Abstract
T-cell alloreactivity directed against non–self-HLA molecules has been assumed to be less peptide specific than conventional T-cell reactivity. A large variation in degree of peptide specificity has previously been reported, including single peptide specificity, polyspecificity, and peptide degeneracy. Peptide polyspecificity was illustrated using synthetic peptide-loaded target cells, but in the absence of confirmation against endogenously processed peptides this may represent low-avidity T-cell reactivity. Peptide degeneracy was concluded based on recognition of Ag-processing defective cells. In addition, because most investigated alloreactive T cells were in vitro activated and expanded, the previously determined specificities may have not been representative for alloreactivity in vivo. To study the biologically relevant peptide specificity and avidity of alloreactivity, we investigated the degree of peptide specificity of 50 different allo-HLA–reactive T-cell clones which were activated and expanded in vivo during GVHD. All but one of the alloreactive T-cell clones, including those reactive against Ag-processing defective T2 cells, recognized a single peptide allo-HLA complex, unique for each clone. Down-regulation of the expression of the recognized Ags using silencing shRNAs confirmed single peptide specificity. Based on these results, we conclude that biologically relevant alloreactivity selected during in vivo immune response is peptide specific.
Introduction
Alloreactive T cells directed against allogeneic HLA (allo-HLA) are involved in the development of GVHD and graft rejection after HLA-mismatched transplantation. In conventional T-cell reactivity directed against pathogens, each T cell is specific for a single foreign peptide presented in a self-HLA molecule.1 During thymic development, T cells that are reactive against self-peptides presented in self-HLA molecules undergo negative selection. Both T cells reactive against a single peptide as well as T cells exerting polyreactivity against > 1 peptide presented in self-HLA molecules will be deleted.2,3 T cells leaving the thymus are therefore tolerant for self-peptide–HLA complexes. Because T cells never encounter allo-HLA molecules during thymic development, T cells polyspecific for peptides presented in allogeneic HLA are not removed, and, therefore T-cell allo-HLA reactivity has been assumed to be less peptide specific than conventional T-cell reactivity.
The degree of peptide specificity of alloreactive T cells has been extensively studied. Most investigators used cells defective in Ag processing, like transporter associated with Ag processing (TAP)–deficient human T2 cells or murine RMA-S cells to address the role of peptide in allorecognition.4-11 In these studies, groups of alloreactive T-cell clones or lines were tested against these Ag-processing deficient cells, unloaded or loaded with peptides. Some T cells only recognized a single peptide and were therefore categorized as peptide specific. T cells recognizing > 1 peptide, without sequence homology, were considered polyspecific. In addition, some alloreactive T cells were reactive against Ag-processing deficient cells in the absence of exogenously loaded peptide, which was initially interpreted as peptide independent recognition because it was assumed that these cells expressed empty MHC molecules on the cell surface. However, because it was demonstrated that these cells do express a limited number of peptides, which are independent of TAP to be expressed in MHC molecules on the cell membrane,12 reactivity against Ag-processing deficient cells may also be based on peptide-specific recognition. Reactivity against allo-HLA molecules irrespective of the sequence of the peptide presented has been termed peptide degenerate allorecognition, although it is unclear whether this type of recognition occurs. Together these studies created the assumption that alloreactivity is a combination of different reactivities, ranging from peptide specific to peptide degenerate alloreactivity.
In several studies, the degree of peptide specificity in allorecognition was determined by testing alloreactive T cells against a limited number of defined peptides.7,8,13-15 In these studies, reactivity against target cells exogenously loaded with high concentrations of peptide was interpreted as biologic relevant reactivity, and recognition of different peptides presented in allo-MHC by alloreactive T cells was concluded to represent bona fide polyspecific allorecognition. However, because recognition of exogenously loaded peptides in the absence of reactivity against endogenously processed and presented Ag may represent low-avidity T-cell recognition, biologic relevance of reactivity against synthetic peptide-loaded target cells needs to be confirmed by investigating the T-cell reactivity against endogenously processed and presented Ag. In the absence of such confirmation, the polyspecific alloreactivity against synthetic peptides described in these studies might not be representative for alloreactivity that is exerted by T cells during in vivo GVHD or graft rejection.
During in vivo immune responses, T cells with high avidity are selectively expanded, as indicated by the observation that the memory T-cell repertoire directed against microbial Ags comprises T cells with high avidity for the specific Ags, whereas the naive T-cell repertoire specific for the same Ags contains a broad range of avidities.16,17 In contrast, in vitro priming of T cells results in a T-cell repertoire containing a large variety of avidities,18,19 comparable with the repertoire of naive T cells, indicating that during in vitro activation and expansion no selection for high avidity occurs. Because under normal circumstances allo-HLA molecules are not encountered, alloreactive T cells which can be present within the naive as well as the memory T-cell pool,20,21 will not have undergone selection for T cells with high avidity for allo-HLA molecules. Therefore, the repertoires of alloreactive T cells derived from individuals who have not been exposed to non-self-HLA molecules will contain a high variety of avidities. Most alloreactive T cells used for the study of peptide specificity and avidity of allorecognition were activated and expanded in vitro, and, accordingly, many of these alloreactive T cells have been reported to demonstrate a very broad range of avidities.4,8,22 We hypothesize that only in circumstances in which in vivo selection does occur, like during in vivo allo-HLA–directed immune responses including GVHD and graft rejection after HLA–mismatched transplantations, T cells with high avidity for allo-HLA molecules will be selected.
To study the biologically relevant degree of peptide specificity and avidity of T-cell alloreactivity, we investigated in detail the repertoire of allo-HLA–reactive T cells which were activated and expanded in vivo. From a patient experiencing severe GVHD after an HLA-A2–mismatched DLI, we isolated 50 different in vivo–activated allo-HLA-A2–reactive CD8+ T-cell clones and investigated their peptide-specific alloreactivity. All but one of the alloreactive T-cell clones showed reactivity against a single HPLC fraction of peptides eluted from HLA-A2, including 6 T-cell clones recognizing T2 cells without exogenously loaded peptides. Using multidimensional HPLC fractionation and mass spectrometry (MS), we identified the different peptides recognized by 7 alloreactive T-cell clones, including one T2-reactive T-cell clone, and demonstrated that the alloreactive T cells exerted high-avidity peptide-specific alloreactivity. Down-regulation of the expression of the recognized Ags using silencing shRNAs confirmed single peptide specificity of the alloreactive T-cell clones. These results demonstrate that T cells exerting biologically relevant alloreactivity in vivo exhibited peptide specificity. Based on these results, we conclude that the biologically relevant specificity which is selected during in vivo immune response is single peptide specific, as defined by recognition of endogenously processed and presented Ag.
Methods
Cell collection and preparation
After informed consent, peripheral blood was obtained from different individuals, and PBMCs were isolated by Ficoll-Isopaque separation and cryopreserved. Stable EBV-transformed B-cell lines (EBV-LCLs) were generated using standard procedures. HLA-A2–negative donor EBV-LCLs and K562 cells were transduced with a retroviral vector encoding for HLA-A*0201 as previously described.23 EBV-LCLs, T2 cells (obtained from ATCC), K562 cells, renal cell carcinoma cell line 1774, and melanoma cell line 1.14 were cultured in IMDM (Lonza) and 10% FBS (Lonza). Fibroblasts were cultured from skin biopsies in DMEM (Lonza) and 10% FBS. Drosophila cells expressing HLA-A2, CD54, and CD80 were kindly provided by Dr Hans Stauss (Department of Immunology and Molecular Pathology, Royal Free Hospital, University College London, London, United Kingdom) and cultured in Schneider medium (Invitrogen) supplemented with 5% FBS, at 24°C and atmospheric CO2. Copper sulfate (100μM) was added 48 hours before peptide loading to induce expression of HLA-A2, CD54, and CD80.
Generation of allo-HLA-A2–reactive T-cell clones
Allo-HLA-A2–reactive T-cell clones were isolated from a patient experiencing acute GVHD after single HLA locus mismatch SCT and subsequent donor lymphocyte infusion (DLI). Based on a crossover, the patient was HLA-A*0201 positive and the sibling donor was HLA-A*0201 negative, whereas all other HLA class I and II molecules were completely matched. To isolate the allo-HLA-A2–reactive T cells, PBMCs collected during GVHD were stained with anti–HLA-A2 FITC (BD Pharmingen), anti–HLA-DR allophycocyanin (BD Pharmingen), and anti-CD8 PE mAbs at 4°C for 30 minutes and washed once. Activated (HLA-DR+), donor-derived (HLA-A*0201−) CD8+ T cells were sorted single cell per well into U-bottom microtiter plates containing 100 μL of feeder mixture consisting of IMDM, 5% FBS, 5% human serum (HS), IL-2 (120 IU/mL; Chiron), PHA (0.8 μg/mL; Murex Biotec Limited), and 50 Gy irradiated allogeneic PBMCs (0.5 × 106/mL). Proliferating T-cell clones were selected and further expanded nonspecifically with PHA, IL-2, and irradiated allogeneic PBMCs. For frequency analyses, HLA-DR+ HLA-A*0201− CD8+ T cells were sorted in bulk and nonspecifically expanded with irradiated autologous PBMCs, IL-2, and PHA.
TCR-Vβ chain analysis
For determination of TCR Vβ usage of T-cell clones, the TCR Vβ kit (Beckman Coulter) was used. TCRβ chains which could not be stained with mAbs were determined using multiplex amplification as previously described.24 By sequence analyses, the variable region and CDR3 region of the TCR Vβ chains of T-cell clones was determined.25
Peptide identification by peptide elution, HPLC, and MS
A total of 3 × 1010 EBV-LCLs or T2 cells were lysed with lysis buffer, composed of 50mM Tris, 150mM NaCl, 5mM EDTA, 0.5% Nonidet P-40, and protease inhibitor mix (Complete; Roche). Lysates were centrifuged for 30 minutes at 11 000g and precleared for 60 minutes with CL4B sepharose beads. HLA-A2 immunoaffinity chromatography was performed with anti-HLA-A2–purified BB7.2 mAb covalently coupled to protein A beads. Subsequently, HLA/peptide complexes were eluted with 10% acetic acid. High molecular mass material (HLA heavy chain and light chain) was removed by size filtration through Centriprep filtration units with a cutoff value of 10 kDa. After freeze drying, the peptide mixture was subjected to multidimentional HPLC fractionation, followed by peptide identification using MS (see supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). To confirm peptide specificity, the allo-HLA–reactive T-cell clones were tested against T2 or HLA-A2+Drosophila cells loaded with titrated concentrations of characterized peptides. In addition, tandem mass spectrum of the synthetic peptides was recorded and compared with its eluted counterpart as additional confirmation.
Functional assays and flow cytometry
HLA-A2 restriction of allo-HLA–reactive T-cell clones was determined in cytotoxicity assays in which T-cell clones were tested in a 4-hour 51Cr-release assay against patient EBV-LCLs, donor EBV-LCLs, and donor EBV-LCLs transduced with HLA-A*0201 at an E:T ratio of 10:1. In addition, blocking studies were performed using BB7.2 (anti–HLA-A2), W6.32 (anti–HLA class I), and B1.23.2 (anti–HLA-B and -C) mAbs. Patient EBV-LCLs were preincubated with saturating concentrations of mAbs for 1 hour at 20°C before addition of T cells.
Tetramer staining was performed at 37°C for 20 minutes using tetramers composed of HLA-A*0201 and peptides CMV-IE1-VLE, CMV-pp65-NLV, EBV-BMLF1-GLC, EBV-BRLF1-YVL, EBV-LMP2-CLG, EBV-LMP2-FLY, and FLU-IMP-GIL. In addition, tetramers composed of HLA-A*0201 and newly identified epitopes HLA-DRA-FID, ATXN10-QVF, VPS13B-SLW, FDPS-YLD, USP11-FTW, and SHIP-GFP were generated as described previously.26 Functionality of tetramers was controlled by staining of the specific T-cell clones; all tetramers were functional with the exception of ATXN10-QVF.
Allo-HLA–reactive T-cell clones were tested against T2 cells loaded with a mixture of HLA-A2–binding peptides derived from CMV, EBV, and Flu at a final concentration of 1 μg/mL. In the stimulation assays, 5000 T cells were stimulated with 20 000 EBV-LCLs, T2 cells, K562+A2 cells, melanoma 1.14 cells, or RCC1774 cells or with 5000 fibroblasts or with 100 000 HLA-A2+Drosophila cells. All functional assays were performed in IMDM containing 5% FBS, 5% HS, and 100 IU/mL IL-2 at a total volume of 150 μL in 96-well plates. After 18 hours of incubation, supernatant was harvested, and IFNγ production was measured by standard ELISA.
Alanine substitution experiment
Of the 2 peptides recognized by clone HSS11 (FIDKFTPPV derived from HLA-DRA and FLLKLTPLL derived from THRAP4) modified peptides were generated with alanine substitutions at the amino acid positions identical between the 2 peptides (1, 4, 6, and 7). In addition, modified peptides were synthesized in which the anchor residues at position 2 and 9 between the 2 peptides were exchanged. To determine the peptide concentrations required for half maximum IFNγ production (EC50), clone HSS11 was incubated with T2 cells loaded with titrated concentrations of wild-type (wt) and modified peptides. EC50 of the modified peptides was compared with the EC50 of wt peptides.
Inhibition of gene expression by silencing shRNA
Lentiviral vectors encoding for shRNA sequences specific for the genes coding for the proteins from which the identified peptides were derived in combination with the puromycin resistance gene were used from the Sigma-Aldrich shRNA library. The following were used: for USP11 shRNA TRCN0000007360, for FDPS TRCN0000036294, for VPS13B TRCN0000083955, for ATXN10 TRCN0000084093, for DRA TRCN0000057309, and for THRAP4 TRCN0000019649. Fibroblasts were transduced with the lentiviral vectors and, after 3 days, puromycin was added. Lentiviral-transduced fibroblasts were cultured for 2 weeks with 4 μg/mL puromycin before testing. The lentiviral vector encoding the shRNAs for USP11, FDPS, VPS13B, and ATXN10 were transduced into HLA-A*0201 fibroblasts derived from healthy donor. The lentiviral vectors encoding the shRNAs for DRA and THRAP4 were transduced into SV40-transformed HLA-A*0201 fibroblasts derived from a bare lymphocyte syndrome patient (BLS; EBA) and its HLA identical healthy sibling (CBA), that were kindly provided by Dr P. van den Elsen (Department of Immunohematology and Blood Transfusion, Leiden University Medical Center, Leiden, The Netherlands). The fibroblasts from BLS patients were previously demonstrated to be DRA deficient.27 Cells were tested in 384-well plates with 3000 fibroblasts/well and 1000 T cells/well. The fibroblasts transduced with shRNA for DRA and THRAP were stimulated for 72 hours with 100 IU of IFN-γ/mL to increase DRA expression.
To confirm shRNA down-regulation of expression of the different genes, quantitative real-time PCR (TaqMan) for the different genes was performed. Total RNA was isolated from cells using the microRNaqueous kit (Ambion). First-strand cDNA synthesis was performed with oligo dT primers using M-MLV reverse transcriptase (Invitrogen). Samples were run with EvaGreen on an ABI 7500 (Applied Biosystems). Primers specific for the different genes are depicted in Table 1.
Primers used for quantitative real-time PCR
| Gene . | Position . | Sequence . |
|---|---|---|
| USP11 | Forward | GGGCCTGTATAACGTCCTGA |
| Reverse | CTCTGGCTCCCAGTCGATAG | |
| FDPS | Forward | TGAGATCTGTGGGGGTCTTC |
| Reverse | TCCCGGAATGCTACTACCAC | |
| VPS13B | Forward | ATGATTGTGTGTGCCTTGGA |
| Reverse | TTGGCACTGAATGTTCCAGA | |
| ATXN10 | Forward | CAAGCCATGTTTCCCAAACT |
| Reverse | ACCCGCAAAAGATCAATCAC | |
| HLA-DRA | Forward | CATCCAGGCCGAGTTCTATC |
| Reverse | CTGGTGGGGTGAACTTGTCT | |
| THRAP4 | Forward | CTGCACATCGCCAAACTAGA |
| Reverse | TACCCTCGGGAGACTCAATG |
| Gene . | Position . | Sequence . |
|---|---|---|
| USP11 | Forward | GGGCCTGTATAACGTCCTGA |
| Reverse | CTCTGGCTCCCAGTCGATAG | |
| FDPS | Forward | TGAGATCTGTGGGGGTCTTC |
| Reverse | TCCCGGAATGCTACTACCAC | |
| VPS13B | Forward | ATGATTGTGTGTGCCTTGGA |
| Reverse | TTGGCACTGAATGTTCCAGA | |
| ATXN10 | Forward | CAAGCCATGTTTCCCAAACT |
| Reverse | ACCCGCAAAAGATCAATCAC | |
| HLA-DRA | Forward | CATCCAGGCCGAGTTCTATC |
| Reverse | CTGGTGGGGTGAACTTGTCT | |
| THRAP4 | Forward | CTGCACATCGCCAAACTAGA |
| Reverse | TACCCTCGGGAGACTCAATG |
Results
Isolation and characterization of allo-HLA-A2–reactive T cells activated during acute GVHD
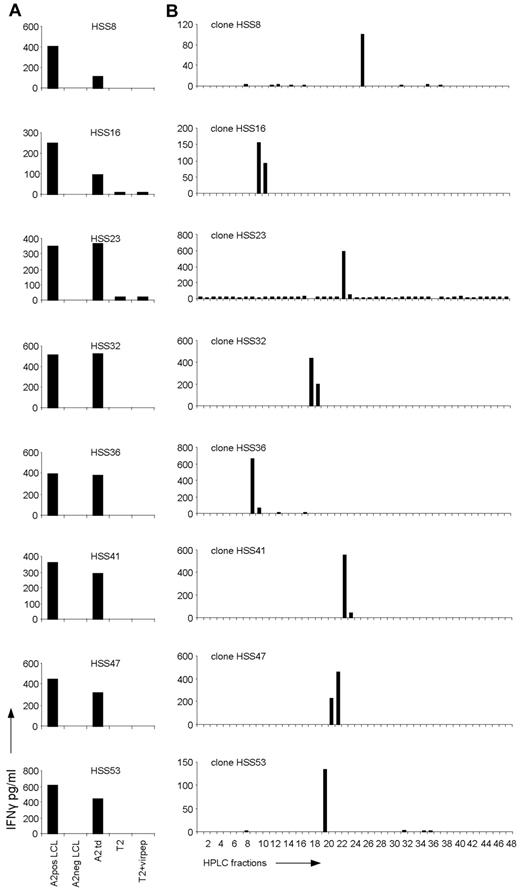
To be able to characterize the peptide-HLA ligand specificity of allo-HLA–reactive T cells exerting allo-immune responses in vivo, we isolated activated donor-derived CD8 T cells from a patient experiencing severe acute GVHD after single HLA-A2 locus mismatch SCT and subsequent DLI. The activated, HLA-DR–positive CD8 T cells of donor origin (HLA-A2 negative) were sorted single cell per well from peripheral blood collected at the time of GVHD and expanded. Fifty of 56 isolated CD8 T-cell clones were shown to be alloreactive because these T cells recognized patient but not donor EBV-LCLs. All 50 alloreactive T-cell clones were allo-HLA-A2 reactive because clones were reactive against donor EBV-LCL retrovirally transduced with HLA-A2, as shown for 8 representative clones (Figure 1A). In addition, recognition of HLA-A2–positive EBV-LCLs could be blocked by anti-HLA class I as well as anti–HLA-A2 mAbs (data not shown). By flow cytometric analysis using Vβ mAbs and multiplex PCR followed by sequencing of the different TCR Vβ chains (data not shown), we demonstrated that all 50 allo-HLA-A2–reactive CD8 T-cell clones were of different clonal origin. In addition, a high variety in TCR Vβ chain usage was observed, and no sequence homology between the allo-HLA–reactive clones was detected.
Recognition of HPLC fractions of HLA-A2–eluted peptides by the allo-HLA–reactive T-cell clones. (A) The allo-HLA–reactive T-cell clones were stimulated with HLA-A2–positive EBV-LCLs (A2pos LCL), HLA-A2–negative EBV-LCLs (A2neg LCL), HLA-A2–negative EBV-LCLs transduced with HLA-A2 (A2 td), T2 cells (T2), and T2 cells loaded with a mixture of different HLA-A2–binding peptides from CMV, EBV, and Flu (T2+virpep). Supernatants were harvested after 18 hours of stimulation and IFNγ was measured by standard ELISA. (B) Peptides were eluted from the HLA-A2 molecules of EBV-LCLs and fractionated by RP-HPLC using a water/acetonitrile/TFA gradient. Twenty-six of the 44 non-T2 recognizing allo-HLA-A2–reactive T-cell clones, of which 8 are shown in this figure, were stimulated with T2 cells loaded with the HPLC fractions.
Recognition of HPLC fractions of HLA-A2–eluted peptides by the allo-HLA–reactive T-cell clones. (A) The allo-HLA–reactive T-cell clones were stimulated with HLA-A2–positive EBV-LCLs (A2pos LCL), HLA-A2–negative EBV-LCLs (A2neg LCL), HLA-A2–negative EBV-LCLs transduced with HLA-A2 (A2 td), T2 cells (T2), and T2 cells loaded with a mixture of different HLA-A2–binding peptides from CMV, EBV, and Flu (T2+virpep). Supernatants were harvested after 18 hours of stimulation and IFNγ was measured by standard ELISA. (B) Peptides were eluted from the HLA-A2 molecules of EBV-LCLs and fractionated by RP-HPLC using a water/acetonitrile/TFA gradient. Twenty-six of the 44 non-T2 recognizing allo-HLA-A2–reactive T-cell clones, of which 8 are shown in this figure, were stimulated with T2 cells loaded with the HPLC fractions.
The allo-HLA reactivity of GVHD-inducing T cells is dependent on peptide-specific recognition
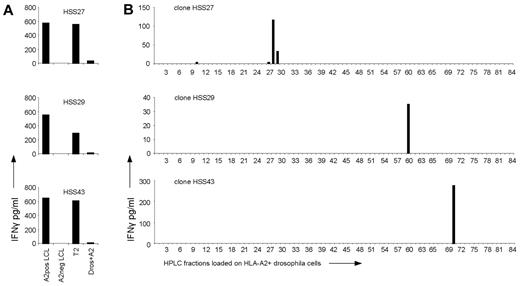
To determine whether allo-HLA-A2–reactive T cells recognized HLA-A2 irrespective of the peptide presented in the peptide-binding groove, T-cell clones were stained with a mix of tetramers composed of HLA-A2 and different virus-specific peptides derived from CMV, EBV, and Flu. None of the T-cell clones stained with the viral HLA-A2 tetramers (data not shown). Next, the 50 allo-HLA-A2–reactive T-cell clones were tested against T2 cells and T2 cells loaded with the different CMV, EBV, and Flu HLA-A2–binding peptides. Forty-four of the 50 T-cell clones showed no reactivity against peptide-loaded or unloaded T2 cells (Figure 1A). six of the 50 T-cell clones were reactive against unloaded T2 cells (reactivity of 3 T-cell clones is shown in Figure 2A) and showed comparable reactivity against peptide-loaded T2 cells (data not shown). Because T2 cells express HLA-A2 peptide complexes, although with restricted peptide variation, the ability of allo-HLA–reactive T cells to recognize allo-HLA-A2 could still be based on peptide-specific recognition. To determine whether these T-cell clones recognized allo-HLA-A2 independent of the peptide presented in HLA-binding groove, the clones were tested against HLA-A2–expressing Drosophila cells loaded with the different HLA-A2–binding peptides. The T2-reactive T-cell clones showed no reactivity against unloaded (Figure 2A) or peptide loaded HLA-A2+Drosophila cells (data not shown). These results indicate that for all 50 alloreactive T-cell clones activation was not only dependent on the interaction of the TCR with allo-HLA-A2 molecule but that specific peptides were required.
Investigation of the peptide specificity of T2-reactive T-cell clones. (A) T2-reactive T-cell clones HSS27, HSS29, and HSS43 were stimulated with HLA-A2–positive EBV-LCLs (A2pos LCL), HLA-A2–negative EBV-LCLs (A2neg LCL), T2 cells (T2), and Drosophila cells expressing HLA-A2, CD80, and CD54 (Dros+A2). (B) Peptides were eluted from the HLA-A2 molecules of T2 cells and fractionated by RP-HPLC. The 6 allo-HLA–reactive T-cell clones which recognized T2 cells were tested for IFNγ production against the HPLC fractionations loaded on Drosophila cells expressing HLA-A2, CD80, and CD54; 3 representative clones are shown.
Investigation of the peptide specificity of T2-reactive T-cell clones. (A) T2-reactive T-cell clones HSS27, HSS29, and HSS43 were stimulated with HLA-A2–positive EBV-LCLs (A2pos LCL), HLA-A2–negative EBV-LCLs (A2neg LCL), T2 cells (T2), and Drosophila cells expressing HLA-A2, CD80, and CD54 (Dros+A2). (B) Peptides were eluted from the HLA-A2 molecules of T2 cells and fractionated by RP-HPLC. The 6 allo-HLA–reactive T-cell clones which recognized T2 cells were tested for IFNγ production against the HPLC fractionations loaded on Drosophila cells expressing HLA-A2, CD80, and CD54; 3 representative clones are shown.
To investigate the degree of peptide specificity exerted by the in vivo selected allo-HLA-A2–reactive T cells, 26 T-cell clones, randomly selected from the 44 allo-HLA-A2–reactive T-cell clones not recognizing T2 cells, were tested against T2 cells loaded with HPLC fractions of HLA-A2–eluted peptides. For this purpose, HLA-A2–positive EBV-LCLs were lysed and their peptide-HLA-A2 complexes were purified. Subsequently, the peptides were eluted from HLA-A2, and separated from the HLA-A2 heavy chain and light chain. The peptide mixture was then subjected to reverse phase (RP)–HPLC and fractions were collected. The T-cell clones were tested against T2 cells loaded with a small sample of each fraction, cultured for 18 hours, after which IFNγ was measured by ELISA. Twenty-one of the 26 tested T-cell clones, of which 8 are shown in Figure 1B, showed recognition of 1 or 2 subsequent HPLC fractions. Almost all clones recognized a different HPLC fraction, indicating that different peptides presented in allo-HLA-A2 were recognized. One of 26 tested clones, clone HSS11, showed recognition of 2 not-adjacent HPLC fractions (see Figure 4A). Four of 26 tested clones showed no reactivity against the HPLC fractions (data not shown).
To investigate the degree of peptide specificity exerted by the 6 allo-HLA–reactive T-cell clones recognizing T2 cells, clones were tested against HLA-A2–expressing Drosophila cells loaded with HPLC fractions of HLA-A2–eluted peptides derived from T2 cells. All 6 clones, of which 3 are shown in Figure 2B, showed recognition of 1 or 2 adjacent fractions, indicating that T2-reactive T-cell clones also exhibited peptide specific allo-HLA reactivity.
Identification of the peptides recognized in the context of allo-HLA-A2
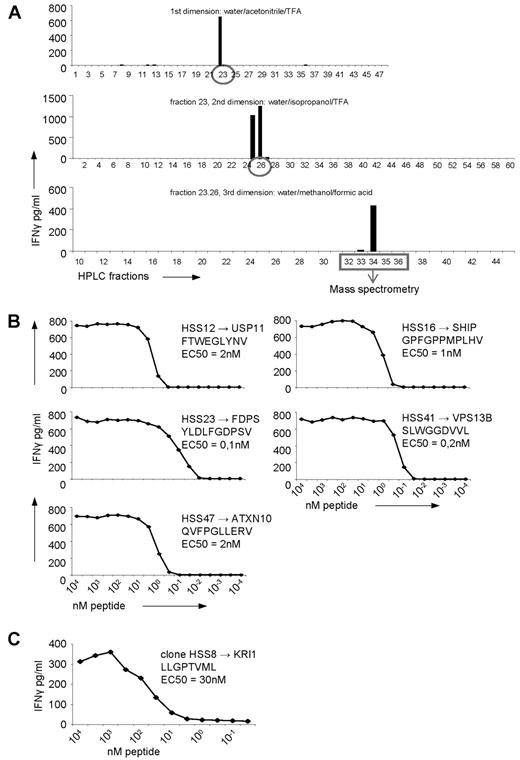
To determine whether one or multiple peptides were recognized by the allo-HLA–reactive T-cell clones, the peptides recognized by 5 randomly selected T-cell clones were identified using multidimensional RP-HPLC fractionation and MS. Identification of the peptide recognized by clone HSS12 is representative for the other 4 clones and is shown in Figure 3A. All 5 clones recognized a single, but unique, peptide (Figure 3B, Table 2).
Identification of the peptides recognized by the allo-HLA–reactive T-cell clones using multidimensional HPLC fractionation and MS. (A) Five non-T2-recognizing allo-HLA–reactive T-cell clones were stimulated for 18 hours with T2 cells loaded with the first dimension HPLC fractions of peptides eluted from HLA-A2 derived from EBV-LCLs, and IFNγ was measured in the supernatant by standard ELISA. The recognized HPLC fractions were subjected to a second fractionation using a water/isopropanol/TFA gradient, loaded on T2 cells, and subsequently tested for recognition by the T-cell clones. The recognized fractions were fractionated a third time, using a water/methanol/formic acid gradient and tested for recognition. After the third fractionation, the peptide masses present in the recognized and in the adjacent not-recognized fractions were analyzed by MS and the sequences of the peptides were identified. The identification of the peptide recognized by clone HSS12, which is representative for the identification of the peptides recognized by the 4 other clones, is shown. (B) The 5 allo-HLA–reactive T-cell clones, for which the recognized peptides were identified, were tested for their affinity for the respective peptides. The T-cell clones were stimulated with T2 cells loaded with titrated concentrations of the peptides for 18 hours, and IFNγ was measured in the supernatant by standard ELISA. (C) By MS it was determined that the peptide recognized by the T2-reactive T-cell clone HSS8 was derived from KRI1. To determine the affinity of the T-cell clone for this peptide, the clone was tested for IFNγ production against Drosophila cells expressing HLA-A2, CD80 and CD54 loaded with titrated concentrations of the peptide. EC50 represents the peptide concentration needed for half of the maximum IFNγ production by the representative T-cell clone.
Identification of the peptides recognized by the allo-HLA–reactive T-cell clones using multidimensional HPLC fractionation and MS. (A) Five non-T2-recognizing allo-HLA–reactive T-cell clones were stimulated for 18 hours with T2 cells loaded with the first dimension HPLC fractions of peptides eluted from HLA-A2 derived from EBV-LCLs, and IFNγ was measured in the supernatant by standard ELISA. The recognized HPLC fractions were subjected to a second fractionation using a water/isopropanol/TFA gradient, loaded on T2 cells, and subsequently tested for recognition by the T-cell clones. The recognized fractions were fractionated a third time, using a water/methanol/formic acid gradient and tested for recognition. After the third fractionation, the peptide masses present in the recognized and in the adjacent not-recognized fractions were analyzed by MS and the sequences of the peptides were identified. The identification of the peptide recognized by clone HSS12, which is representative for the identification of the peptides recognized by the 4 other clones, is shown. (B) The 5 allo-HLA–reactive T-cell clones, for which the recognized peptides were identified, were tested for their affinity for the respective peptides. The T-cell clones were stimulated with T2 cells loaded with titrated concentrations of the peptides for 18 hours, and IFNγ was measured in the supernatant by standard ELISA. (C) By MS it was determined that the peptide recognized by the T2-reactive T-cell clone HSS8 was derived from KRI1. To determine the affinity of the T-cell clone for this peptide, the clone was tested for IFNγ production against Drosophila cells expressing HLA-A2, CD80 and CD54 loaded with titrated concentrations of the peptide. EC50 represents the peptide concentration needed for half of the maximum IFNγ production by the representative T-cell clone.
Identified peptides recognized by the alloreactive T-cell clones
| Clone . | Protein . | Peptide . | RefSeq ID . |
|---|---|---|---|
| HSS8 | KRI1 | LLGPTVML | NM_023008 |
| HSS11 | HLA-DR1α | FIDKFTPPV | NM_019111 |
| HSS11 | THRAP4 | FLLKLTPLL | NM_001079518 |
| HSS12 | USP11 | FTWEGLYNV | NM_004651 |
| HSS16 | SHIP | GPFGPPMPLHV | NM_001017915 |
| HSS23 | FDPS | YLDLFGDPSV | NM_002004 |
| HSS41 | VPS13B | SLWGGDVVL | NM_181661 |
| HSS47 | ATXN10 | QVFPGLLERV | NM_013236 |
| Clone . | Protein . | Peptide . | RefSeq ID . |
|---|---|---|---|
| HSS8 | KRI1 | LLGPTVML | NM_023008 |
| HSS11 | HLA-DR1α | FIDKFTPPV | NM_019111 |
| HSS11 | THRAP4 | FLLKLTPLL | NM_001079518 |
| HSS12 | USP11 | FTWEGLYNV | NM_004651 |
| HSS16 | SHIP | GPFGPPMPLHV | NM_001017915 |
| HSS23 | FDPS | YLDLFGDPSV | NM_002004 |
| HSS41 | VPS13B | SLWGGDVVL | NM_181661 |
| HSS47 | ATXN10 | QVFPGLLERV | NM_013236 |
For 7 T-cell clones (HSS8, 11, 12, 16, 23, 41, and 47), including one T2-reactive clone (HSS8), the recognized peptides were identified. Clone HSS11 showed recognition of peptides with sequence similarity. The protein of which the peptides are derived from, the sequence of the peptides, and the Reference Sequence identification number (http://srs.bioinformatics.nl/hspv/search.php) of the peptides are shown.
To test the affinity of the 5 allo-HLA–reactive T cells for their respective recognized peptide presented in allo-HLA-A2, the allo-HLA–reactive T-cell clones were tested against T2 cells loaded with titrated concentrations of the identified peptides. The concentration of peptide needed for half maximum IFNγ production (EC50) ranged between 3nM and 100pM for all T-cell clones.
In addition, for one of the T2-reactive T-cell clones, clone HSS8, peptide identification was performed with the HLA-A2–eluted peptides of T2 by multidimensional HPLC fractionation and MS (Table 2). Peptide titration demonstrate that 30nM of this peptide LLGPTVML derived from KRI-1 homolog, was needed for half maximum IFNγ production (EC50) by clone HSS8 (Figure 3C). This illustrates that the recognition of TAP-deficient T2 cells by allo-HLA–reactive T cells is also based on peptide-specific allo-HLA recognition.
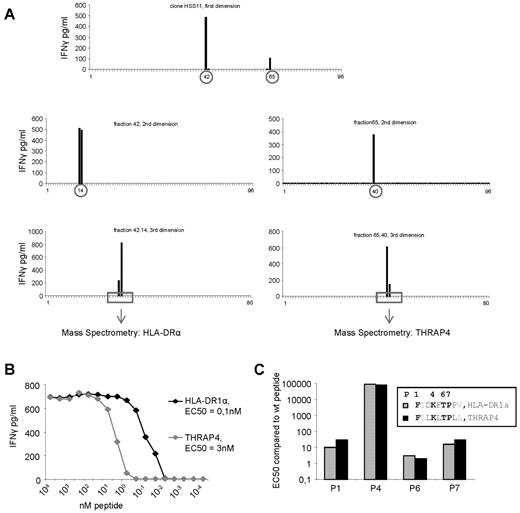
One of the allo-HLA–reactive T-cell clones, clone HSS11, recognized 2 fractions after first HPLC fractionation (Figure 4A). To determine whether this recognition pattern was based on recognition of 2 different peptides or based on recognition of different length variants of the same peptide, these 2 fractions were subsequently subjected to RP-HPLC for multidimensional fractionation, and peptides present in the positive fractions after 3 dimensions were identified by MS (Figure 4A, Table 2). Testing the T-cell clone against the identified peptides loaded on T2 cells showed that clone HSS11 recognized epitope FIDKFTPPV derived from HLA-DRA, and epitope FLLKLTPLL derived from THRAP4. Peptide titration, shown in Figure 4B, demonstrated that 100pM HLA-DRA peptide or 3nM THRAP4 peptides was needed for half maximum IFNγ production (EC50) of T-cell clone HSS11, illustrating that the T-cell clone HSS11 was specific for 2 peptides. The 2 recognized peptides share sequence homology. The amino acids at position 2 and 9 are different between the 2 peptides; however, these amino acids form the anchor residues which provide binding to HLA but do not interact with the TCR. Exchange of the amino acids at position 2 and 9 between the 2 peptides demonstrated no change in EC50 for both peptides (data not shown), indicating that the 2 anchor residues were similarly involved in HLA binding and did not influence the TCR interaction of the other amino acids. Of the 7 remaining amino acids, 4 were identical. To investigate whether these identical amino acids at positions 1, 4, 6, and 7 were involved in TCR interaction, these amino acids were substituted by an alanine residue. As illustrated in Figure 4C, all 4 amino acids were involved in TCR interaction, because alanine substitution at all positions increased EC50, demonstrating a lower binding affinity between the TCR and peptide/HLA-A2 complex. In addition, an identical change in EC50 for both peptides at all 4 positions was observed. Substitution at position 1 and 7 resulted in an intermediate 10- to 30-fold increase in EC50, substitution at position 4 resulted in a dramatic increase in EC50, and substitution at position 6 resulted only in a small change in EC50 of 3-fold. Together, these data suggest that the 2 peptide/HLA-A2 complexes recognized by clone HSS11 are conformational look-alikes. These results illustrate that allo-HLA–reactive T cells, including T2-reactive T cells, derived from an in vivo GVH response recognize one or a restricted number of peptides with high affinity in the context of allo-HLA.
Identification of the peptides recognized by allo-HLA–reactive T-cell clone HSS11. (A) Clone HSS11 recognized 2 fractions after the first HPLC fractionation of peptides eluted from HLA-A2 derived from EBV-LCLs. These 2 recognized fractions were subjected to a second and a third fractionation. The peptide masses present in the fractions recognized by clone HSS11 after the third fractionation as well as in the adjacent not recognized fractions were determined by MS. Sequence analysis by tandem MS was performed on those peptide masses that correlated with the recognition pattern of the T-cell clone. (B) To determine the affinity of clone HSS11 for the 2 identified peptides derived from HLA-DRA and THRAP4, the clone was stimulated with T2 cells loaded with titrated concentrations of the 2 peptides. EC50 represents the peptide concentration needed for half of the maximum IFNγ production by the T-cell clone. (C) To determine whether the 4 amino acids identical between the 2 peptides recognized by clone HSS11, at position P1, P4, P6, and P7, were involved in the TCR interaction, these amino acids were substituted by alanine residues. EC50 of the wt peptides and the modified peptides was determined by stimulating clone HSS11 with T2 cells loaded with titrated concentrations of the different peptides. The EC50 of the modified peptides was divided by the EC50 of the corresponding wt peptide.
Identification of the peptides recognized by allo-HLA–reactive T-cell clone HSS11. (A) Clone HSS11 recognized 2 fractions after the first HPLC fractionation of peptides eluted from HLA-A2 derived from EBV-LCLs. These 2 recognized fractions were subjected to a second and a third fractionation. The peptide masses present in the fractions recognized by clone HSS11 after the third fractionation as well as in the adjacent not recognized fractions were determined by MS. Sequence analysis by tandem MS was performed on those peptide masses that correlated with the recognition pattern of the T-cell clone. (B) To determine the affinity of clone HSS11 for the 2 identified peptides derived from HLA-DRA and THRAP4, the clone was stimulated with T2 cells loaded with titrated concentrations of the 2 peptides. EC50 represents the peptide concentration needed for half of the maximum IFNγ production by the T-cell clone. (C) To determine whether the 4 amino acids identical between the 2 peptides recognized by clone HSS11, at position P1, P4, P6, and P7, were involved in the TCR interaction, these amino acids were substituted by alanine residues. EC50 of the wt peptides and the modified peptides was determined by stimulating clone HSS11 with T2 cells loaded with titrated concentrations of the different peptides. The EC50 of the modified peptides was divided by the EC50 of the corresponding wt peptide.
Confirmation of single peptide specificity using inhibition of gene expression by silencing shRNA
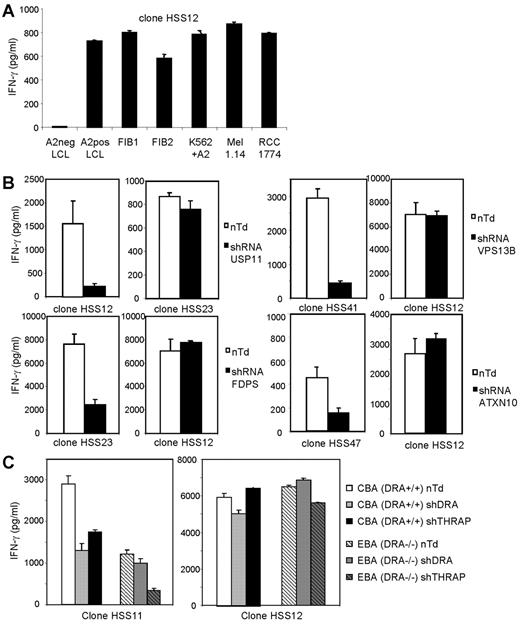
The allo-HLA-A2–reactive T-cell clones recognized EBV-LCLs as well as other HLA-A2–positive target cells including tumor cells and fibroblasts, as shown for one representative clone, clone HSS12 (Figure 5A). To confirm the single peptide specificity of the allo-HLA–reactive T cells and biologic relevance of the identified specificities, and to demonstrate that the reactivity directed against the other HLA-A2–positive target cells was also mediated by recognition of the same HLA-A2/peptide-complex, we transduced fibroblasts with silencing shRNAs specific for the genes coding for the proteins from which the identified peptides were derived. By quantitative real-time PCR, we confirmed that all shRNAs down-regulated the mRNA expression, varying between 5- and 31-fold (see legend, Figure 5B-C). The results in Figure 5B demonstrate that silencing with the specific shRNA almost completely blocked the recognition of the corresponding allo-HLA-A2–reactive T-cell clones whereas the reactivity of other allo-HLA-A2–reactive T-cell clones was unaltered. In Figure 5C, we demonstrate that the double peptide specificity of HSS11 directed against DRA and THRAP4 could be confirmed by transduction of fibroblast CBA with shRNA for either DRA or THRAP4. Both shRNAs were able to down-regulate the recognition of clone HSS11, whereas the reactivity of control T-cell clone HSS12 was unaltered. To confirm that the reactivity of HSS11 was mediated by recognition of both DRA and THRAP4, DRA−/− fibroblasts (EBA) were transduced with shRNA for DRA or THRAP4. The reactivity of HSS11 directed against DRA−/− cells was reduced compared with DRA+/+ cells. Transduction with shRNA for THRAP4 almost completely blocked reactivity, and transduction with shRNA for DRA as expected did not alter reactivity. These results clearly demonstrate that all allo-reactive T-cell clones recognized unique allo-HLA/peptide complexes.
Recognition of different HLA-A2–positive target cells by allo-HLA–reactive T-cell clones and confirmation of their single peptide specificity by transduction of shRNA specific for their respective recognized Ags. (A) The allo-HLA–reactive T-cell clones were stimulated with HLA-A2–negative EBV-LCLs (A2neg LCL), HLA-A2–positive EBV-LCLs (A2pos LCL), HLA-A2+ fibroblasts derived from 2 different individuals (FIB1 and FIB2), K562 cells transduced with HLA-A2 (K562+A2), HLA-A2–positive melanoma cell line 1.14 (Mel 1.14), and HLA-A2–positive renal cell carcinoma cell line 1774 (RCC 1774). Reactivity of one representative clone, clone HSS12, is shown. (B) Allo-HLA–reactive T-cell clones HSS12, HSS23, HSS41, and HSS47 were tested against fibroblasts transduced with lentiviral vectors encoding shRNAs specific for the genes of USP11, FDPS, VPS13B, or ATXN10 in combination with the puromycin resistance gene. shRNA-transduced cells were cultured for 14 days with puromycin (4 μg/mL) and used as stimulator cells for the corresponding allo-reactive T-cell clone and a control allo-reactive T-cell clone. Nontransduced cells (nTd) were used as control stimulator cells. By quantitative real-time PCR, the down-regulation of mRNA of the different genes compared with nontransduced cells was analyzed, the fold decrease for USP11 = 5, FDPS = 31, VPS13B = 5, ATXN10 = 10. (C) HSS11 and HSS12 were tested against DRA+/+ (CBA) and DRA−/− (EBA) fibroblasts transduced with lentiviral vectors encoding shRNAs specific for the genes of DRA or THRAP4, selected for 14 days with puromycin and cultured for 72 hours with 100 IU of IFNγ/mL. Down-regulation of mRNA of DRA and THRAP4 was analyzed by qRT-PCR, the decrease for THRAP4 in CBA and EBA was 10-fold, the decrease for DRA in CBA was 4-fold. No expression of DRA was measured in EBA.
Recognition of different HLA-A2–positive target cells by allo-HLA–reactive T-cell clones and confirmation of their single peptide specificity by transduction of shRNA specific for their respective recognized Ags. (A) The allo-HLA–reactive T-cell clones were stimulated with HLA-A2–negative EBV-LCLs (A2neg LCL), HLA-A2–positive EBV-LCLs (A2pos LCL), HLA-A2+ fibroblasts derived from 2 different individuals (FIB1 and FIB2), K562 cells transduced with HLA-A2 (K562+A2), HLA-A2–positive melanoma cell line 1.14 (Mel 1.14), and HLA-A2–positive renal cell carcinoma cell line 1774 (RCC 1774). Reactivity of one representative clone, clone HSS12, is shown. (B) Allo-HLA–reactive T-cell clones HSS12, HSS23, HSS41, and HSS47 were tested against fibroblasts transduced with lentiviral vectors encoding shRNAs specific for the genes of USP11, FDPS, VPS13B, or ATXN10 in combination with the puromycin resistance gene. shRNA-transduced cells were cultured for 14 days with puromycin (4 μg/mL) and used as stimulator cells for the corresponding allo-reactive T-cell clone and a control allo-reactive T-cell clone. Nontransduced cells (nTd) were used as control stimulator cells. By quantitative real-time PCR, the down-regulation of mRNA of the different genes compared with nontransduced cells was analyzed, the fold decrease for USP11 = 5, FDPS = 31, VPS13B = 5, ATXN10 = 10. (C) HSS11 and HSS12 were tested against DRA+/+ (CBA) and DRA−/− (EBA) fibroblasts transduced with lentiviral vectors encoding shRNAs specific for the genes of DRA or THRAP4, selected for 14 days with puromycin and cultured for 72 hours with 100 IU of IFNγ/mL. Down-regulation of mRNA of DRA and THRAP4 was analyzed by qRT-PCR, the decrease for THRAP4 in CBA and EBA was 10-fold, the decrease for DRA in CBA was 4-fold. No expression of DRA was measured in EBA.
Determination of the relative frequency of the identified allo-HLA/peptide specificities
To determine the frequency of the different allo-HLA-A2 specificities at the time of acute GVHD, the activated CD8+ donor T cells were isolated, expanded, and used for frequency analyses. Frequency analyses were performed by staining the pool of donor T cells with tetramers composed of HLA-A2 in complex with the different newly identified alloepitopes. As shown in Figure 6, all allo-HLA–reactive T-cell specificities could be observed, and the frequency of these different specificities varied between 0.07% and 1.16% of activated donor CD8+ T cells.
Variable frequencies of the different identified allo-HLA/peptide reactivities. Activated CD8+ donor T cells were sorted from PBMCs at the time of GVHD, expanded nonspecifically, and stained with the different functional tetramers conjugated with either PE or allophycocyanin, in combination with anti-CD8 Alexa 700. As a control, the CMV-pp65-NLV tetramer was included. In the dot plots, the CD8+ T cells are shown, and numbers in the different quadrants represent the percentage tetramer-positive CD8+ T cells.
Variable frequencies of the different identified allo-HLA/peptide reactivities. Activated CD8+ donor T cells were sorted from PBMCs at the time of GVHD, expanded nonspecifically, and stained with the different functional tetramers conjugated with either PE or allophycocyanin, in combination with anti-CD8 Alexa 700. As a control, the CMV-pp65-NLV tetramer was included. In the dot plots, the CD8+ T cells are shown, and numbers in the different quadrants represent the percentage tetramer-positive CD8+ T cells.
Discussion
In this study, we demonstrate that alloreactive T cells isolated from a patient with GVHD after an HLA-A2–mismatched transplantation exerted peptide-specific allo-HLA reactivity. The alloreactive T-cell clones derived from the in vivo alloimmune response, including those recognizing TAP-deficient T2 cells, were reactive against 1 or 2 subsequent HPLC fractions of peptides eluted from HLA-A2. Identification of the different peptides recognized in the context of the allo-HLA molecule followed by confirmation of the single peptide specificity by down-regulation of the respective Ags using silencing shRNAs, demonstrated that the alloreactive T cells exert high-avidity recognition for a single endogenously processed and presented peptide.
Similar to other studies, we observed that part of the allo-HLA–reactive T cells recognized TAP-deficient T2 cells. Rammensee and colleagues eluted and identified multiple peptides presented in the context of the HLA molecules expressed at the cell surfaces of TAP-deficient T2 cells.12 Most of these peptides could also be found on TAP-expressing cells, indicating that many peptides can enter the endoplasmic reticulum and be presented in HLA molecules at the cell surface independent of TAP. It is therefore not surprising that part of the alloreactive T cells recognized TAP-independent peptides and were reactive against TAP-deficient cells. Our results illustrate that high-avidity reactivity against T2 cells is not based on peptide degenerate allorecognition, but on peptide-specific allorecognition.
One of 26 T-cell clones tested against the HPLC fractions, clone HSS11, showed recognition of 2 different fractions, and peptide identification demonstrated that HSS11 recognized 2 different peptides. The 2 peptides share relevant sequence homology because 4 of 7 amino acids involved in interaction with the TCR are identical. Alanine substitutions of these 4 amino acids suggested that the 2 HLA-A2/peptide complexes are conformational look-alikes, most likely exhibiting a similar 3 dimensional structure. This degree of peptide cross-reactivity has also been described for T cells specific for foreign peptides presented in self-HLA molecules.28,29
In most studies reporting polyspecific allorecognition, alloreactive T cells were tested against a limited number of defined peptides loaded onto allo-HLA–expressing target cells.2,8,13,14 Many of the alloreactive T cells tested in this manner showed reactivity against several different peptides. However, because recognition of different peptides was not confirmed by testing the T cells against endogenously processed and presented peptides, it is possible that the demonstrated T-cell reactivity directed against synthetic peptides loaded target cells represented low-avidity T-cell reactivity. It was previously demonstrated that low-avidity T-cell recognition does not lead to effective T-cell reactivity in vivo.30 In our study, peptides recognized by the alloreactive T cells were identified using multidimensional HPLC fractionations of peptides eluted from HLA-A2 of EBV-LCLs. In this method large numbers of peptides (10 000-20 000)31 are tested at relatively low concentrations, especially after second and third fractionation. Therefore, by testing T cells against HPLC fractions, only peptides recognized with high avidity were identified and low-avidity T-cell recognition of other peptides was not determined. Down-regulation of the identified Ags using silencing shRNAs almost completely blocked the reactivity of the corresponding allo-HLA–reactive T cells, demonstrating that the identified peptides, represented the actual biologically relevant specificities of investigated alloreactive T cells. During in vivo immune responses directed against pathogens, T cells with high avidity against a single antigenic peptide presented in the context of self-HLA molecules are selectively expanded.16,17 Similarly, our results demonstrate that T cells which are selectively activated and expanded during in vivo allo-HLA–directed immune responses, apparently also exhibit high-avidity recognition against single peptides presented in allo-HLA molecules.
Our results illustrate that high-avidity allo-HLA recognition is as peptide specific as conventional T-cell recognition of foreign peptides presented in self-HLA molecules. These findings appear to be in conflict with the hypothesis that allorecognition is less peptide specific than conventional T-cell reactivity because allo-MHC molecules are not encountered during thymic development and therefore T cells crossreactive against different peptides presented in allo-MHC molecules are not depleted.2,3 This hypothesis is based on the assumption that the prethymic T-cell repertoire contains large numbers of MHC and peptide cross-reactive T cells and that thymic selection is responsible for removal of these T cells. This assumption was already disputed by Zerrahn et al, who demonstrated that prethymic T cells are as MHC cross-reactive as T cells after normal thymic selection, indicating that there may not be a large prethymic pool of highly MHC and peptide cross-reactive T cells which is removed during thymic selection.32 Our results of peptide-specific allo-HLA-A2 reactivity of 50 different in vivo–activated and –expanded alloreactive T cells did also not provide evidence for the existence of T cells cross-reactive against multiple endogenously processed and presented peptides in the context of allo-HLA molecules not encountered during thymic development. We therefore speculate that highly cross-reactive prethymic T cells are either rare or nonexistent.
In summary, the results in this study demonstrate that T cells exerting biologically relevant alloreactivity in vivo exhibit high-avidity recognition against single peptide–allo-HLA complexes. Based on the fact that down-regulation of the identified Ags almost completely blocked the reactivity of the corresponding T cells, we conclude that only these high-avidity interactions, as defined by reactivity against endogenously processed and presented Ag, are biologically relevant in vivo. We hypothesize that during in vivo allo-HLA–directed immune responses, only T cells exhibiting high-avidity recognition against single peptides presented in the context of allo-HLA molecules are selectively expanded.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Guido de Roo and Menno van der Hoorn for expert technical assistance. They thank Martijn Rabelink and Prof Dr Rob Hoeben for providing us the lentiviral constructs encoding the different shRNAs. Drosophila cells expressing HLA-A2, CD54, and CD80 were kindly provided by Prof Dr Hans Stauss, and Dr Peter van den Elsen provided us with the DRA−/− fibroblasts derived from BLS patients.
This work was supported by the Landsteiner Foundation for Blood Transfusion (LSBR0611) and the Dutch Cancer Society (UL2005-3251).
Authorship
Contribution: A.L.A. designed and performed research, and wrote the manuscript; D.M.v.d.S., R.S.H., M.G.D.K., C.A.M.v.B., and A.H.d.R. performed research; J.W.D. provided essential reagents; J.H.F.F. wrote the manuscript; P.A.v.V. and M.H.M.H. designed research and wrote the manuscript; and all authors reviewed and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mirjam H. M. Heemskerk, Department of Hematology, Leiden University Medical Center, PO Box 9600, 2300RC Leiden, The Netherlands; e-mail: mhmheemskerk@lumc.nl.
References
Author notes
P.A.v.V. and M.H.M.H. share senior authorship.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal