Abstract
Abstract 3564
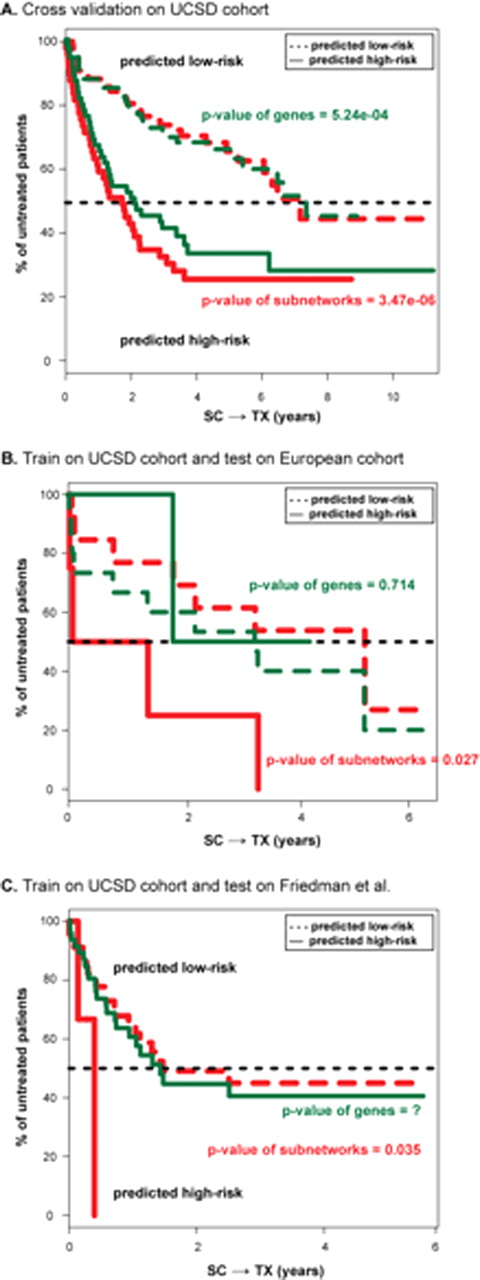
Use of expression levels of genes versus subnetworks to stratify patient samples. (A) Five-fold cross validation on the 130 patients from UCSD. Survival analyses on SC→TX are shown for both the low (dashed lines) and high (solid lines) risk groups predicted by subnetwork signatures (red lines) or by gene signatures (green lines). (B-C) Survival curves on SC→TX for the 17 European patients (B) or for the patient cohort in Friedman et al (2009) (C). The two risk groups are predicted by two sets of markers developed on the UCSD cohort, including the 38 subnetworks (red lines) and the top 230 genes (green lines).
Use of expression levels of genes versus subnetworks to stratify patient samples. (A) Five-fold cross validation on the 130 patients from UCSD. Survival analyses on SC→TX are shown for both the low (dashed lines) and high (solid lines) risk groups predicted by subnetwork signatures (red lines) or by gene signatures (green lines). (B-C) Survival curves on SC→TX for the 17 European patients (B) or for the patient cohort in Friedman et al (2009) (C). The two risk groups are predicted by two sets of markers developed on the UCSD cohort, including the 38 subnetworks (red lines) and the top 230 genes (green lines).
Foa:Roche: Consultancy, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal