Abstract
Abstract 1191
The metalloprotease ADAMTS13 inhibits the growth of platelet thrombi by cleaving the Tyr1605-Met1606 peptide bond in the A2 domain of von Willerbrand factor (VWF). Previous studies have shown that when subjected to tensile stress in solution, bound to platelets, or on endothelial cell surfaces, VWF changes conformation and interacts with multiple exosites on ADAMTS13 through a combination of structural features. These close contacts enhance the highly specific interaction between ADAMTS13 and VWF in vivo. Like VWF, ADAMTS13 is a large multidomain protein that could be regulated by large-scale conformational changes induced by substrate binding. Here we report the use of small-angle X-ray scattering (SAXS) to study the structure of ADAMTS13 and its interactions with VWF peptides in solution.
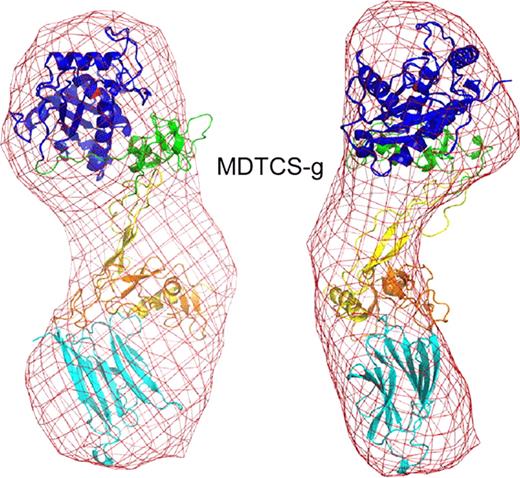
ADAMTS13 truncated after the spacer domain, consisting of metalloprotease (M), disintegrin-like (D), thrombospondin type 1 (TSP1, T), Cys-rich (C) and spacer (S) domains (MDTCS), was produced in either T-Rex 293 cells (MDTCS-t) or HEK293S GNTI cells (MDTCS-g). GNTI cells do not have N-acetylglucosaminyltransferase I activity and expressed proteins lack complex N-glycans. In addition, a variant with the mutation E225Q in the M domain was produced in T-Rex 293 cells (MqDTCS-t). Construct MqDTCS-t lacks enzymatic activity but binds normally to VWF. Two VWF peptides were prepared: cVWF63 is a C-terminal A2 domain cleavage product consisting of VWF residues Met1606-Arg1668, and VWF71 consists of VWF residues Gln1599-Arg1668 with an additional Gly at the N-terminus. SAXS data were collected at the SIBYLS beamline (Lawrence Berkeley National Laboratory) for MDTCS variants without and with bound cVWF63 or VWF71. The quality of the SAXS data was evaluated by comparison to an atomic model of ADAMTS13. The M domain was modeled on ADAMTS4, DTCS domains were from the corresponding crystal structure (PDB: 3ghm).
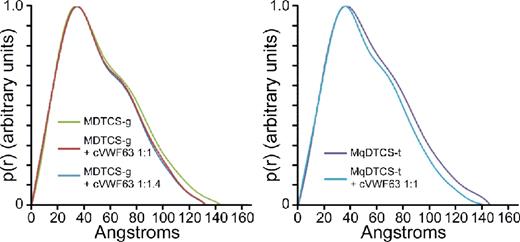
Pair distance distribution function of MDTCS with or without cVWF63.
These results show that SAXS clearly distinguishes the structure of ADAMTS13 with or without bound product or substrate analog. ADAMTS13 domains distal to the spacer domain also contribute to VWF binding, and stepwise addition of these TSP-1 and CUB domains to MDTCS will allow the visualization of conformational changes induced by interaction with larger substrate analogs, and construction of a model for a physiologically relevant enzyme-substrate complex.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal