control RNU6B. 2 + 12 / 2 represents that the fold changes in mRNA levels were calculated by comparing the value of IL-2+IL-12–cultured cells (2 + 12) to that of IL-2 cultured samples2 in parallel. The results are mean ± SEM of representative experiment with NK cells from 3 different donors. *P < .05, **P < .01 vs 0 hours untreated cells.
Pretreatment of NK cells with IL-12 reduces IFN-γ production linked to miR-132, -212, and -200a overexpression
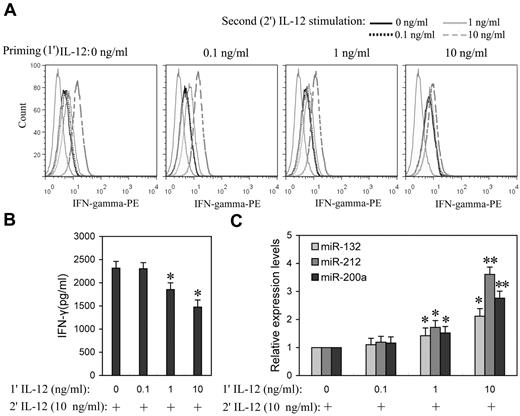
Prolonged IL-12 stimulation has been reported to dramatically lower IFN-γ levels,15 suggesting that IL-12 induces tolerance in NK cells. To better understand the impact of the priming concentration in relation to miRNA expression during the induction of IL-12 tolerance in vitro, NK cells were “primed” with a range of concentrations of IL-12 (0.1, 1.0, and 10 ng/mL) for 24 hours, followed by washing with PBS, and then further stimulated with various concentration of IL-12. After 8 hours of incubation with the second dose of IL-12, IFN-γ expression levels were measured by flow cytometry or ELISA. After priming with 0.1 ng/mL IL-12, cells subsequently treated with even 10 ng/mL IL-12 were not significantly different from controls in IFN-γ expression, whereas cells that were primed with 1 and 10 ng/mL IL-12 showed significantly decreased IFN-γ expression in response to subsequent exposure to IL-12, especially at higher concentrations (Figure 4A and supplemental Figure 4). In response to the second 10 ng/mL IL-12 stimulation, cells primed with 1 ng/mL IL-12 showed ∼ 20% decrease (P < .05) of IFN-γ production, while cells primed with 10 ng/mL IL-12 resulted in ∼ 37% decrease (P < .05) of IFN-γ production (Figure 4B), suggesting that NK cells primed with higher concentrations of IL-12 are more refractory to subsequent IL-12 stimulation. Consistent with tolerance, priming with 1 and 10 ng/mL IL-12 significantly induced miR-132, -212, and -200a expression (Figure 4C). These results emphasized the correlation of IL-12 priming concentration with up-regulation of miR-132, -212, and -200a and IL-12 tolerance.
Higher priming concentration of IL-12 induced higher miR-132, -212, and -200a levels and more efficient suppression of IFN-γ production in subsequent IL-12 challenge. (A) NK cells were primed with 0, 0.1, 1, or 10 ng/mL IL-12 (1′ IL-12) continuously for 24 hours in the presence of IL-2, washed twice with PBS, and challenged with 0, 0.1, 1, or 10 ng/mL of IL-12 (2′ IL-12) for another 8 hours in the presence of IL-2. The expression of IFN-γ was measured by flow cytometry. (B) NK cells were primed with 0, 0.1, 1, or 10 ng/mL IL-12 (1′ IL-12) continuously for 24 hours in the presence of IL-2, washed twice with PBS, and challenged with 10 ng/mL of IL-12 (2′ IL-12) for another 8 hours in the presence of IL-2. The IFN-γ levels in the medium were quantified by ELISA. (C) NK cells were treated as in panel B, qRT-PCR analysis of total small RNA for miR-132, miR-212 and miR-200a expression analysis in total small RNA. The results are mean ± SEM of representative experiment with NK cells from 3 different donors. *P < .05, **P < .01 versus IL-12–unprimed cells.
Higher priming concentration of IL-12 induced higher miR-132, -212, and -200a levels and more efficient suppression of IFN-γ production in subsequent IL-12 challenge. (A) NK cells were primed with 0, 0.1, 1, or 10 ng/mL IL-12 (1′ IL-12) continuously for 24 hours in the presence of IL-2, washed twice with PBS, and challenged with 0, 0.1, 1, or 10 ng/mL of IL-12 (2′ IL-12) for another 8 hours in the presence of IL-2. The expression of IFN-γ was measured by flow cytometry. (B) NK cells were primed with 0, 0.1, 1, or 10 ng/mL IL-12 (1′ IL-12) continuously for 24 hours in the presence of IL-2, washed twice with PBS, and challenged with 10 ng/mL of IL-12 (2′ IL-12) for another 8 hours in the presence of IL-2. The IFN-γ levels in the medium were quantified by ELISA. (C) NK cells were treated as in panel B, qRT-PCR analysis of total small RNA for miR-132, miR-212 and miR-200a expression analysis in total small RNA. The results are mean ± SEM of representative experiment with NK cells from 3 different donors. *P < .05, **P < .01 versus IL-12–unprimed cells.
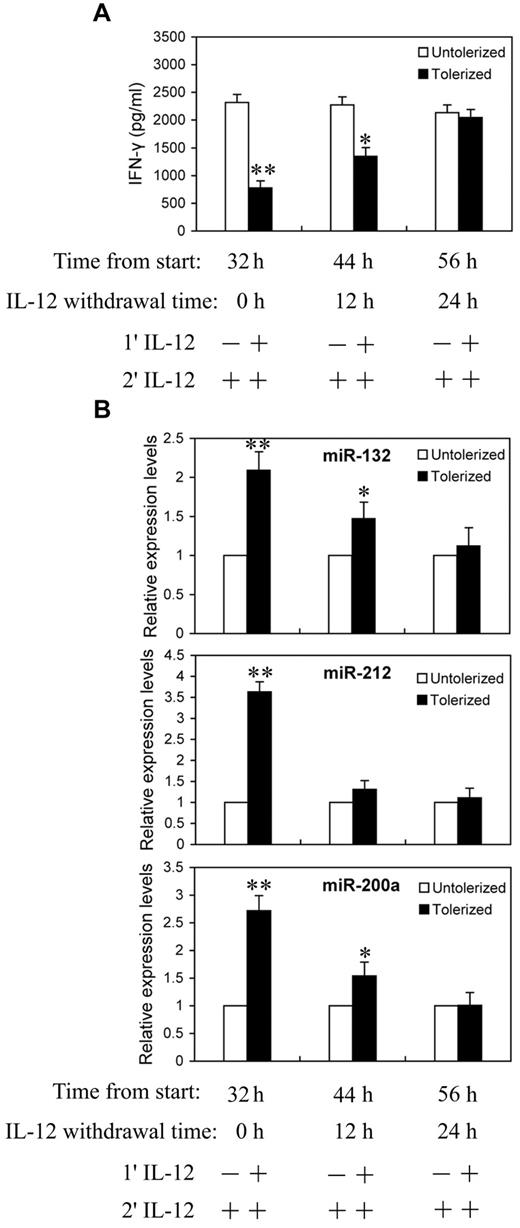
We next asked whether the continuing increases in miR-132, -212, and -200a levels require continued exposure to IL-12. NK cells were primed with 10 ng/mL of IL-12 continuously for 24 hours, then washed twice with PBS and cultured in complete growth medium for an additional 0, 12, or 24 hours (IL-12 withdrawal). At each time point after IL-12 withdrawal, cells were challenged with 10 ng/mL of IL-12 for 8 hours before analysis for IFN-γ protein production in culture supernatant by ELISA (Figure 5A) and qRT-PCR analysis of miR-132, -212, and -200a expression (Figure 5B). With 24 hours of continuous IL-12 priming and 8 hours of IL-12 challenge, significant reductions in IFN-γ secretion and highly elevated miR-132, -212, and -200a levels were observed, again as expected. Interestingly, after 12 hours of IL-12 withdrawal, cells started to regain IL-12 responsiveness to produce IFN-γ and almost completely recovered from tolerance after 24 hours of IL-12 withdrawal (Figure 5A). Figure 5B shows the expression of miR-132, -212, and -200a decreased at 12 and 24 hours after IL-12 withdrawal. At 24 hours after IL-12 withdrawal, the levels of IFN-γ and miR-132, -212, and -200a were no longer significantly different from the unprimed controls. Thus, the kinetics of miR-132, -212, and -200a induction by IL-12 priming correlated with the induction of IL-12 tolerance.
Reduction in miR-132, -212, and -200a expression in IL-12 tolerized NK cells inversely correlated with IFN-γ production. (A) NK cells were cultured with (tolerized) or without (untolerized) 10 ng/mL IL-12 continuously for 24 hours in the presence of IL-2. Cells were then washed twice with PBS and cultured in complete growth medium with IL-2 for additional 0, 12, or 24 hours (IL-12 withdrawal). At each time point after IL-12 withdrawal, cells were challenged with 10 ng/mL of IL-12 for 8 hours in the presence of IL-2 before analysis of IFN-γ protein production in culture supernatant by ELISA. (B) qRT-PCR analysis of total small RNA for miR-132, miR-212 and miR-200a expression. NK cells were cultured as in panel A. The results are mean ± SEM of representative experiment with NK cells from 3 different donors. *P < .05, **P < .01 versus IL-12 untolerized cells.
Reduction in miR-132, -212, and -200a expression in IL-12 tolerized NK cells inversely correlated with IFN-γ production. (A) NK cells were cultured with (tolerized) or without (untolerized) 10 ng/mL IL-12 continuously for 24 hours in the presence of IL-2. Cells were then washed twice with PBS and cultured in complete growth medium with IL-2 for additional 0, 12, or 24 hours (IL-12 withdrawal). At each time point after IL-12 withdrawal, cells were challenged with 10 ng/mL of IL-12 for 8 hours in the presence of IL-2 before analysis of IFN-γ protein production in culture supernatant by ELISA. (B) qRT-PCR analysis of total small RNA for miR-132, miR-212 and miR-200a expression. NK cells were cultured as in panel A. The results are mean ± SEM of representative experiment with NK cells from 3 different donors. *P < .05, **P < .01 versus IL-12 untolerized cells.
Up-regulation of miR-132, -212, and -200a can mimic IL-12 priming in the induction of IL-12 tolerance in NK cells
To further identify whether miR-132, miR-212 and miR-200a expression can mimic IL-12 priming to induce IL-12 tolerance in NK cells, we investigated the potential effects of these miRNAs on STAT4 and IL-12–induced IFN-γ production in primary human NK cells. NK cells were transfected with miR-491-5p, miR-132, miR-212, miR-200a, or an equimolar mixture of miR-132, miR-212, and miR-200a mimics. The levels of transfected miRNAs were confirmed by QRT-PCR comparison of each miRNA in pre-transfection and post-transfection cells (data not shown). As predicted, miR-132, miR-212 and miR-200a mimics decreased the expression of STAT4 in NK cells transfected with miR-132, miR-212, miR-200a or miR mix, compared with that of the cells transfected with the control miRNA or the miR-491-5p mimic (Figure 6A-B). Consistent with STAT4 protein level changes, the levels of phospho-STAT4 relatively decreased in NK cells transfected with miR-132, miR-212, miR-200a or miR mix, compared with those in the cells transfected with the control miRNA mimics (Figure 6C-D). IL-12–induced IFN-γ production also noticeably decreased in the cells transfected with miR-132, miR-212, miR-200a or miR mix compared with that in cells transfected with control miRNA mimics. By way of comparison there was no significant difference between the IFN-γ production of cells transfected with miR-491-5p mimics and that of cells transfected with negative control mimics (Figure 6E-F). In addition, over-expression of miR-132, miR-212, and miR-200a did not significantly affect NK cell growth (supplemental Figure 5), excluding the possibility that the lower STAT4 and IFN-γ production resulted from apoptosis or inhibition of NK cell proliferation. These results suggest that miR-132, -212, and -200a could contribute to the induction of IL-12 tolerance (hyporesponsiveness) via specific negative regulation of STAT4 in NK cells.
miR-132, miR-212, and miR-200a negatively regulate STAT4 expression and IL-12–induced IFN-γ production in NK cells. (A) Human NK cells (2 × 105) were transfected with control mimics (miR ctrl), miR-491-5p, miR-132, miR-212, miR-200a, or an equimolar mixture of miR-132, miR-212, and miR-200a (miR mix) mimics as indicated at an aggregate final concentration of 50nM. After 48 hours culture in the presence of IL-2, the expression of STAT4 was measured by flow cytometry. Data are representative of 3 independent experiments with NK cells from different donors. (B) Summary of data from panel A, mean of STAT4 levels in indicated miRNA overexpression cells were analyzed to determine the relative effects of different miRNA on STAT4 protein levels. The results are STAT4 MFI mean ± SEM of representative experiment with NK cells from 3 different donors. *P < .05 versus miR ctrl. (C) Cells treated as in panel A (but not harvested) were washed with complete growth medium and cultured with or without 10 ng/mL of IL-12 for another 12 hours in the presence of IL-2. Cellular expression of phosphorylated STAT4 was measured by flow cytometry. Data are representative of 3 independent experiments with NK cells from different donors. (D) Summary of data from panel C. Mean of p-STAT4 levels in indicated miRNA overexpression cells was analyzed to determine the relative effects of different miRNA on IL-12–induced STAT4 phosphorylation. The results are p-STAT4 MFI mean ± SEM of representative experiment with NK cells from 3 different donors. *P < .05 versus miR ctrl. (E) Cells treated as in panel C. Cellular expression of IFN-γ was measured by flow cytometry. Data are representative of 3 independent experiments with NK cells from different donors. (F) Summary of data from panel E. Mean of IL-12–induced IFN-γ expression in indicated miRNA overexpression cells (IFN-γ MFI with IL-12 – IFN-γ MFI without IL-12) was calculated to determine the relative effects of different miRNA on IL-12–induced IFN-γ expression. The results are increased IFN-γ MFI mean ± SEM of representative experiment with NK cells from 3 different donors. *P < .05, **P < .01 versus miR ctrl. The gray dotted histogram represents the isotype control staining of NK cells.
miR-132, miR-212, and miR-200a negatively regulate STAT4 expression and IL-12–induced IFN-γ production in NK cells. (A) Human NK cells (2 × 105) were transfected with control mimics (miR ctrl), miR-491-5p, miR-132, miR-212, miR-200a, or an equimolar mixture of miR-132, miR-212, and miR-200a (miR mix) mimics as indicated at an aggregate final concentration of 50nM. After 48 hours culture in the presence of IL-2, the expression of STAT4 was measured by flow cytometry. Data are representative of 3 independent experiments with NK cells from different donors. (B) Summary of data from panel A, mean of STAT4 levels in indicated miRNA overexpression cells were analyzed to determine the relative effects of different miRNA on STAT4 protein levels. The results are STAT4 MFI mean ± SEM of representative experiment with NK cells from 3 different donors. *P < .05 versus miR ctrl. (C) Cells treated as in panel A (but not harvested) were washed with complete growth medium and cultured with or without 10 ng/mL of IL-12 for another 12 hours in the presence of IL-2. Cellular expression of phosphorylated STAT4 was measured by flow cytometry. Data are representative of 3 independent experiments with NK cells from different donors. (D) Summary of data from panel C. Mean of p-STAT4 levels in indicated miRNA overexpression cells was analyzed to determine the relative effects of different miRNA on IL-12–induced STAT4 phosphorylation. The results are p-STAT4 MFI mean ± SEM of representative experiment with NK cells from 3 different donors. *P < .05 versus miR ctrl. (E) Cells treated as in panel C. Cellular expression of IFN-γ was measured by flow cytometry. Data are representative of 3 independent experiments with NK cells from different donors. (F) Summary of data from panel E. Mean of IL-12–induced IFN-γ expression in indicated miRNA overexpression cells (IFN-γ MFI with IL-12 – IFN-γ MFI without IL-12) was calculated to determine the relative effects of different miRNA on IL-12–induced IFN-γ expression. The results are increased IFN-γ MFI mean ± SEM of representative experiment with NK cells from 3 different donors. *P < .05, **P < .01 versus miR ctrl. The gray dotted histogram represents the isotype control staining of NK cells.
Down-regulation of miR-132, -212, and -200a in IL-12–treated NK cells restores responsiveness to subsequent IL-12 stimulation
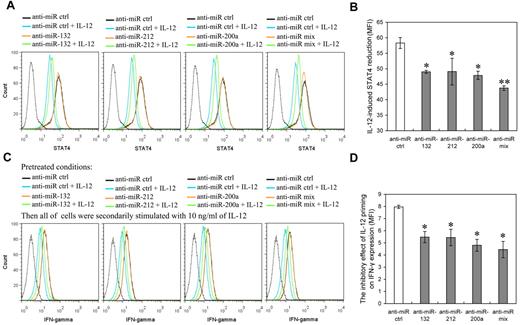
To further corroborate the role of miR-132, -212, and -200a in mediation of IL-12 hyporesponsiveness in NK cells, we investigated the effects of blocking IL-12–induced miR-132, -212, and -200a on IL-12 tolerance through suppressing their activity with specific miRNA inhibitors. NK cells transfected with miR-132, miR-212, miR-200a, or miR mix inhibitor were primed with (or without) 10 ng/mL IL-12 for 48 hours followed by washing with PBS and challenged with the same dose of IL-12 for another 12 hours. The levels of miR-132, miR-212, and miR-200a and the expression of STAT4 were measured after the miRNA-transfected cells were primed with IL-12 for 48 hours. IL-12–induced IFN-γ expression was measured after the subsequent 12 hours incubation with IL-12 (“secondary stimulation”). In the cells transfected with miR-132, miR-212, or miR-200a inhibitors, the expression of the respective miRNAs was down-regulated over 2 fold, compared with mock transfected cells (supplemental Figure 6). Inhibition of miR-132, miR-212, or miR-200a induction suppressed the IL-12–induced reduction of STAT4 (Figure 7A-B) and IFN-γ (Figure 7C-D), while having no discernible effect on their levels in the absence of IL-12. These results suggested that inhibition of miR-132, -212, and -200a induction in IL-12–treated NK cells helped cells to regain STAT4 levels and IL-12 responsiveness. Taken together, our data suggested that IL-12–induced miR-132, -212, and -200a up-regulation contribute to IL-12 tolerance (hyporesponsiveness).
Inhibition of IL-12–induced miR-132, miR-212, or miR-200a blocks the reduction of STAT4 induced by IL-12 and increases the IFN-γ production induced by further IL-12 stimulation. (A) Human NK cells (2 × 105) were transfected with control inhibitor (anti-miR ctrl), anti–miR-132, anti–miR-212, anti–miR-200a, or an equimolar mixture of anti–miR-132, miR-212, and miR-200a inhibitor (anti-miR mix) as indicated at an aggregate final concentration of 50nM. These transfected cells were treated with or without IL-12 (10 ng/mL) for 48 hours in the presence of IL-2, and the expression of STAT4 was measured by flow cytometry. Data are representative of 3 independent experiments with NK cells from different donors. The gray dotted line represents the isotype control staining of NK cells. (B) Summary of data from panel A, mean of IL-12–induced STAT4 reduction in cells transfected with anti-miR as indicated (STAT4 MFI without IL-12 – STAT4 MFI with IL-12) was calculated to determine the relative inhibitory effects of different miRNA inhibitors on IL-12–induced STAT4 reduction. The results are STAT4 MFI decreased mean ± SEM of representative experiment with NK cells from 3 different donors. *P < .05, **P < .01 versus anti-miR ctrl. (C) Cells treated with IL-12 and the indicated mir inhibitors as in panel A (but not harvested) were washed with complete growth medium and secondarily stimulated with 10 ng/mL of IL-12 for 12 hours in the presence of IL-2. Cellular expression of IFN-γ was measured by flow cytometry. Data are representative of 3 independent experiments with NK cells from different donors. The gray dotted line represents the isotype control staining of NK cells. (D) Summary of data from panel C, mean of the inhibitory effect of IL-12 pretreatment on secondary IL-12–induced IFN-γ production in indicated anti-miR–transfected cells (IFN-γ MFI without IL-12 priming – IFN-γ MFI with IL-12 priming) was calculated to determine the relative inhibitory effects of different miRNA inhibitors on the inhibitory effect of IL-12 pretreatment on secondary IL-12–induced IFN-γ production. The results are decreased IFN-γ MFI mean ± SEM of representative experiment with NK cells from 3 different donors. *P < .05 versus anti-miR ctrl.
Inhibition of IL-12–induced miR-132, miR-212, or miR-200a blocks the reduction of STAT4 induced by IL-12 and increases the IFN-γ production induced by further IL-12 stimulation. (A) Human NK cells (2 × 105) were transfected with control inhibitor (anti-miR ctrl), anti–miR-132, anti–miR-212, anti–miR-200a, or an equimolar mixture of anti–miR-132, miR-212, and miR-200a inhibitor (anti-miR mix) as indicated at an aggregate final concentration of 50nM. These transfected cells were treated with or without IL-12 (10 ng/mL) for 48 hours in the presence of IL-2, and the expression of STAT4 was measured by flow cytometry. Data are representative of 3 independent experiments with NK cells from different donors. The gray dotted line represents the isotype control staining of NK cells. (B) Summary of data from panel A, mean of IL-12–induced STAT4 reduction in cells transfected with anti-miR as indicated (STAT4 MFI without IL-12 – STAT4 MFI with IL-12) was calculated to determine the relative inhibitory effects of different miRNA inhibitors on IL-12–induced STAT4 reduction. The results are STAT4 MFI decreased mean ± SEM of representative experiment with NK cells from 3 different donors. *P < .05, **P < .01 versus anti-miR ctrl. (C) Cells treated with IL-12 and the indicated mir inhibitors as in panel A (but not harvested) were washed with complete growth medium and secondarily stimulated with 10 ng/mL of IL-12 for 12 hours in the presence of IL-2. Cellular expression of IFN-γ was measured by flow cytometry. Data are representative of 3 independent experiments with NK cells from different donors. The gray dotted line represents the isotype control staining of NK cells. (D) Summary of data from panel C, mean of the inhibitory effect of IL-12 pretreatment on secondary IL-12–induced IFN-γ production in indicated anti-miR–transfected cells (IFN-γ MFI without IL-12 priming – IFN-γ MFI with IL-12 priming) was calculated to determine the relative inhibitory effects of different miRNA inhibitors on the inhibitory effect of IL-12 pretreatment on secondary IL-12–induced IFN-γ production. The results are decreased IFN-γ MFI mean ± SEM of representative experiment with NK cells from 3 different donors. *P < .05 versus anti-miR ctrl.
Discussion
IL-12 is a relatively large heterodimeric molecule that is not rapidly degraded or excreted. The relatively long half-life of infused recombinant IL-12 leads to the persistence of physiologic levels of active cytokine for prolonged periods after each administration.28 However, most clinical studies have shown that repeated IL-12 administration has a profound abrogating effect on IL-12–induced IFN-γ production in vivo.29-32 Our studies demonstrate that prolonged or repeated IL-12 treatment of NK cells in vitro leads to a state of hyporesponsiveness to subsequent IL-12 stimulation.15 This phenomenon, referred to as IL-12 hyporesponsiveness, or IL-12 tolerance, may contribute to the ineffectiveness of IL-12 in clinical trials. Although this hyporesponsiveness to IL-12 in vivo is undoubtedly because of multiple factors, prolonged IL-12 treatment in vitro also induces IL-12 hyporesponsiveness in T and NK cells, and leads to a reduction of IFN-γ production after subsequent IL-12 stimulation.14,15 Taken together, results from in vivo and in vitro studies indicate that IL-12 signaling is tightly auto-regulated by elaborate mechanisms to control its onset and termination.
In a previous study we observed higher concentrations of IL-12 induce NK cell apoptosis, which undoubtedly contributes to the response of NK cells to subsequent IL-12 stimulation.15 However, the reduction of IFN-γ at early stages (Figure 2A) is disproportionate to the kinetics of cell apoptosis (supplemental Figure 7). Therefore, we think that the IL-12 signaling pathway down-regulation is the predominant cause of reduced IFN-γ production, especially in the early stages.
In this study, we investigated the expression profiles of miRNAs in IL-12–treated NK cells and the effects of IL-12–inducible miRNAs on regulation of IL-12 signaling, and demonstrated that miR-132, -212, and -200a, are up-regulated during prolonged or repeated IL-12 treatment. Further, these miRNAs negatively regulate the IL-12 signaling pathway by down-regulating STAT4 expression in human NK cells. This finding appears to represent a novel autoregulatory mechanism whereby IL-12 down-regulates the responsiveness of NK cells to further IL-12 stimulation.
IL-12 acts by binding to receptors at the cell surface to activate complex signal transduction pathways including the JAK2-STAT4 pathway in the IL-12 induction of IFN-γ production by T cells and NK cells.6,7 However, rampant cytokine signal transduction can have disastrous biologic consequences, and for this reason, signaling pathways are tightly controlled at multiple points.33 Three families of proteins, the SH2-containing phosphatases (SHP), the protein inhibitors of activated STATs (PIAS), and the suppressors of cytokine signaling (SOCS), have previously been described to inhibit specific and distinct aspects of cytokine signal transduction.33-36 SHP proteins are constitutively expressed, and can attenuate cytokine signal transduction by dephosphorylating signaling intermediates such as JAK and its receptor.33,34 PIAS proteins, also constitutively expressed, sumoylate STATs to inhibit transcriptional activation.35 SOCS proteins are induced in response to cytokine signaling and can inhibit JAK activity or target signaling components for ubiquitination and subsequent proteolysis. SOCS-3 inhibits IL-12–induced STAT4 activation by binding through its SH2 domain to the STAT4 docking site in the IL-12 receptor β2 subunit.36 None of these mechanisms reduce the levels of STAT protein,35,36 whereas the miRNA-based mechanism described in our work does. Although STAT4 mRNA gradually decreases after continued IL-12 treatment, the mRNA levels of STAT4 are not affected as significantly as the protein levels of STAT4 in IL-12–treated NK cells. Moreover, the kinetics of miRNA down-regulation correlate better with STAT4 protein expression than STAT4 mRNA in IL-12 treated cells. This suggests that control of steady state mRNA levels may contribute to the STAT4 down-regulation induced by IL-12, but the mechanisms downstream of steady state mRNA (eg, translation) play a quantitatively greater role.
miRNA control has emerged as a critical regulatory principle in our understanding of the mammalian immune system.37,38 Key features of this control are a dose-dependent regulation of target protein concentrations over a mostly modest range and the targeting of multiple functionally related proteins.39 STAT4, a critical IL-12 signaling component, is constitutively degraded in NK cells.15 The presence of IL-2 can maintain the levels of STAT4 in NK cells by up-regulation of STAT4 expression, and enhances the response of NK cells to IL-12.22 However, STAT4 protein levels decrease noticeably after prolonged IL-12 treatment in the presence of IL-2, in a proteasome pathway-independent manner in NK cells.15 In this study, miR-132, miR-212, and miR-200a, were shown to be progressively reduced with time in NK cells cultured with IL-2 alone, whereas they progressively increase with time in NK cells cultured with IL-2 plus IL-12 (Figure 3). Taken together, these data suggest that the expressions of miR-132, miR-212, and miR-200a are regulated by both IL-2 and IL-12 signaling, and are involved in the regulation of IL-12 signaling by regulating STAT4. IL-2 probably up-regulates and maintains the expression of STAT4 by inhibition of the expression of miR-132, miR-212, and miR-200a to enhance the responsiveness of cells to IL-12 stimulation, though more work needs to be done on this phenomenon. Conversely IL-12 down-regulates the expression of STAT4 at least in part by inducing miR-132, miR-212, and miR-200a to induce IL-12 tolerance in NK cells. These results further demonstrate that small changes in the concentration of key cellular proteins affected by miRNA can have significant biologic consequences. miRNAs may have evolved to modulate the concentrations of such key cellular proteins and affect signal transduction. Transfection of miR-132, miR-212, and miR-200a mimics into NK cells results in the reduction of STAT4, a key signal molecules in the IL-12 signaling pathway, indicating the negative regulatory consequences of these miRNAs on STAT4 expression. Although we did not observe that the cytotoxicity of NK cells transfected with miR-132, -212, or -200a was significantly different from that of cells transfected with miR control in the presence or absence of IL-12 (supplemental Figure 8), transfection of miR-132, miR-212, and miR-200a mimics into either NK cells or T cells results in reduced IL-12 induction of IFN-γ (supplemental Figure 9). This suggests that IFN-γ production is more sensitive than cytotoxicity to the changes of STAT4 levels affected by miRNA regulation. Conversely, transfection of miR-132, miR-212, and miR-200a inhibitors into NK cells results in the recovery of cells from IL-12 tolerance and restores IFN-γ secretion in response to IL-12 challenge. Taken together, these findings support an important regulatory role for miR-132, miR-212, and miR-200a in IL-12 tolerance, and suggest these miRNAs could be negative feedback regulators of IL-12 signaling through targeting of STAT4.
miRNAs have been thought to target multiple mRNAs to regulate gene expression. A single miRNA might tune protein synthesis from thousands of genes by direct or indirect effects, only a few of which may be critical for a particular biologic process.40 In the present study, miR-132, -212, and -200a have been identified as negative regulators of the IL-12 signaling pathway by targeting STAT4 in human NK cells. It is probable that we are far from unveiling the complete mechanisms underlying IL-12 auto-regulation and other signaling components are involved. Ongoing work in our laboratory is addressing these issues.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the National Institutes of Health Blood Bank for providing healthy human blood, Xue Wang for experimental design and discussion, and Peng Wang for help in statistical analysis.
Authorship
Contribution: Y.H., M.Z., and A.I.D. designed the experiments, interpreted the data and wrote the paper; and Y.H. performed the experiments with assistance and advice from Y.L., H.Z. and L.H.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andrew I. Dayton, Laboratory of Molecular Virology, Center for Biologics Evaluation and Research, Food and Drug Administration, 1401 Rockville Pike, Rockville, MD 20892; e-mail:andrew.dayton@fda.hhs.gov; or Mingjie Zhang, Laboratory of Molecular Virology, Center for Biologics Evaluation and Research, Food and Drug Administration, 1401 Rockville Pike, Rockville, MD 20892; e-mail: ming.zhang@fda.hhs.gov.
References
Author notes
Y.H. and Y.L. contributed equally to this article.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal