Abstract
Endoplasmic reticulum (ER) stress triggers a homeostatic cellular response in mammalian cells to ensure efficient folding, sorting, and processing of client proteins. In lytic-permissive lymphoblastoid cell lines (LCLs), pulse exposure to the chemical ER-stress inducer thapsigargin (TG) followed by recovery resulted in the activation of the EBV immediate-early (BRLF1, BZLF1), early (BMRF1), and late (gp350) genes, gp350 surface expression, and virus release. The protein phosphatase 1 a (PP1a)–specific phosphatase inhibitor Salubrinal (SAL) synergized with TG to induce EBV lytic genes; however, TG treatment alone was sufficient to activate EBV lytic replication. SAL showed ER-stress–dependent and –independent antiviral effects, preventing virus release in human LCLs and abrogating gp350 expression in 12-O-tetradecanoylphorbol-13-acetate (TPA)–treated B95-8 cells. TG resulted in sustained BCL6 but not BLIMP1 or CD138 expression, which is consistent with maintenance of a germinal center B-cell, rather than plasma-cell, phenotype. Microarray analysis identified candidate genes governing lytic replication in LCLs undergoing ER stress.
Introduction
EBV is the causative agent of acute infectious mononucleosis and most posttransplantation lymphomas. The virus is also associated with a variety of lymphoid and epithelial malignancies, such as Burkitt lymphoma, nasopharyngeal carcinoma, and gastric lymphomas. The latent form of infection is observed in rare circulating B lymphocytes from patients who have recovered from acute EBV infection (latency I) and during initial infection of B lymphocytes (latency III), whereas lytic EBV gene expression occurs during passage of B lymphocytes (mostly plasma cells and tonsillar B lymphocytes) within epithelial tissue.1
The conversion of B lymphocytes from latent to lytic gene expression is controlled by the EBV DNA-binding, immediate-early transactivator proteins, ZTA (Z) and RTA (R). During lytic reactivation, Z is initially expressed, binding to and activating expression of R, via 3 Z-response elements in the R promoter. Together, Z and R are capable of activating the entire set of EBV lytic gene promoters.2-4 However, the cellular events responsible for the initiation of lytic replication remain unknown. The histone deacetylase inhibitors butyric and valproic acid induce lytic replication, as does the protein kinase C (PKC) activator 12-O-tetradecanoylphorbol-13-acetate (TPA) in certain cell lines. Recently, there has been growing interest in the effects of various chemotherapeutic drugs on EBV replication.5-7
Data derived from the use of these chemicals, however, provide limited insight regarding the specific pathways required for conversion of EBV-immortalized lymphoblastoid cell lines (LCLs) from latency to the lytic phase. For example, histone deacetylase inhibitors result in histone rearrangements and gene activation at multiple loci, many of which may be unrelated to physiologic lytic cues, and therefore cannot be used to study EBV lytic gene activation and replication.
Studies using reporter assays show that ZTA and RTA promoters are very weakly activated by the unfolded protein response (UPR)–activated form of XBP1, XBP1(s), and more robustly in combination with protein kinase D in EBV-positive epithelial and LCLs,8,9 suggesting the possibility that the UPR is a trigger for EBV lytic reactivation. XBP1 is required for both plasma cell differentiation and UPR induction.10
EBV may mount a UPR during initial infection because of the buildup of viral proteins in the ER or via direct eIF2α phosphorylation through PKR activation secondary to viral RNA (including EBV-encoded RNA) accumulation, resulting in a “lytic burst” of viral amplification. During lytic replication in the oropharynx, the UPR may occur when infected lymphocytes are exposed to bacterial TLR agonists such as lipopolysaccharides or methylated CpG nucleotides produced by colonizing bacteria,9,11-15 or during expansion of the ER in the course of activated B lymphocyte to plasma-cell differentiation in the germinal center reaction. The latter hypothesis is supported by studies suggesting that the lytic pool of B lymphocytes may consist mainly of memory B and plasma cells.11
To investigate the effects of the UPR on the lytic cycle in vitro, we used the UPR inducer thapsigargin (TG), a noncompetitive inhibitor of the endoplasmic reticulum Ca2+ ATPase (SERCA2), which depletes luminal ER calcium stores and is thought to induce ER stress by disabling Ca2+-dependent resident ER chaperone proteins such as calnexin and calreticulin. We also used tunicamycin (TM), which induces the UPR by preventing protein glycosylation in the Golgi and secondarily causes protein accumulation in the ER. Salubrinal (SAL), a protein-phosphatase 1 inhibitor identified in a chemical screen for agents that prevent TG-induced apoptosis,16 was used to antagonize ER stress in our experiments.
We conducted a series of experiments with these agents, measuring: (1) expression of immediate-early, early, and late EBV lytic mRNAs; (2) cell-surface gp350 expression; and (3) EBV copies in supernatants of permissive LCLs and EBV particles in electron micrography (EM) sections after chemical induction of ER stress by TG or protection against ER stress by SAL.
To specifically identify genes involved in ER-stress–induced lytic replication, we performed gene-expression profiling at early (before viral lytic protein expression) and late time points, resulting in the identification of previously characterized and novel cellular proteins.
Methods
Cell lines
LCLs derived from B95-8 infection of PBMCs, were contributed by David Rowe (Graduate School of Public Health, University of Pittsburgh) and cultured in RPMI 1640 plus GlutaMax (Invitrogen), supplemented with 10% FBS. B95-8 marmoset B cells were acquired from ATCC. The experiments shown in Figure 3A through D were conducted with LCL-6, which contains > 400 copies of episomal DNA per cell.
Chemical treatments
LCLs were split at 2 × 105 cells/mL in complete medium 24 hours before treatment with 50μM TG (Sigma-Aldrich), 5 μg/μL of TM (Sigma-Aldrich), 40μM SAL (Thermo Fisher Scientific), or 22.5 μg/mL of acyclovir (ACY; Sigma-Aldrich). T24 and T72 cells were treated with TG for 3 or 6 hours, washed with complete medium, and cultured in fresh complete medium for an additional 18 or 66 hours for a total of 24 (T24) or 72 (T72) hours. In dual treatments with either TG and SAL (TS24 and TS72) or TM and SAL, the cells were treated with both chemicals for 6 hours, then washed in complete medium, and subsequently cultured in growth medium supplemented with SAL for an additional 18 or 66 hours. S24 and S72 cells were treated with SAL continuously for either 24 or 72 hours. In control experiments, cells were cultured in ACY continuously for 24 or 72 hours.
Western blot analysis
LCLs were harvested 24 hours after chemical treatment and lysed in 100 μL of RIPA buffer supplemented 1× with protease and phosphatase inhibitors. Samples were run on 8% SDS-PAGE gels with equal protein loading and transferred to PVDF membranes. Membranes were blocked in 5% (wt/vol) BSA and probed with either eIF2a (total), eIF2α pS51, (Cell Signaling Technology), anti-ZTA (Santa Cruz Biotechnologies), or anti-LMP1 Abs (Dako) and anti–rabbit IgG-HRP secondary Abs (Jackson ImmunoResearch Laboratories).
RT-PCR and qPCR
Cell pellets were harvested at 24 or 72 hours after chemical treatment. RNA was isolated from the cell pellets using the RNeasy Mini kit (QIAGEN) following the manufacturer's instructions. cDNA was synthesized using 1-5 μg of purified RNA, the SuperScript II First Strand Synthesis Kit (Invitrogen), and random hexamers. Quantitative PCR (qPCR) was performed in triplicate for each data point shown using an ABI 7500, Maxima SYBR green/ROX mix (Thermo Fisher Scientific) and 1 μL of cDNA with the following primers: BRLF1 5′-AATTTACAGCCGGGAGTGTG-3′ (sense) and 5′-AGCCCGTCTTCTTACCCTGT-3′ (antisense), BZLF1 5′-CATGTTTCAACCGCTCCGACTGG-3′ (sense) and 5′-GCGCAGCCTGTCATTTTCAGATG-3′ (antisense), BMRF1 5′-CTAGCCGTCCTGTCCAAGTGC-3′ (sense) and 5′-AGCCAAACGCTCCTTGCCCA-3′ (antisense), gp350 5′-GTCAGTACACCATCCAGAGCC-3′ (sense) and 5′-TTGGTAGACAGCCTTCGTATG-3′ (antisense), BLIMP1 5′ - TCT TTGGGACATTCTTTGGG-3′ (sense), and 5′ - CGGAGAGCTGACAATGATGA-3′ (antisense), BCL6 5′ - CAATGCCTTGCTTCACAGTC-3′ (sense) and 5′ - TGGGGTTCTTAGAAGTGGTGA-3′ (antisense), CD138 5′ - AGCCATCTTGATCTTCAGGG-3′ (sense) and 5′ - CTCTGGCTCTGGCTGTGC-3′ (antisense), and GAPDH 5′ - GAGTCAACGGATTTGGTCGT-3′ (sense) and 5′ - TTGATTTTGGAGGGATCTCG-3′ (antisense). Relative mRNA abundance was calculated using GAPDH as the internal control using the 2ΔΔCT method. For each experiment, DMSO was normalized to 1 and the mRNA changes were represented relative to DMSO.
Affymetrix data analysis
cDNA synthesis, labeling, hybridization to Affymetrix U133 plus 2.0 chips, and signal-intensity analysis was performed by the University of Pittsburgh Genomics and Proteomics Core Laboratories (24- and 72-hour samples) or by Asuragen (3-hour samples). Data from each treatment condition were derived from independent experiments, independent labeling, and hybridization steps, and the fluorescence intensity was averaged over 2 replicates. Median normalization for fold change was used throughout, filtering for signal intensity > 20. For early (3-hour) gene-expression analysis, LCL-03 and LCL-06 cells were treated in duplicate with either TG or DMSO. Clustering was performed filtering on signal intensity > 20 and an absolute fold change > 2, and considering coordinately regulated gene sets at 3, 24, or 72 hours using the Venn diagram utility in GeneSpring GX software (Agilent Technologies).
EBV copy number determinations
DNA was extracted from 200-μL portions of cell culture supernatants using the QIAsymphony instrument (QIAGEN), and eluted into 50-μL aliquots for downstream PCR. EBV copy number was normalized per milliliter of cell culture supernatant. A primer-probe set for a 90-bp target within the EBV DNA polymerase gene (BALF-5) was used in this TaqMan PCR assay. Details of the assay have been described previously.17
Determination of EBV-gp350 expression by FACS analysis
EBV-positive, permissive marmoset B cells (B95-8) were plated at 0.5 × 106 cells/well in 6-well tissue-culture plates 24 hours before treatment. B958 cells were treated with DMSO (0.01%), SAL (40μM), and ACY (22.5 μg/mL) with or without TPA (50 ng/mL). Cells were also treated with SAL 0, 24, 48, and 72 hours after TPA treatment. LCLs were treated with DMSO (0.01%), TG (50μM), SAL (40μM), or TG + SAL. Cells were harvested 5 days after chemical treatment, counted, and washed with FACS medium (DMEM with 1% FBS, 20mM HEPES, penicillin 10 U/mL, and streptomycin 10 μg/mL), and incubated with monoclonal anti-gp350/250 Ab (clone 2L10; Millipore) for 60 minutes on ice with constant shaking. Cells were washed twice with FACS medium, stained with goat anti–mouse FITC-conjugated secondary Abs for 45 minutes on ice in the dark, washed 3 times, fixed, and analyzed by flow cytometry (FACSCalibur; BD Biosciences).
Transmission EM
Cells treated with DMSO or TG were fixed in 2.5% glutaraldehyde, processed, and sectioned.42 Sections were observed on a JEM 1210 electron microscope (JEOL).
Results
Effects of TG and SAL on eIF2α phosphorylation and expression of lytic and latent genes
ER stress causes translational arrest via protein kinase RNA-like endoplasmic reticulum kinase (PERK)–mediated phosphorylation of eIF2α. To determine the effects of TG and SAL on this pathway in LCLs, we applied short (6-hour) exposure to TG (50μM), followed by 18 hours of washout with medium. This series of treatments resulted in increased eIF2α phosphorylation (Figure 1A lane 3 vs lane 1). Likewise, continuous (24-hour) exposure to SAL (40mM) also resulted in eIF2α phosphorylation (Figure 1A lane 2 vs lane 1). Short exposure to TG in the continuous presence of SAL also resulted in eIF2α phosphorylation, with little synergy observed with the combination of treatments (Figure 1A lane 4 vs lane 1). These data indicate that in LCLs, continuous protein phosphatase 1 a (PP1a) activity is required to maintain dephosphorylated eIF2α, and that both SAL and TG exposure result in increased levels of eIF2α serine 31 phosphorylation with little cooperativity.
ER stress induces phosphorylation of eIF2α and expression of ZTA and LMP1. (A) SAL and ER stress result in increased levels of eIF2α S51 phosphorylation. LCLs were treated with SAL (40μM for 24 hours), TG (50μM for 6 hours), or SAL and TG as described. Western blot analysis for total eIF2α and pS51 eIF2α were performed 24 hours after treatment. (B) ER stress induces ZTA expression in LCLs. LCLs were treated with TG, SAL, or TG + SAL as described and at 6 or 24 hours after treatment, the cells were lysed in 1× RIPA buffer. Equal amounts of protein were analyzed by Western blot for ZTA. (C) Time-course experiment of ZTA and LMP1 protein expression after the induction of ER stress. LCLs were treated with TG and cells were lysed at the indicated times after TG treatment in 1× RIPA buffer. Equal amounts of protein were analyzed by Western blot for ZTA and LMP1.
ER stress induces phosphorylation of eIF2α and expression of ZTA and LMP1. (A) SAL and ER stress result in increased levels of eIF2α S51 phosphorylation. LCLs were treated with SAL (40μM for 24 hours), TG (50μM for 6 hours), or SAL and TG as described. Western blot analysis for total eIF2α and pS51 eIF2α were performed 24 hours after treatment. (B) ER stress induces ZTA expression in LCLs. LCLs were treated with TG, SAL, or TG + SAL as described and at 6 or 24 hours after treatment, the cells were lysed in 1× RIPA buffer. Equal amounts of protein were analyzed by Western blot for ZTA. (C) Time-course experiment of ZTA and LMP1 protein expression after the induction of ER stress. LCLs were treated with TG and cells were lysed at the indicated times after TG treatment in 1× RIPA buffer. Equal amounts of protein were analyzed by Western blot for ZTA and LMP1.
To determine the effects of these treatments on EBV genes, we selected candidate lytic (ZTA) and latency III (LMP1) proteins. Twenty-four hours after the start of TG exposure, ZTA protein levels were increased 5- to 6-fold (Figure 1B lane 2 bottom panel). SAL alone had little effect on ZTA levels (Figure 1B lane 3 top and bottom panels), but antagonized TG induced ZTA induction (Figure 1B lane 4 bottom panel). A time-course experiment indicated that ZTA protein levels were minimally increased at 3, 6, and after TG exposure, but increased dramatically starting at 24 hours. Levels were also elevated as early as 3 hours after TG exposure, and continued to increase over the course of the experiment (Figure 1C).
ER stress induces EBV lytic genes at 24 and 72 hours and SAL has both inhibitory and additive effects
Our preliminary data indicated that ER stress could increase ZTA protein levels. To determine whether TG activates EBV lytic gene promoters, we performed qRT-PCR from total RNA purified after 24 or 72 hours from LCLs that had been exposed to TG, SAL, or both (Figure 2B-C). TG was applied to cells for 6 hours, and the medium was replaced with either DMSO or with SAL for the remaining 18 or 66 hours of the assay, as indicated in Figure 2A. Cells maintained > 80% viability by trypan blue exclusion with all chemical treatments except TS24 and TS72 (cell viability, 50%). TG reproducibly increased mRNA abundance relative to DMSO of immediate-early (BRLF1 ∼ 7.3-fold), early (BMRF1 ∼ 3.2-fold), and late mRNAs (gp350 ∼ 7.6-fold) 24 hours after treatment, whereas DMSO alone or SAL alone had little effect. When cells were allowed to recover for 24 hours in the presence of SAL, EBV lytic gene activation was reduced: BRLF1 activation decreased from ∼ 7.3-fold to ∼ 2.6-fold; BZLF1 from ∼ 7.3-fold to ∼ 2.7-fold; BMRF1 from ∼ 3.2-fold to ∼ 1.4-fold; and gp350 from ∼ 7.6-fold to ∼ 3.0-fold (Figure 2B).
ER stress induces EBV lytic genes in LCLs. (A) Schematic representation of drug treatments in LCLs. Cells were treated with DMSO (panel 1) or 40μM SAL (panel 3) for the total length of the experiment; with a 6-hour pulse of 50μM TG and then cultured in media lacking TG for the remaining length of the experiment (panel 2); or with a 6-hour pulse of 50μM TG as in panel 2 but in the presence of 40μM SAL for the total length of the experiment (panel 4). (B) Induction of EBV lytic genes 24 hours after chemical treatment. At 24 hours after drug treatment, LCLs were harvested, RNA isolated, and gene expression of immediate-early (BRLF1 and BZLF1), early (BMRF1), and late (gp350) EBV lytic genes was evaluated by real-time qPCR. Comparison of DMSO versus TG, DMSO versus TG + SAL, and TG versus TG + SAL were all statistically significant at P < .05. (C) Induction of EBV lytic genes 72 hours after drug treatment, when LCLs were harvested and treated as in panel B. Comparison of DMSO versus TG, DMSO versus TG + SAL, and TG versus TG + SAL were all statistically significant at P < .05. (D) TM treatment also induces EBV lytic genes. LCLs were treated with a 4-hour pulse of TM (5 μg/mL) or TM + SAL and then cultured for a total of 72 hours. At 72 hours after drug treatment, LCLs were harvested as described and EBV lytic gene expression was analyzed by real-time qPCR. All comparisons were statistically significant.
ER stress induces EBV lytic genes in LCLs. (A) Schematic representation of drug treatments in LCLs. Cells were treated with DMSO (panel 1) or 40μM SAL (panel 3) for the total length of the experiment; with a 6-hour pulse of 50μM TG and then cultured in media lacking TG for the remaining length of the experiment (panel 2); or with a 6-hour pulse of 50μM TG as in panel 2 but in the presence of 40μM SAL for the total length of the experiment (panel 4). (B) Induction of EBV lytic genes 24 hours after chemical treatment. At 24 hours after drug treatment, LCLs were harvested, RNA isolated, and gene expression of immediate-early (BRLF1 and BZLF1), early (BMRF1), and late (gp350) EBV lytic genes was evaluated by real-time qPCR. Comparison of DMSO versus TG, DMSO versus TG + SAL, and TG versus TG + SAL were all statistically significant at P < .05. (C) Induction of EBV lytic genes 72 hours after drug treatment, when LCLs were harvested and treated as in panel B. Comparison of DMSO versus TG, DMSO versus TG + SAL, and TG versus TG + SAL were all statistically significant at P < .05. (D) TM treatment also induces EBV lytic genes. LCLs were treated with a 4-hour pulse of TM (5 μg/mL) or TM + SAL and then cultured for a total of 72 hours. At 72 hours after drug treatment, LCLs were harvested as described and EBV lytic gene expression was analyzed by real-time qPCR. All comparisons were statistically significant.
When cells were harvested 72 hours after the start of TG pulse, a pattern of EBV lytic gene activation similar to the 24-hour TG treatment was observed (Figure 2C). An ∼ 7-fold increase in the mRNA abundances of BRLF1, BZLF1, and gp350 and a 3-fold increase in the mRNA abundance of BMRF1 was observed relative to DMSO. However, in contrast to the 24-hour data series, SAL augmented TG-induced EBV lytic gene activation. Relative to TG alone, combined treatment with SAL further increased the mRNA abundance of BRLF 7.3-fold to 6.5-fold; BZLF1 7.3-fold to 21.4-fold; BMRF1 3.2-fold to 4.8-fold; and gp350 7.6-fold to 15.6-fold (Figure 2C). Remarkably, TG activation and TG + SAL synergy were maintained as long as 5 days after the 6-hour TG pulse (Figure 3E). The 3-hour TG pulse resulted in a similar pattern of EBV lytic gene expression (data not shown).
ER stress induces late events in EBV lytic production. (A-B). TG treatment results in gp350 expression at the plasma membrane. Five days after chemical treatment, LCLs were stained for gp350 and analyzed by flow cytometry. The percentage of gp350-positive cells is plotted (A). The shift in fluorescence intensity of gp350-positive cells from DMSO-treated (pink trace) to TG-treated (green trace) LCLs can be seen (B). The background fluorescence of LCLs is shown as the filled trace. (C) TG treatment results in increased EBV copy number in high-copy-number LCLs (> 400 episomes/cell). Five days after chemical treatment of LCLs, supernatant was collected and EBV copy number determined by qPCR. (D) TG treatment does not increase EBV copy number in low-copy-number LCLs (< 50 episomes/cell). Five days after chemical treatment of LCLs, supernatant was collected and EBV copy number determined by qPCR. (E) Induction of EBV lytic genes 5 days after chemical treatment, when LCLs were harvested, RNA isolated, and gene expression of immediate-early (BRLF1 and BZLF1), early (BMRF1), and late (gp350) EBV lytic genes was evaluated by real-time qPCR.
ER stress induces late events in EBV lytic production. (A-B). TG treatment results in gp350 expression at the plasma membrane. Five days after chemical treatment, LCLs were stained for gp350 and analyzed by flow cytometry. The percentage of gp350-positive cells is plotted (A). The shift in fluorescence intensity of gp350-positive cells from DMSO-treated (pink trace) to TG-treated (green trace) LCLs can be seen (B). The background fluorescence of LCLs is shown as the filled trace. (C) TG treatment results in increased EBV copy number in high-copy-number LCLs (> 400 episomes/cell). Five days after chemical treatment of LCLs, supernatant was collected and EBV copy number determined by qPCR. (D) TG treatment does not increase EBV copy number in low-copy-number LCLs (< 50 episomes/cell). Five days after chemical treatment of LCLs, supernatant was collected and EBV copy number determined by qPCR. (E) Induction of EBV lytic genes 5 days after chemical treatment, when LCLs were harvested, RNA isolated, and gene expression of immediate-early (BRLF1 and BZLF1), early (BMRF1), and late (gp350) EBV lytic genes was evaluated by real-time qPCR.
EBV lytic gene activation is achieved with a different class of UPR inducer
Because a TG pulse induces ER stress by transiently altering calcium concentrations in the ER, we used TM, a drug that induces ER stress by a different mechanism (inhibition of protein N-glycosylation), to determine whether ER stress per se could cause lytic gene activation. Pulse TM treatment (for 6 hours) also caused consistent increases in lytic gene activation with moderate additive effects of SAL over a 72-hour experiment (P < .05 for all comparisons, Figure 2D).
Plasma cell marker expression in cells exposed to TG
Because plasma cell differentiation has been implicated in lytic replication, we measured the levels of mRNAs encoding the plasma cell differentiation proteins BLIMP1 and BCL6 and the plasma cell marker CD138 in LCLs, activating lytic genes 1, 3, 6, and 72 hours after the TG pulse. At 1 hour (Figure 4A), BLIMP1 and BCL6 mRNA levels were increased 2- to 3-fold in TG- and TG +SAL–treated cells relative to DMSO-treated cells; however, by 3 hours (Figure 4B) and 6 hours (Figure 4C), only BCL6 remained elevated. Seventy-two hours after the TG pulse (Figure 4D), we observed no effect on BLIMP1 mRNA levels, slightly increased BCL6 mRNA levels, and decreased mRNA levels for the plasma cell marker CD138/Syndecan (P < .05 for TG vs DMSO for BCL6 and CD138 only). However, these effects were not observed with TG + SAL under conditions that result in synergistic up-regulation of EBV lytic mRNAs (Figure 2C). These data indicate that TG and TG + SAL treatments result in transcriptional responses more consistent with germinal center B cells, rather than a postgerminal center B- or plasma-cell phenotype.
ER stress effects on germinal center B- and plasma-cell markers. Gene expression of plasma cell markers Blimp1, CD138, and BCL6 were evaluated by real-time qPCR. LCLs were treated with TG and SAL for 1 hour (A), 3 hours (B), or 6 hours (C) before gene expression of plasma cell markers was evaluated by real-time qPCR. (D) LCLs were treated with TG and SAL as described and plasma cell markers were evaluated at 72 hours after drug treatment by real-time qPCR.
ER stress effects on germinal center B- and plasma-cell markers. Gene expression of plasma cell markers Blimp1, CD138, and BCL6 were evaluated by real-time qPCR. LCLs were treated with TG and SAL for 1 hour (A), 3 hours (B), or 6 hours (C) before gene expression of plasma cell markers was evaluated by real-time qPCR. (D) LCLs were treated with TG and SAL as described and plasma cell markers were evaluated at 72 hours after drug treatment by real-time qPCR.
Effects of UPR induction and UPR protection on cell-surface gp350 staining and EBV episome copy number in cell-culture supernatants
To exclude the possibility that the UPR induced a transient activation of EBV lytic genes without true viral assembly and release, we monitored for the appearance of surface gp350 by FACS in LCLs treated with a variety of agents (Figure 3A-B), as well as the appearance of EBV DNA in cell supernatants of high (> 400 episomes/cell) and low (< 50 episomes/cell) copy number LCLs by qPCR (Figure 3C-D). In high-copy-number LCLs, we observed an ∼ 5% rate of spontaneous gp350 expression in both DMSO-treated and -untreated cells (Figure 3A DMSO and NEG). Remarkably, 5 days after the 3- or 6-hour TG pulse, gp350 expression increased from 5% to 26% (Figure 3A-B). Although SAL strongly increased TG activation of lytic genes in this assay, doubling or tripling mRNA abundances of lytic genes relative to TG alone (Figure 3E), this did not translate into increased gp350 expression, which remained at 25%-26% under TG + SAL conditions, suggesting that, despite increased lytic mRNAs, other factors were limiting for virus maturation or assembly. TM treatment did not result in increased gp350 membrane staining, likely because of inhibition of EBV glycoprotein glycosylation, indicating that this method of UPR induction is incompatible with viral production. As expected, by blocking viral DNA synthesis and late gene expression, ACY blocked spontaneous gp350 membrane expression (Figure 3A).
To determine the effects of these agents on cell-free virus in high-copy-number LCLs, EBV DNA was measured by PCR in cell supernatants (Figure 3C) and the effects on EBV lytic genes were assayed by qRT-PCR in parallel (Figure 3E). Induction of the UPR by TG resulted in increased titers of free virus, with EBV DNA copies increasing from 66 × 106/mL to 171 × 106/mL (Figure 3C), which is consistent with TG transcriptional effects on lytic genes (Figure 3D). Whereas SAL alone had little activity in activating EBV lytic genes, it potently reduced EBV replication. In the representative experiment shown, incubation with SAL alone resulted in a reduction of EBV copies from ∼ 66 × 106/mL to ∼ 35 × 106/mL (an ∼ 50% reduction), indicating a lytic inhibitory effect even in the absence of exogenous ER stress. For comparison, under the same conditions, ACY treatment reduced copy number from ∼ 66 × 106/mL to 15 × 106/mL (a 78% reduction). Furthermore, whereas the TG pulse resulted in a near doubling of EBV copies (66 × 106/mL to 171 × 106/mL), this effect was completely abrogated when SAL was continuously present in the culture medium after the TG pulse, resulting in a decrease of EBV copies from 171 × 106/mL to 28 × 106/mL (an 84% reduction). Therefore, SAL inhibited both basal and ER-stress–induced lytic replication. Similar results were not observed in low-copy-number LCLs (Figure 3D), in which UPR induction by TG did not result in increased EBV DNA copies (1.6 × 106/mL to 1.2 × 106/mL). However, SAL treatment did result in a reduction of EBV copies from ∼ 1.5 × 106/mL to ∼ 0.7 × 106/mL (an ∼ 50% reduction), as was observed in high-copy-number LCLs.
SAL prevents lytic replication independently of ER stress induction
Treatment of B95-8 marmoset B cells with TPA results in robust lytic replication.18 To confirm that SAL could inhibit lytic replication under non–ER-stress conditions (Figure 3C-D), we induced lytic replication with an alternative method (TPA-induced PKC activation) in B95-8 cells and applied SAL at different time points after TPA treatment (Figure 5). When SAL was applied at the same time as TPA, lytic replication was inhibited by > 90% with an efficacy comparable to ACY. Inhibition was reduced when SAL was applied at later time points. When SAL was applied 24 hours after TPA, inhibition was reduced to ∼ 65%, and only a ∼ 25% inhibition was observed when SAL was applied at 48 or 72 hours. These data suggest an immediate-early or early effect of SAL on viral replication in TPA-stimulated B95-8 cells. The ability of SAL to inhibit lytic replication in B95-8 cells exposed to TPA indicates an effect independent of ER stress.
SAL prevents TPA-induced virus production in B95-8 cells. The permissive EBV-positive marmoset B-cell line B95-8 was treated with SAL 0, 24, 48, and 72 hours after TPA treatment or with TPA, ACY, or DMSO alone. Cells were incubated for a total of 5 days after TPA treatment and assayed for gp350 expression, as described in “Methods.”
SAL prevents TPA-induced virus production in B95-8 cells. The permissive EBV-positive marmoset B-cell line B95-8 was treated with SAL 0, 24, 48, and 72 hours after TPA treatment or with TPA, ACY, or DMSO alone. Cells were incubated for a total of 5 days after TPA treatment and assayed for gp350 expression, as described in “Methods.”
TG treatment of LCLs results in the production of infectious viral particles
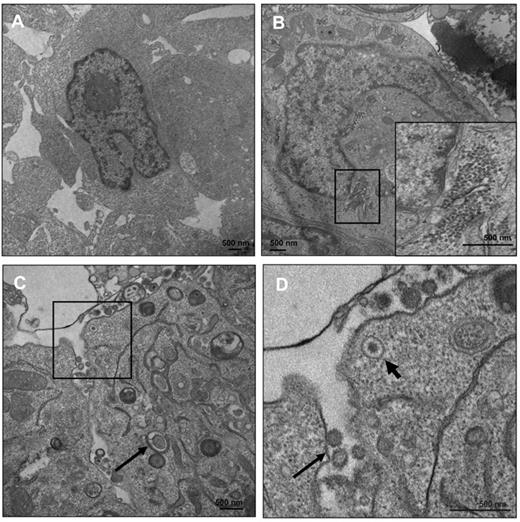
Whereas TG treatment causes increases in EBV DNA in supernatants of treated LCLs, the possibility that these supernatants consist of naked viral DNA due to cell lysis of noninfectious particles remained. We performed EM on these cells 5 days after the TG pulse (Figure 6A-D). Exhaustive searching of multiple EM fields failed to show any nucleocapsid or viral particles (Figure 6A), with largely intact nuclear structure in DMSO-treated cells. In contrast, in TG-treated cells, we observed massive accumulation of nucleocapsids (Figure 6B boxed area and inset) adjacent to the disrupted inner and outer nuclear laminae. Numerous fields indicated enveloped, cell-free virus that was visible because it had been trapped within intercellular regions (Figure 6C-D). To determine whether TG treatment resulted in the production of infectious virus, we infected EBV-negative BJAB cells with the supernatants of TG-treated LCLs, induced lytic replication in BJABs, isolated RNA, and quantified expression of lytic genes BZLF1, BRLF1, and BMRF1 (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). These genes failed to amplify with mock-infected BJABs. The mRNA abundance relative to supernatants from DMSO-treated LCLs (which have an ∼ 5% rate of lytic activity by gp350 staining) was measured and indicated production of lytic viral RNAs in infected BJAB cells. These data indicate that TG treatment results in the production of infectious viral particles.
ER stress induces virus production. LCLs were treated with DMSO (A) or TG (B-D) and at 5 days after treatment the cells were processed for EM. (B) Accumulation of nucleocapsid cores near the nuclear membrane with an inset of a higher magnification of the nucleocapsid cores. (C) EBV-enveloped virus transport and budding at the plasma membrane. Arrow indicates the occurrence of a double-membrane vesicle, a hallmark of autophagy. (D) Higher magnification of the boxed area in panel C. Thick arrow shows an enveloped virion being transported to the plasma membrane and the thin arrow shows a budding virus.
ER stress induces virus production. LCLs were treated with DMSO (A) or TG (B-D) and at 5 days after treatment the cells were processed for EM. (B) Accumulation of nucleocapsid cores near the nuclear membrane with an inset of a higher magnification of the nucleocapsid cores. (C) EBV-enveloped virus transport and budding at the plasma membrane. Arrow indicates the occurrence of a double-membrane vesicle, a hallmark of autophagy. (D) Higher magnification of the boxed area in panel C. Thick arrow shows an enveloped virion being transported to the plasma membrane and the thin arrow shows a budding virus.
Differential gene-expression analysis of ER stress effects in LCLs
Microarray data are available at the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) under accession number GSE 31447. We performed microarray analysis using Affymetrix HG133U_Plus 2 chips containing 53 613 probe sets derived from > 30 000 unique genes and control probe sets. We used mRNA from independent cell lines (LCL-3 and LCL-6) to determine the early (3-hour) effects of TG on gene expression, and compared these patterns with cells harvested at 24 and 72 hours in LCL-6. Because ZTA protein is expressed at 24 but not 6 hours after TG treatment (Figure 1B), early transcriptional changes are kinetically upstream of ZTA.
To prevent the bias that results from hierarchical cluster analysis and curating of multiple gene clusters, we identified regulation clusters by Venn diagram analysis. We filtered on absolute fold change > 2 (ie, fold change > 2 or fold change < −2) to identify 6 gene clusters of interest: (1) sets of mRNAs with absolute fold change > 2 at 3, 24, and 72 hours (sustained activation and sustained repression, Figure 7A-C and Table 1); (2) mRNAs with absolute fold change > 2 at 3 hours but absolute fold change < 2 at 24 and 72 hours (early activation and early repression, Figure 7A-B,D, and Table 2); and (3) mRNAs with absolute fold change < 2 at 3 hours and > 2 absolute fold change at 24 and 72 hours (late activation and late repression, Figure 7A-B,E, and Table 3).
Sustained gene expression changes in stressed LCLs
| Gene . | LCL-6, 3 h . | LCL-3, 3 h . | LCL-6, 24 h . | LCL-06, 72 h . | Average . | RefSeq transcript ID . |
|---|---|---|---|---|---|---|
| Sustained activation (3, 24, and 72 hours) | ||||||
| TAC1 | 2.8 | 18.3 | 25.1 | 27.5 | 18.4 | NM_003182 |
| IL8 | 21.1 | 30.1 | 8.7 | 11.1 | 17.8 | NM_000584 |
| NR4A2 | 6.9 | 18.0 | 3.2 | 4.4 | 8.1 | NM_006186 |
| INHBE | 4.4 | 5.4 | 9.1 | 12.0 | 7.7 | NM_031479 |
| KLF4 | 9.7 | 3.4 | 5.5 | 6.8 | 6.4 | NM_004235 |
| NR4A3 | 4.4 | 9.9 | 3.6 | 6.3 | 6.1 | NM_006981 |
| PTHLH | 3.2 | 9.0 | 3.6 | 3.0 | 4.7 | NM_002820 |
| EMP1 | 3.9 | 2.1 | 6.5 | 5.9 | 4.6 | NM_001423 |
| NAMPT | 4.3 | 4.9 | 2.1 | 3.4 | 3.7 | NM_005746 |
| AGPAT9 | 3.1 | 3.5 | 3.9 | 3.3 | 3.5 | NM_032717 |
| CCL4 | 3.6 | 3.0 | 3.1 | 4.1 | 3.4 | NM_002984 |
| PHLDA1 | 3.1 | 3.3 | 3.2 | 3.3 | 3.2 | NM_007350 |
| STC2 | 2.4 | 2.5 | 3.1 | 3.8 | 2.9 | NM_003714 |
| RAB11FIP1 | 3.0 | 3.0 | 2.1 | 3.4 | 2.9 | NM_001002233 |
| MAFF | 2.9 | 2.2 | 2.3 | 4.0 | 2.9 | NM_012323 |
| CTH | 3.6 | 3.1 | 2.2 | 2.4 | 2.8 | NM_001902 |
| CEBPB | 2.7 | 3.1 | 2.5 | 2.8 | 2.8 | NM_005194 |
| NFIL3 | 2.0 | 3.4 | 2.2 | 3.0 | 2.7 | NM_005384 |
| VLDLR | 3.2 | 2.5 | 2.2 | 2.2 | 2.5 | NM_001018056 |
| AHNAK | 3.1 | 2.2 | 2.4 | 2.3 | 2.5 | NM_001620 |
| CD55 | 2.3 | 3.0 | 2.2 | 2.4 | 2.5 | NM_000574 |
| ANKRD28 | 2.3 | 2.2 | 2.3 | 2.3 | 2.3 | NM_015199 |
| CHAC1 | 2.5 | 2.0 | 2.3 | 2.1 | 2.2 | NM_001142776 |
| Sustained repression (3, 24, and 72 h) | ||||||
| RNASE6 | 2.3 | 2.3 | 2.2 | 3.6 | 2.6 | NM_005615 |
| Gene . | LCL-6, 3 h . | LCL-3, 3 h . | LCL-6, 24 h . | LCL-06, 72 h . | Average . | RefSeq transcript ID . |
|---|---|---|---|---|---|---|
| Sustained activation (3, 24, and 72 hours) | ||||||
| TAC1 | 2.8 | 18.3 | 25.1 | 27.5 | 18.4 | NM_003182 |
| IL8 | 21.1 | 30.1 | 8.7 | 11.1 | 17.8 | NM_000584 |
| NR4A2 | 6.9 | 18.0 | 3.2 | 4.4 | 8.1 | NM_006186 |
| INHBE | 4.4 | 5.4 | 9.1 | 12.0 | 7.7 | NM_031479 |
| KLF4 | 9.7 | 3.4 | 5.5 | 6.8 | 6.4 | NM_004235 |
| NR4A3 | 4.4 | 9.9 | 3.6 | 6.3 | 6.1 | NM_006981 |
| PTHLH | 3.2 | 9.0 | 3.6 | 3.0 | 4.7 | NM_002820 |
| EMP1 | 3.9 | 2.1 | 6.5 | 5.9 | 4.6 | NM_001423 |
| NAMPT | 4.3 | 4.9 | 2.1 | 3.4 | 3.7 | NM_005746 |
| AGPAT9 | 3.1 | 3.5 | 3.9 | 3.3 | 3.5 | NM_032717 |
| CCL4 | 3.6 | 3.0 | 3.1 | 4.1 | 3.4 | NM_002984 |
| PHLDA1 | 3.1 | 3.3 | 3.2 | 3.3 | 3.2 | NM_007350 |
| STC2 | 2.4 | 2.5 | 3.1 | 3.8 | 2.9 | NM_003714 |
| RAB11FIP1 | 3.0 | 3.0 | 2.1 | 3.4 | 2.9 | NM_001002233 |
| MAFF | 2.9 | 2.2 | 2.3 | 4.0 | 2.9 | NM_012323 |
| CTH | 3.6 | 3.1 | 2.2 | 2.4 | 2.8 | NM_001902 |
| CEBPB | 2.7 | 3.1 | 2.5 | 2.8 | 2.8 | NM_005194 |
| NFIL3 | 2.0 | 3.4 | 2.2 | 3.0 | 2.7 | NM_005384 |
| VLDLR | 3.2 | 2.5 | 2.2 | 2.2 | 2.5 | NM_001018056 |
| AHNAK | 3.1 | 2.2 | 2.4 | 2.3 | 2.5 | NM_001620 |
| CD55 | 2.3 | 3.0 | 2.2 | 2.4 | 2.5 | NM_000574 |
| ANKRD28 | 2.3 | 2.2 | 2.3 | 2.3 | 2.3 | NM_015199 |
| CHAC1 | 2.5 | 2.0 | 2.3 | 2.1 | 2.2 | NM_001142776 |
| Sustained repression (3, 24, and 72 h) | ||||||
| RNASE6 | 2.3 | 2.3 | 2.2 | 3.6 | 2.6 | NM_005615 |
Early gene expression changes in stressed LCLs
| Gene . | LCL-6, 3 h . | LCL3, 3 h . | Average . | Refseq transcript ID . |
|---|---|---|---|---|
| Early activation (3 h only) | ||||
| EGR3 | 6.7 | 6.4 | 6.6 | NM_004430 |
| MYADM | 5.6 | 4.7 | 5.2 | NM_001020818 |
| FOS | 3.2 | 6.3 | 4.8 | NM_005252 |
| TC2N | 5.9 | 2.3 | 4.1 | NM_001128595 |
| PTGS2 | 3.1 | 4.3 | 3.7 | NM_000963 |
| SIAH2 | 2.8 | 4.2 | 3.5 | NM_005067 |
| OTUD1 | 3.6 | 3.2 | 3.4 | NM_001145373 |
| RGS2 | 2.0 | 4.7 | 3.4 | NM_002923 |
| FAM62B | 2.9 | 3.7 | 3.3 | NM_020728 |
| KLF6 | 2.9 | 3.2 | 3.0 | NM_001160124 |
| TUBE1 | 2.8 | 3.3 | 3.0 | NM_016262 |
| FOSB | 2.8 | 3.3 | 3.0 | NM_001114171 |
| PSAT1 | 2.7 | 3.3 | 3.0 | NM_021154 |
| TMEM87A | 2.6 | 3.2 | 2.9 | NM_001110503 |
| CSDA | 3.1 | 2.6 | 2.9 | NM_001145426 |
| TEX9 | 2.0 | 3.6 | 2.8 | NM_198524 |
| JUN | 2.4 | 3.2 | 2.8 | NM_002228 |
| SCHIP1 | 2.8 | 2.8 | 2.8 | NM_014575 |
| ARL4C | 3.3 | 2.3 | 2.8 | NM_005737 |
| B3GNT2 | 3.1 | 2.5 | 2.8 | NM_006577 |
| HIPK1 | 2.9 | 2.6 | 2.8 | NM_152696 |
| LONRF1 | 2.5 | 3.0 | 2.7 | NM_152271 |
| FOSL2 | 2.1 | 3.3 | 2.7 | NM_005253 |
| TMEM70 | 2.7 | 2.6 | 2.6 | NM_001040613 |
| JUND | 2.3 | 2.8 | 2.5 | NM_005354 |
| NAB1 | 2.2 | 2.9 | 2.5 | NM_005966 |
| CDC14B | 2.1 | 2.9 | 2.5 | NM_001077181 |
| EXOC5 | 2.6 | 2.4 | 2.5 | NM_006544 |
| IFRD1 | 2.4 | 2.5 | 2.5 | NM_001007245 |
| ATP1B3 | 2.4 | 2.5 | 2.5 | NM_001679 |
| ANKRD28 | 2.4 | 2.5 | 2.4 | NM_015199 |
| LETM2 | 2.0 | 2.8 | 2.4 | NM_144652 |
| WSB1 | 2.2 | 2.7 | 2.4 | NM_015626 |
| RELL1 | 2.5 | 2.3 | 2.4 | NM_001085399 |
| PARL | 2.3 | 2.5 | 2.4 | NM_001037639 |
| TRIB1 | 2.1 | 2.7 | 2.4 | NM_025195 |
| NFKBID | 2.6 | 2.2 | 2.4 | NM_139239 |
| NUDT4 | 2.3 | 2.5 | 2.4 | NM_019094 |
| MNS1 | 2.1 | 2.7 | 2.4 | NM_018365 |
| ATP2B1 | 2.3 | 2.4 | 2.4 | NM_001001323 |
| PCTK2 | 2.7 | 2.1 | 2.4 | NM_002595 |
| PIP5K1B | 2.3 | 2.4 | 2.4 | NM_003558 |
| EIF5B | 2.2 | 2.5 | 2.3 | NM_015904 |
| TFG | 2.5 | 2.1 | 2.3 | NM_001007565 |
| SDCBP | 2.3 | 2.3 | 2.3 | NM_001007067 |
| SQLE | 2.3 | 2.3 | 2.3 | NM_003129 |
| ST8SIA4 | 2.0 | 2.5 | 2.3 | NM_005668 |
| PCGF5 | 2.3 | 2.2 | 2.2 | NM_032373 |
| RSPH10B2 | 2.4 | 2.1 | 2.2 | NM_001099697 |
| SLC25A13 | 2.3 | 2.2 | 2.2 | NM_001160210 |
| ANKRD12 | 2.3 | 2.1 | 2.2 | NM_001083625 |
| SLC4A7 | 2.0 | 2.4 | 2.2 | NM_003615 |
| ARL2BP | 2.1 | 2.3 | 2.2 | NM_012106 |
| SESN2 | 2.3 | 2.1 | 2.2 | NM_031459 |
| ZMYM6 | 2.1 | 2.2 | 2.2 | NM_007167 |
| ZFP36L1 | 2.3 | 2.0 | 2.2 | NM_004926 |
| SLC3A2 | 2.2 | 2.1 | 2.2 | NM_001012661 |
| AMMECR1 | 2.0 | 2.3 | 2.2 | NM_001025580 |
| GNG2 | 2.3 | 2.0 | 2.1 | NM_053064 |
| FAM107B | 2.0 | 2.2 | 2.1 | NM_031453 |
| TRIB3 | 2.1 | 2.2 | 2.1 | NM_021158 |
| CSNK2A1 | 2.1 | 2.1 | 2.1 | NM_001895 |
| NIPSNAP3B | 2.1 | 2.1 | 2.1 | NM_018376 |
| PPP1R2 | 2.1 | 2.0 | 2.1 | NM_006241 |
| PANK2 | 2.0 | 2.0 | 2.0 | NM_024960 |
| Early repression (3 h only) | ||||
| FAM46C | −3.3 | −2.8 | −3.0 | NM_017701 |
| LCP2 | −2.3 | −2.3 | −2.3 | NM_005565 |
| NOTCH2 | −2.2 | −2.1 | −2.1 | NM_024408 |
| AICDA | −2.2 | −2.0 | −2.1 | NM_020661 |
| Gene . | LCL-6, 3 h . | LCL3, 3 h . | Average . | Refseq transcript ID . |
|---|---|---|---|---|
| Early activation (3 h only) | ||||
| EGR3 | 6.7 | 6.4 | 6.6 | NM_004430 |
| MYADM | 5.6 | 4.7 | 5.2 | NM_001020818 |
| FOS | 3.2 | 6.3 | 4.8 | NM_005252 |
| TC2N | 5.9 | 2.3 | 4.1 | NM_001128595 |
| PTGS2 | 3.1 | 4.3 | 3.7 | NM_000963 |
| SIAH2 | 2.8 | 4.2 | 3.5 | NM_005067 |
| OTUD1 | 3.6 | 3.2 | 3.4 | NM_001145373 |
| RGS2 | 2.0 | 4.7 | 3.4 | NM_002923 |
| FAM62B | 2.9 | 3.7 | 3.3 | NM_020728 |
| KLF6 | 2.9 | 3.2 | 3.0 | NM_001160124 |
| TUBE1 | 2.8 | 3.3 | 3.0 | NM_016262 |
| FOSB | 2.8 | 3.3 | 3.0 | NM_001114171 |
| PSAT1 | 2.7 | 3.3 | 3.0 | NM_021154 |
| TMEM87A | 2.6 | 3.2 | 2.9 | NM_001110503 |
| CSDA | 3.1 | 2.6 | 2.9 | NM_001145426 |
| TEX9 | 2.0 | 3.6 | 2.8 | NM_198524 |
| JUN | 2.4 | 3.2 | 2.8 | NM_002228 |
| SCHIP1 | 2.8 | 2.8 | 2.8 | NM_014575 |
| ARL4C | 3.3 | 2.3 | 2.8 | NM_005737 |
| B3GNT2 | 3.1 | 2.5 | 2.8 | NM_006577 |
| HIPK1 | 2.9 | 2.6 | 2.8 | NM_152696 |
| LONRF1 | 2.5 | 3.0 | 2.7 | NM_152271 |
| FOSL2 | 2.1 | 3.3 | 2.7 | NM_005253 |
| TMEM70 | 2.7 | 2.6 | 2.6 | NM_001040613 |
| JUND | 2.3 | 2.8 | 2.5 | NM_005354 |
| NAB1 | 2.2 | 2.9 | 2.5 | NM_005966 |
| CDC14B | 2.1 | 2.9 | 2.5 | NM_001077181 |
| EXOC5 | 2.6 | 2.4 | 2.5 | NM_006544 |
| IFRD1 | 2.4 | 2.5 | 2.5 | NM_001007245 |
| ATP1B3 | 2.4 | 2.5 | 2.5 | NM_001679 |
| ANKRD28 | 2.4 | 2.5 | 2.4 | NM_015199 |
| LETM2 | 2.0 | 2.8 | 2.4 | NM_144652 |
| WSB1 | 2.2 | 2.7 | 2.4 | NM_015626 |
| RELL1 | 2.5 | 2.3 | 2.4 | NM_001085399 |
| PARL | 2.3 | 2.5 | 2.4 | NM_001037639 |
| TRIB1 | 2.1 | 2.7 | 2.4 | NM_025195 |
| NFKBID | 2.6 | 2.2 | 2.4 | NM_139239 |
| NUDT4 | 2.3 | 2.5 | 2.4 | NM_019094 |
| MNS1 | 2.1 | 2.7 | 2.4 | NM_018365 |
| ATP2B1 | 2.3 | 2.4 | 2.4 | NM_001001323 |
| PCTK2 | 2.7 | 2.1 | 2.4 | NM_002595 |
| PIP5K1B | 2.3 | 2.4 | 2.4 | NM_003558 |
| EIF5B | 2.2 | 2.5 | 2.3 | NM_015904 |
| TFG | 2.5 | 2.1 | 2.3 | NM_001007565 |
| SDCBP | 2.3 | 2.3 | 2.3 | NM_001007067 |
| SQLE | 2.3 | 2.3 | 2.3 | NM_003129 |
| ST8SIA4 | 2.0 | 2.5 | 2.3 | NM_005668 |
| PCGF5 | 2.3 | 2.2 | 2.2 | NM_032373 |
| RSPH10B2 | 2.4 | 2.1 | 2.2 | NM_001099697 |
| SLC25A13 | 2.3 | 2.2 | 2.2 | NM_001160210 |
| ANKRD12 | 2.3 | 2.1 | 2.2 | NM_001083625 |
| SLC4A7 | 2.0 | 2.4 | 2.2 | NM_003615 |
| ARL2BP | 2.1 | 2.3 | 2.2 | NM_012106 |
| SESN2 | 2.3 | 2.1 | 2.2 | NM_031459 |
| ZMYM6 | 2.1 | 2.2 | 2.2 | NM_007167 |
| ZFP36L1 | 2.3 | 2.0 | 2.2 | NM_004926 |
| SLC3A2 | 2.2 | 2.1 | 2.2 | NM_001012661 |
| AMMECR1 | 2.0 | 2.3 | 2.2 | NM_001025580 |
| GNG2 | 2.3 | 2.0 | 2.1 | NM_053064 |
| FAM107B | 2.0 | 2.2 | 2.1 | NM_031453 |
| TRIB3 | 2.1 | 2.2 | 2.1 | NM_021158 |
| CSNK2A1 | 2.1 | 2.1 | 2.1 | NM_001895 |
| NIPSNAP3B | 2.1 | 2.1 | 2.1 | NM_018376 |
| PPP1R2 | 2.1 | 2.0 | 2.1 | NM_006241 |
| PANK2 | 2.0 | 2.0 | 2.0 | NM_024960 |
| Early repression (3 h only) | ||||
| FAM46C | −3.3 | −2.8 | −3.0 | NM_017701 |
| LCP2 | −2.3 | −2.3 | −2.3 | NM_005565 |
| NOTCH2 | −2.2 | −2.1 | −2.1 | NM_024408 |
| AICDA | −2.2 | −2.0 | −2.1 | NM_020661 |
Late gene expression changes in stressed LCLs
| Gene . | LCL-06, 24 h . | LCL-06, 72 h . | Average . | RefSeq Transcript ID . |
|---|---|---|---|---|
| Late activation (24, 72 h only) | ||||
| IL2 | 5.39 | 11.35 | 8.37 | NM_000586 |
| DGKI | 4.25 | 12.48 | 8.37 | NM_004717 |
| FGFBP2 | 3.79 | 12.85 | 8.32 | NM_031950 |
| CTHRC1 | 6.77 | 8.23 | 7.50 | NM_138455 |
| SMPDL3A | 8.45 | 5.79 | 7.12 | NM_006714 |
| GZMB | 2.78 | 10.55 | 6.66 | NM_004131 |
| IFNG | 2.29 | 10.58 | 6.43 | NM_000619 |
| GBP2 | 5.09 | 7.05 | 6.07 | NM_004120 |
| F5 | 5.16 | 6.77 | 5.97 | NM_000130 |
| SERPINA1 | 3.63 | 7.99 | 5.81 | NM_000295 127704 |
| PALLD | 4.90 | 6.40 | 5.65 | NM_016081 |
| SORBS1 | 2.10 | 9.02 | 5.56 | NM_001034954 |
| GLYATL2 | 7.37 | 3.73 | 5.55 | NM_145016 |
| SERPINB2 | 5.84 | 5.14 | 5.49 | NM_001143818 |
| HS3ST1 | 4.70 | 6.02 | 5.36 | NM_005114 |
| MYOF | 4.59 | 6.11 | 5.35 | NM_013451 |
| MAP1B | 6.50 | 4.15 | 5.32 | NM_005909 |
| TNFRSF9 | 2.10 | 8.36 | 5.23 | NM_001561 |
| ASS1 | 4.07 | 5.96 | 5.01 | NM_000050 |
| XCL1 | 3.29 | 6.21 | 4.75 | NM_002995 |
| GPR56 | 4.08 | 5.32 | 4.70 | NM_001145770 |
| FILIP1L | 4.12 | 4.96 | 4.54 | NM_001042459 |
| ETV7 | 3.42 | 5.45 | 4.43 | NM_016135 |
| PRF1 | 2.26 | 6.55 | 4.41 | NM_001083116 |
| CD72 | 4.01 | 4.54 | 4.27 | NM_001782 |
| PDZK1 | 4.31 | 4.08 | 4.19 | NM_002614 |
| DHRS9 | 3.76 | 4.49 | 4.12 | NM_001142270 |
| TMEM100 | 2.94 | 5.05 | 3.99 | NM_001099640 |
| HLF | 2.21 | 5.72 | 3.97 | NM_002126 |
| S100A10 | 5.75 | 2.17 | 3.96 | NM_002966 |
| GSTA4 | 4.21 | 3.58 | 3.89 | NM_001512 |
| LAG3 | 2.78 | 4.83 | 3.80 | NM_002286 |
| LBH | 2.28 | 5.24 | 3.76 | NM_030915 |
| CD109 | 3.73 | 3.69 | 3.71 | NM_001159587 |
| IL1R2 | 2.08 | 5.26 | 3.67 | NM_004633 |
| CCDC144A | 3.55 | 3.77 | 3.66 | NM_014695 |
| SHC4 | 2.01 | 5.24 | 3.62 | NM_203349 |
| ENC1 | 2.44 | 4.68 | 3.56 | NM_003633 |
| ATP9A | 2.02 | 5.01 | 3.51 | NM_006045 |
| ABAT | 3.43 | 3.40 | 3.41 | NM_000663 |
| RUNDC3B | 4.35 | 2.32 | 3.34 | NM_001134405 |
| GABRB2 | 4.48 | 2.13 | 3.30 | NM_000813 |
| COL5A2 | 3.71 | 2.86 | 3.29 | NM_000393 |
| ITK | 2.35 | 4.15 | 3.25 | NM_005546 |
| AMY1A | 3.11 | 3.39 | 3.25 | NM_000699 |
| ALDH1L2 | 3.43 | 3.01 | 3.22 | NM_001034173 |
| NKG7 | 2.40 | 4.05 | 3.22 | NM_005601 |
| DKK4 | 3.57 | 2.81 | 3.19 | NM_014420 |
| IGF2R | 2.33 | 3.95 | 3.14 | NM_000876 |
| COL9A3 | 2.73 | 3.55 | 3.14 | NM_001853 |
| CAV1 | 2.14 | 4.11 | 3.12 | NM_001753 |
| PLEKHA7 | 3.96 | 2.25 | 3.10 | NM_175058 |
| CBS | 3.24 | 2.96 | 3.10 | NM_000071 |
| CD160 | 3.44 | 2.74 | 3.09 | NM_007053 |
| SYNPO2L | 2.45 | 3.70 | 3.07 | NM_001114133 |
| FYN | 2.96 | 3.13 | 3.05 | NM_002037 |
| ITPRIPL2 | 2.58 | 3.45 | 3.01 | NM_001034841 |
| FNIP1 | 3.73 | 2.25 | 2.99 | NM_001008738 |
| RASSF6 | 3.54 | 2.39 | 2.97 | NM_177532 |
| GLT8D2 | 3.00 | 2.86 | 2.93 | NM_031302 |
| UGT2B4 | 3.71 | 2.14 | 2.93 | NM_021139 |
| ABCB4 | 2.60 | 3.16 | 2.88 | NM_000443 |
| GPT2 | 3.21 | 2.55 | 2.88 | NM_001142466 |
| PABPC1L | 2.54 | 3.22 | 2.88 | NM_001124756 |
| PDE4DIP | 2.40 | 3.35 | 2.87 | NM_001002810 |
| LRRC50 | 2.31 | 3.40 | 2.85 | NM_178452 |
| CACNA1A | 2.49 | 3.18 | 2.84 | NM_000068 |
| BEND6 | 3.63 | 2.04 | 2.84 | NM_152731 |
| DDR2 | 2.76 | 2.91 | 2.84 | NM_001014796 |
| ASZ1 | 3.06 | 2.58 | 2.82 | NM_130768 |
| MYLK | 2.44 | 3.10 | 2.77 | NM_053025 |
| SERPINB1 | 2.06 | 3.48 | 2.77 | NM_030666 |
| TMEM149 | 2.73 | 2.78 | 2.76 | NM_024660 |
| CACNA1D | 3.16 | 2.17 | 2.67 | NM_000720 |
| ANK2 | 2.83 | 2.50 | 2.67 | NM_001127493 |
| NDFIP1 | 2.58 | 2.75 | 2.66 | NM_030571 |
| TNFRSF10D | 2.60 | 2.70 | 2.65 | NM_003840 |
| FRY | 2.02 | 3.24 | 2.63 | NM_023037 |
| ZDHHC11 | 2.69 | 2.53 | 2.61 | NM_024786 |
| TOX2 | 2.62 | 2.55 | 2.59 | NM_001098796 |
| BEX2 | 2.25 | 2.91 | 2.58 | NM_032621 |
| CCL5 | 2.93 | 2.17 | 2.55 | NM_002985 |
| PAM | 2.25 | 2.84 | 2.55 | NM_000919 |
| ANKRD29 | 2.67 | 2.38 | 2.53 | NM_173505 |
| ZFYVE21 | 2.33 | 2.69 | 2.51 | NM_024071 |
| ATF5 | 2.43 | 2.53 | 2.48 | NM_012068 |
| TSC22D1 | 2.47 | 2.48 | 2.47 | NM_006022 |
| TPBG | 2.35 | 2.55 | 2.45 | NM_006670 |
| SLC19A2 | 2.76 | 2.14 | 2.45 | NM_006996 |
| DBNDD2 | 2.05 | 2.83 | 2.44 | NM_001048221 |
| ABCB1 | 2.18 | 2.67 | 2.42 | NM_000443 |
| CCPG1 | 2.16 | 2.66 | 2.41 | NM_004748 |
| JAG1 | 2.27 | 2.54 | 2.41 | NM_000214 |
| PLA1A | 2.23 | 2.58 | 2.40 | NM_015900 |
| BSPRY | 2.47 | 2.33 | 2.40 | NM_017688 |
| NLRP1 | 2.10 | 2.67 | 2.39 | NM_001033053 |
| TM6SF1 | 2.46 | 2.31 | 2.38 | NM_001144903 |
| LPAR5 | 2.21 | 2.56 | 2.38 | NM_001142961 |
| TRIM2 | 2.29 | 2.46 | 2.37 | NM_001130067 |
| PHLDA1 | 2.48 | 2.25 | 2.37 | NM_007350 |
| TMEM22 | 2.14 | 2.56 | 2.35 | NM_001097599 |
| CCDC88A | 2.61 | 2.05 | 2.33 | NM_001135597 |
| SPAG1 | 2.40 | 2.24 | 2.32 | NM_003114 |
| B3GNT7 | 2.30 | 2.33 | 2.31 | NM_145236 |
| ALDH8A1 | 2.48 | 2.14 | 2.31 | NM_022568 |
| GPR133 | 2.25 | 2.37 | 2.31 | NM_198827 |
| SPAG9 | 2.36 | 2.25 | 2.30 | NM_001130528 |
| PELI1 | 2.16 | 2.44 | 2.30 | NM_020651 |
| CEACAM21 | 2.33 | 2.24 | 2.29 | NM_001098506 |
| SNX18 | 2.32 | 2.23 | 2.27 | NM_001102575 |
| PCGF5 | 2.29 | 2.25 | 2.27 | NM_032373 |
| KIF1B | 2.15 | 2.39 | 2.27 | NM_015074 |
| UNC84A | 2.46 | 2.09 | 2.27 | NM_001130965 |
| CRIP1 | 2.34 | 2.20 | 2.27 | NM_001311 |
| IFT57 | 2.24 | 2.30 | 2.27 | NM_018010 |
| NINJ2 | 2.33 | 2.19 | 2.26 | NM_016533 |
| N4BP2L2 | 2.16 | 2.35 | 2.25 | NM_014887 |
| LTBP3 | 2.43 | 2.06 | 2.25 | NM_001130144 |
| MLLT11 | 2.11 | 2.37 | 2.24 | NM_006818 |
| PCGF5 | 2.40 | 2.08 | 2.24 | NM_032373 |
| ZBTB32 | 2.27 | 2.18 | 2.23 | NM_014383 |
| MBD6 | 2.05 | 2.40 | 2.22 | NM_052897 |
| WARS | 2.05 | 2.38 | 2.22 | NM_004184 |
| CARS | 2.22 | 2.20 | 2.21 | NM_001014437 |
| GCG | 2.23 | 2.19 | 2.21 | NM_002054 |
| UBE2F | 2.07 | 2.31 | 2.19 | NM_080678 |
| PEX5L | 2.18 | 2.17 | 2.17 | NM_016559 |
| SUPT3H | 2.30 | 2.03 | 2.17 | NM_003599 |
| FAM169A | 2.04 | 2.26 | 2.15 | NM_015566 |
| ALG13 | 2.25 | 2.01 | 2.13 | NM_018466 |
| FRMD6 | 2.17 | 2.07 | 2.12 | NM_001042481 |
| ZNF79 | 2.05 | 2.20 | 2.12 | NM_007135 |
| TWSG1 | 2.11 | 2.11 | 2.11 | NM_020648 |
| FCHO2 | 2.10 | 2.11 | 2.11 | NM_001146032 |
| ERMP1 | 2.09 | 2.12 | 2.10 | NM_024896 |
| CNR1 | 2.19 | 2.02 | 2.10 | NM_001160226 |
| RAB3B | 2.10 | 2.10 | 2.10 | NM_002867 |
| MPZL3 | 2.03 | 2.16 | 2.09 | NM_198275 |
| GBP1 | 2.04 | 2.13 | 2.09 | NM_002053 |
| DFNB59 | 2.05 | 2.12 | 2.09 | NM_001042702 |
| LOXL3 | 2.05 | 2.12 | 2.09 | NM_032603 |
| NEU1 | 2.08 | 2.09 | 2.08 | NM_000434 |
| RTTN | 2.09 | 2.08 | 2.08 | NM_173630 |
| TESK1 | 2.11 | 2.05 | 2.08 | NM_006285 |
| RAPGEF2 | 2.00 | 2.13 | 2.07 | NM_014247 |
| PCGF5 | 2.06 | 2.06 | 2.06 | NM_032373 |
| DNASE2 | 2.02 | 2.08 | 2.05 | NM_001375 |
| PARP11 | 2.03 | 2.06 | 2.04 | NM_020367 |
| TWSG1 | 2.05 | 2.03 | 2.04 | NM_020648 |
| CTTN | 2.02 | 2.05 | 2.04 | NM_005231 |
| Late repression (24, 72 h only) | ||||
| MCM4 | −2.61 | −4.96 | −3.78 | NM_005914 |
| FCRL4 | −3.33 | −3.77 | −3.55 | NM_031282 |
| PRKCB | −3.88 | −2.84 | −3.36 | NM_002738 |
| STAT1 | −3.31 | −2.99 | −3.15 | NM_007315 |
| CCNE2 | −3.44 | −2.57 | −3.01 | NM_057749 |
| CT45A5 | −3.98 | −2.01 | −2.99 | NM_001007551 |
| LMNB1 | −3.08 | −2.71 | −2.89 | NM_005573 |
| BUB1 | −2.79 | −2.86 | −2.83 | NM_004336 |
| NUCKS1 | −2.82 | −2.83 | −2.83 | NM_022731 |
| RASSF5 | −3.23 | −2.39 | −2.81 | NM_182663 |
| SFPQ | −2.79 | −2.57 | −2.68 | NM_005066 |
| RAD51L3 | −2.76 | −2.34 | −2.55 | NM_001142571 |
| SLC29A1 | −2.38 | −2.62 | −2.50 | NM_001078174 |
| SFRS1 | −2.82 | −2.17 | −2.50 | NM_001078166 |
| TMPO | −2.25 | −2.70 | −2.48 | NM_001032283 |
| SCYL2 | −2.52 | −2.42 | −2.47 | NM_017988 |
| WNT6 | −2.42 | −2.46 | −2.44 | NM_006522 |
| CDV3 | −2.56 | −2.25 | −2.40 | NM_001134422 |
| ESCO2 | −2.43 | −2.30 | −2.36 | NM_001017420 |
| MRPL30 | −2.14 | −2.56 | −2.35 | NM_145212 |
| CXCR7 | −2.34 | −2.29 | −2.31 | NM_020311 |
| PDE7A | −2.50 | −2.10 | −2.30 | NM_002603 |
| EXOSC3 | −2.13 | −2.45 | −2.29 | NM_001002269 |
| ATP6V0D2 | −2.55 | −2.03 | −2.29 | NM_152565 |
| BAT2D1 | −2.30 | −2.25 | −2.27 | NM_015172 |
| UNG | −2.10 | −2.41 | −2.26 | NM_003362 |
| RFC3 | −2.28 | −2.20 | −2.24 | NM_002915 |
| NEXN | −2.22 | −2.22 | −2.22 | NM_144573 |
| POLH | −2.36 | −2.08 | −2.22 | NM_006502 |
| PKN2 | −2.38 | −2.01 | −2.19 | NM_006256 |
| PNN | −2.32 | −2.05 | −2.18 | NM_002687 |
| SKP2 | −2.27 | −2.07 | −2.17 | NM_005983 |
| GART | −2.02 | −2.13 | −2.08 | NM_000819 |
| PHTF2 | −2.07 | −2.06 | −2.06 | NM_001127357 |
| POLE3 | −2.06 | −2.04 | −2.05 | NM_017443 |
| PRPF18 | −2.02 | −2.02 | −2.02 | NM_003675 |
| Gene . | LCL-06, 24 h . | LCL-06, 72 h . | Average . | RefSeq Transcript ID . |
|---|---|---|---|---|
| Late activation (24, 72 h only) | ||||
| IL2 | 5.39 | 11.35 | 8.37 | NM_000586 |
| DGKI | 4.25 | 12.48 | 8.37 | NM_004717 |
| FGFBP2 | 3.79 | 12.85 | 8.32 | NM_031950 |
| CTHRC1 | 6.77 | 8.23 | 7.50 | NM_138455 |
| SMPDL3A | 8.45 | 5.79 | 7.12 | NM_006714 |
| GZMB | 2.78 | 10.55 | 6.66 | NM_004131 |
| IFNG | 2.29 | 10.58 | 6.43 | NM_000619 |
| GBP2 | 5.09 | 7.05 | 6.07 | NM_004120 |
| F5 | 5.16 | 6.77 | 5.97 | NM_000130 |
| SERPINA1 | 3.63 | 7.99 | 5.81 | NM_000295 127704 |
| PALLD | 4.90 | 6.40 | 5.65 | NM_016081 |
| SORBS1 | 2.10 | 9.02 | 5.56 | NM_001034954 |
| GLYATL2 | 7.37 | 3.73 | 5.55 | NM_145016 |
| SERPINB2 | 5.84 | 5.14 | 5.49 | NM_001143818 |
| HS3ST1 | 4.70 | 6.02 | 5.36 | NM_005114 |
| MYOF | 4.59 | 6.11 | 5.35 | NM_013451 |
| MAP1B | 6.50 | 4.15 | 5.32 | NM_005909 |
| TNFRSF9 | 2.10 | 8.36 | 5.23 | NM_001561 |
| ASS1 | 4.07 | 5.96 | 5.01 | NM_000050 |
| XCL1 | 3.29 | 6.21 | 4.75 | NM_002995 |
| GPR56 | 4.08 | 5.32 | 4.70 | NM_001145770 |
| FILIP1L | 4.12 | 4.96 | 4.54 | NM_001042459 |
| ETV7 | 3.42 | 5.45 | 4.43 | NM_016135 |
| PRF1 | 2.26 | 6.55 | 4.41 | NM_001083116 |
| CD72 | 4.01 | 4.54 | 4.27 | NM_001782 |
| PDZK1 | 4.31 | 4.08 | 4.19 | NM_002614 |
| DHRS9 | 3.76 | 4.49 | 4.12 | NM_001142270 |
| TMEM100 | 2.94 | 5.05 | 3.99 | NM_001099640 |
| HLF | 2.21 | 5.72 | 3.97 | NM_002126 |
| S100A10 | 5.75 | 2.17 | 3.96 | NM_002966 |
| GSTA4 | 4.21 | 3.58 | 3.89 | NM_001512 |
| LAG3 | 2.78 | 4.83 | 3.80 | NM_002286 |
| LBH | 2.28 | 5.24 | 3.76 | NM_030915 |
| CD109 | 3.73 | 3.69 | 3.71 | NM_001159587 |
| IL1R2 | 2.08 | 5.26 | 3.67 | NM_004633 |
| CCDC144A | 3.55 | 3.77 | 3.66 | NM_014695 |
| SHC4 | 2.01 | 5.24 | 3.62 | NM_203349 |
| ENC1 | 2.44 | 4.68 | 3.56 | NM_003633 |
| ATP9A | 2.02 | 5.01 | 3.51 | NM_006045 |
| ABAT | 3.43 | 3.40 | 3.41 | NM_000663 |
| RUNDC3B | 4.35 | 2.32 | 3.34 | NM_001134405 |
| GABRB2 | 4.48 | 2.13 | 3.30 | NM_000813 |
| COL5A2 | 3.71 | 2.86 | 3.29 | NM_000393 |
| ITK | 2.35 | 4.15 | 3.25 | NM_005546 |
| AMY1A | 3.11 | 3.39 | 3.25 | NM_000699 |
| ALDH1L2 | 3.43 | 3.01 | 3.22 | NM_001034173 |
| NKG7 | 2.40 | 4.05 | 3.22 | NM_005601 |
| DKK4 | 3.57 | 2.81 | 3.19 | NM_014420 |
| IGF2R | 2.33 | 3.95 | 3.14 | NM_000876 |
| COL9A3 | 2.73 | 3.55 | 3.14 | NM_001853 |
| CAV1 | 2.14 | 4.11 | 3.12 | NM_001753 |
| PLEKHA7 | 3.96 | 2.25 | 3.10 | NM_175058 |
| CBS | 3.24 | 2.96 | 3.10 | NM_000071 |
| CD160 | 3.44 | 2.74 | 3.09 | NM_007053 |
| SYNPO2L | 2.45 | 3.70 | 3.07 | NM_001114133 |
| FYN | 2.96 | 3.13 | 3.05 | NM_002037 |
| ITPRIPL2 | 2.58 | 3.45 | 3.01 | NM_001034841 |
| FNIP1 | 3.73 | 2.25 | 2.99 | NM_001008738 |
| RASSF6 | 3.54 | 2.39 | 2.97 | NM_177532 |
| GLT8D2 | 3.00 | 2.86 | 2.93 | NM_031302 |
| UGT2B4 | 3.71 | 2.14 | 2.93 | NM_021139 |
| ABCB4 | 2.60 | 3.16 | 2.88 | NM_000443 |
| GPT2 | 3.21 | 2.55 | 2.88 | NM_001142466 |
| PABPC1L | 2.54 | 3.22 | 2.88 | NM_001124756 |
| PDE4DIP | 2.40 | 3.35 | 2.87 | NM_001002810 |
| LRRC50 | 2.31 | 3.40 | 2.85 | NM_178452 |
| CACNA1A | 2.49 | 3.18 | 2.84 | NM_000068 |
| BEND6 | 3.63 | 2.04 | 2.84 | NM_152731 |
| DDR2 | 2.76 | 2.91 | 2.84 | NM_001014796 |
| ASZ1 | 3.06 | 2.58 | 2.82 | NM_130768 |
| MYLK | 2.44 | 3.10 | 2.77 | NM_053025 |
| SERPINB1 | 2.06 | 3.48 | 2.77 | NM_030666 |
| TMEM149 | 2.73 | 2.78 | 2.76 | NM_024660 |
| CACNA1D | 3.16 | 2.17 | 2.67 | NM_000720 |
| ANK2 | 2.83 | 2.50 | 2.67 | NM_001127493 |
| NDFIP1 | 2.58 | 2.75 | 2.66 | NM_030571 |
| TNFRSF10D | 2.60 | 2.70 | 2.65 | NM_003840 |
| FRY | 2.02 | 3.24 | 2.63 | NM_023037 |
| ZDHHC11 | 2.69 | 2.53 | 2.61 | NM_024786 |
| TOX2 | 2.62 | 2.55 | 2.59 | NM_001098796 |
| BEX2 | 2.25 | 2.91 | 2.58 | NM_032621 |
| CCL5 | 2.93 | 2.17 | 2.55 | NM_002985 |
| PAM | 2.25 | 2.84 | 2.55 | NM_000919 |
| ANKRD29 | 2.67 | 2.38 | 2.53 | NM_173505 |
| ZFYVE21 | 2.33 | 2.69 | 2.51 | NM_024071 |
| ATF5 | 2.43 | 2.53 | 2.48 | NM_012068 |
| TSC22D1 | 2.47 | 2.48 | 2.47 | NM_006022 |
| TPBG | 2.35 | 2.55 | 2.45 | NM_006670 |
| SLC19A2 | 2.76 | 2.14 | 2.45 | NM_006996 |
| DBNDD2 | 2.05 | 2.83 | 2.44 | NM_001048221 |
| ABCB1 | 2.18 | 2.67 | 2.42 | NM_000443 |
| CCPG1 | 2.16 | 2.66 | 2.41 | NM_004748 |
| JAG1 | 2.27 | 2.54 | 2.41 | NM_000214 |
| PLA1A | 2.23 | 2.58 | 2.40 | NM_015900 |
| BSPRY | 2.47 | 2.33 | 2.40 | NM_017688 |
| NLRP1 | 2.10 | 2.67 | 2.39 | NM_001033053 |
| TM6SF1 | 2.46 | 2.31 | 2.38 | NM_001144903 |
| LPAR5 | 2.21 | 2.56 | 2.38 | NM_001142961 |
| TRIM2 | 2.29 | 2.46 | 2.37 | NM_001130067 |
| PHLDA1 | 2.48 | 2.25 | 2.37 | NM_007350 |
| TMEM22 | 2.14 | 2.56 | 2.35 | NM_001097599 |
| CCDC88A | 2.61 | 2.05 | 2.33 | NM_001135597 |
| SPAG1 | 2.40 | 2.24 | 2.32 | NM_003114 |
| B3GNT7 | 2.30 | 2.33 | 2.31 | NM_145236 |
| ALDH8A1 | 2.48 | 2.14 | 2.31 | NM_022568 |
| GPR133 | 2.25 | 2.37 | 2.31 | NM_198827 |
| SPAG9 | 2.36 | 2.25 | 2.30 | NM_001130528 |
| PELI1 | 2.16 | 2.44 | 2.30 | NM_020651 |
| CEACAM21 | 2.33 | 2.24 | 2.29 | NM_001098506 |
| SNX18 | 2.32 | 2.23 | 2.27 | NM_001102575 |
| PCGF5 | 2.29 | 2.25 | 2.27 | NM_032373 |
| KIF1B | 2.15 | 2.39 | 2.27 | NM_015074 |
| UNC84A | 2.46 | 2.09 | 2.27 | NM_001130965 |
| CRIP1 | 2.34 | 2.20 | 2.27 | NM_001311 |
| IFT57 | 2.24 | 2.30 | 2.27 | NM_018010 |
| NINJ2 | 2.33 | 2.19 | 2.26 | NM_016533 |
| N4BP2L2 | 2.16 | 2.35 | 2.25 | NM_014887 |
| LTBP3 | 2.43 | 2.06 | 2.25 | NM_001130144 |
| MLLT11 | 2.11 | 2.37 | 2.24 | NM_006818 |
| PCGF5 | 2.40 | 2.08 | 2.24 | NM_032373 |
| ZBTB32 | 2.27 | 2.18 | 2.23 | NM_014383 |
| MBD6 | 2.05 | 2.40 | 2.22 | NM_052897 |
| WARS | 2.05 | 2.38 | 2.22 | NM_004184 |
| CARS | 2.22 | 2.20 | 2.21 | NM_001014437 |
| GCG | 2.23 | 2.19 | 2.21 | NM_002054 |
| UBE2F | 2.07 | 2.31 | 2.19 | NM_080678 |
| PEX5L | 2.18 | 2.17 | 2.17 | NM_016559 |
| SUPT3H | 2.30 | 2.03 | 2.17 | NM_003599 |
| FAM169A | 2.04 | 2.26 | 2.15 | NM_015566 |
| ALG13 | 2.25 | 2.01 | 2.13 | NM_018466 |
| FRMD6 | 2.17 | 2.07 | 2.12 | NM_001042481 |
| ZNF79 | 2.05 | 2.20 | 2.12 | NM_007135 |
| TWSG1 | 2.11 | 2.11 | 2.11 | NM_020648 |
| FCHO2 | 2.10 | 2.11 | 2.11 | NM_001146032 |
| ERMP1 | 2.09 | 2.12 | 2.10 | NM_024896 |
| CNR1 | 2.19 | 2.02 | 2.10 | NM_001160226 |
| RAB3B | 2.10 | 2.10 | 2.10 | NM_002867 |
| MPZL3 | 2.03 | 2.16 | 2.09 | NM_198275 |
| GBP1 | 2.04 | 2.13 | 2.09 | NM_002053 |
| DFNB59 | 2.05 | 2.12 | 2.09 | NM_001042702 |
| LOXL3 | 2.05 | 2.12 | 2.09 | NM_032603 |
| NEU1 | 2.08 | 2.09 | 2.08 | NM_000434 |
| RTTN | 2.09 | 2.08 | 2.08 | NM_173630 |
| TESK1 | 2.11 | 2.05 | 2.08 | NM_006285 |
| RAPGEF2 | 2.00 | 2.13 | 2.07 | NM_014247 |
| PCGF5 | 2.06 | 2.06 | 2.06 | NM_032373 |
| DNASE2 | 2.02 | 2.08 | 2.05 | NM_001375 |
| PARP11 | 2.03 | 2.06 | 2.04 | NM_020367 |
| TWSG1 | 2.05 | 2.03 | 2.04 | NM_020648 |
| CTTN | 2.02 | 2.05 | 2.04 | NM_005231 |
| Late repression (24, 72 h only) | ||||
| MCM4 | −2.61 | −4.96 | −3.78 | NM_005914 |
| FCRL4 | −3.33 | −3.77 | −3.55 | NM_031282 |
| PRKCB | −3.88 | −2.84 | −3.36 | NM_002738 |
| STAT1 | −3.31 | −2.99 | −3.15 | NM_007315 |
| CCNE2 | −3.44 | −2.57 | −3.01 | NM_057749 |
| CT45A5 | −3.98 | −2.01 | −2.99 | NM_001007551 |
| LMNB1 | −3.08 | −2.71 | −2.89 | NM_005573 |
| BUB1 | −2.79 | −2.86 | −2.83 | NM_004336 |
| NUCKS1 | −2.82 | −2.83 | −2.83 | NM_022731 |
| RASSF5 | −3.23 | −2.39 | −2.81 | NM_182663 |
| SFPQ | −2.79 | −2.57 | −2.68 | NM_005066 |
| RAD51L3 | −2.76 | −2.34 | −2.55 | NM_001142571 |
| SLC29A1 | −2.38 | −2.62 | −2.50 | NM_001078174 |
| SFRS1 | −2.82 | −2.17 | −2.50 | NM_001078166 |
| TMPO | −2.25 | −2.70 | −2.48 | NM_001032283 |
| SCYL2 | −2.52 | −2.42 | −2.47 | NM_017988 |
| WNT6 | −2.42 | −2.46 | −2.44 | NM_006522 |
| CDV3 | −2.56 | −2.25 | −2.40 | NM_001134422 |
| ESCO2 | −2.43 | −2.30 | −2.36 | NM_001017420 |
| MRPL30 | −2.14 | −2.56 | −2.35 | NM_145212 |
| CXCR7 | −2.34 | −2.29 | −2.31 | NM_020311 |
| PDE7A | −2.50 | −2.10 | −2.30 | NM_002603 |
| EXOSC3 | −2.13 | −2.45 | −2.29 | NM_001002269 |
| ATP6V0D2 | −2.55 | −2.03 | −2.29 | NM_152565 |
| BAT2D1 | −2.30 | −2.25 | −2.27 | NM_015172 |
| UNG | −2.10 | −2.41 | −2.26 | NM_003362 |
| RFC3 | −2.28 | −2.20 | −2.24 | NM_002915 |
| NEXN | −2.22 | −2.22 | −2.22 | NM_144573 |
| POLH | −2.36 | −2.08 | −2.22 | NM_006502 |
| PKN2 | −2.38 | −2.01 | −2.19 | NM_006256 |
| PNN | −2.32 | −2.05 | −2.18 | NM_002687 |
| SKP2 | −2.27 | −2.07 | −2.17 | NM_005983 |
| GART | −2.02 | −2.13 | −2.08 | NM_000819 |
| PHTF2 | −2.07 | −2.06 | −2.06 | NM_001127357 |
| POLE3 | −2.06 | −2.04 | −2.05 | NM_017443 |
| PRPF18 | −2.02 | −2.02 | −2.02 | NM_003675 |
To prevent cell-line–specific changes, only genes differentially expressed (using a fold cutoff of 2.0) in both cell lines, LCL-03 and LCL-06, were included in the 3-hour analysis. Genes induced or repressed > 2.0-fold in LCL-06 at 3 hours and in LCL-06 at 24 and 72 hours (Figure 7A set m) were overlapped with genes induced or repressed > 2.0-fold in LCL-03 at 3 hours and LCL-06 at 24 and 72 hours (Figure 7B set q). The intersection of sets m and q (plotted in Figure 7C) therefore represents sustained activation or repression over the entire time course of TG-induced lytic replication and consists of 79 probe sets. After eliminating uncharacterized open reading frames and control and redundant probe sets, we determined that 23 genes were up-regulated and 1 gene was down-regulated at all time points (Figure 7C and Table 1). Because Z and LMP1 protein levels are increased by TG at 24 but not 6 hours after TG, these genes are all kinetically upstream of Z and LMP1. The UPR chaperone CHOP/DDIT3 was up-regulated 3.0-fold in LCL-6 and 1.8-fold in LCL-03.
Genes regulated in a sustained manner include 2 members of the nuclear hormone receptor family (NR4A2 and NR4A3). NR4A1 and NR4A3 are induced by lytic replication secondary to BCR cross-linking in AKATA cells before ZTA expression,19 a transcription factor (CEBPβ) induced by proteasome inhibition and involved in Z activation,5 as well as candidate transcription factors of interest such as NFIL3, KLF4, MAFF, cytokines such as IL8 and CCL4, and genes involved in adjustment to calcium stress (eg, STC2). Only RNASE6 was repressed at all time points.
Early up-regulated genes included EGR3, a transcription factor induced by BCR cross-linking, and early-response genes such as c-Jun, JunD, FOS, FOSB, and KLF6.19 Tribbles 1 and 3, previously reported to be induced by ER stress, were up-regulated 2- to 3-fold. Notch2 was down-regulated 2-fold, suggesting a contribution of Notch/EBNA2 signaling to the maintenance of latency. Activation-induced cytidine deaminase (AICDA) was also repressed. AICDA is involved in the somatic hypermutation, gene conversion, and class-switch recombination of immunoglobulin genes required for several crucial steps of B-cell terminal differentiation that is necessary for efficient Ab responses, which is consistent with our observations that ER stress is associated with inhibition of the plasma cell differentiation pathway.
Late-regulated genes included those known to be induced by ER stress (eg, ASS1, ATF5, and DHRS9), antiviral genes (IFNG), and calcium-binding proteins (eg, S100A). DHRS9, a retinol-metabolizing enzyme, is activated directly by ZTA binding during lytic infection in epithelial cells.20 XCL1 was up-regulated 3.3- and 6.2-fold at 24 and 72 hours, respectively (Table 3), so it may be important for B cells undergoing lytic infection to migrate to epithelial surfaces for viral release into saliva.
As shown in Figure 4A, at 24 hours, SAL incubation reduced the amplitude of EBV lytic gene induction by TG pulse. We therefore hypothesized that genes that are induced by T24 but in which the mRNA expression level is reduced to near DMSO levels in TS24 experiments might be specifically associated with EBV lytic gene expression, either causing or resulting from activation of EBV lytic genes (Table 4 and supplemental Figure 2).
Unique gene expression changes in T24 versus TS24
| Gene . | -Fold change . | RefSeq Transcript ID . |
|---|---|---|
| Genes > 3-fold up in T24 but not TS24 | ||
| CTHRC1 | 7.6 | NM_138455 |
| EMP1 | 7.2 | NM_001423 |
| GLYATL2 | 6.5 | NM_145016 |
| MYLK | 6.1 | NM_053025 |
| PALLD | 5.3 | NM_016081 |
| SERPINB2 | 5.2 | NM_001143818 |
| GBP2 | 5.1 | NM_004120 |
| CD24 | 5.0 | NM_013230 |
| ANXA1 | 4.7 | NM_000700 |
| XCL1 | 4.7 | NM_002995 |
| MUC4 | 4.5 | NM_004532 |
| PLEKHA7 | 4.4 | NM_175058 |
| LY6K | 4.4 | NM_017527 |
| GABRB2 | 4.3 | NM_000813 |
| DHRS9 | 4.3 | NM_001142270 |
| DKK4 | 4.3 | NM_014420 |
| CTNNA2 | 4.3 | NM_004389 |
| KLF5 | 4.2 | NM_001730 |
| FHL5 | 4.2 | NM_020482 |
| ARAP3 | 4.2 | NM_022481 |
| DCHS1 | 4.0 | NM_003737 |
| SFN | 4.0 | NM_006142 |
| DHRS9 | 4.0 | NM_001142270 |
| CD72 | 3.8 | NM_001782 |
| FGFBP2 | 3.7 | NM_031950 |
| SERPIND1 | 3.6 | NM_000185 |
| TNS3 | 3.6 | NM_022748 |
| ANKRD29 | 3.5 | NM_173505 |
| FNIP1 | 3.5 | NM_001008738 |
| GAP43 | 3.5 | NM_001130064 |
| NR4A3 | 3.4 | NM_006981 |
| RAB3B | 3.3 | NM_002867 |
| EVI2A | 3.3 | NM_001003927 |
| RUNDC3B | 3.3 | NM_001134405 |
| CITED2 | 3.3 | NM_006079 |
| IL2 | 3.3 | NM_000586 |
| IL4 | 3.2 | NM_000589 |
| DNAH12 | 3.2 | NM_178504 |
| CCDC88A | 3.1 | NM_001135597 |
| MYADM | 3.1 | NM_001020818 |
| TEX9 | 3.1 | NM_198524 |
| ENDOD1 | 3.1 | NM_015036 |
| MPEG1 | 3.0 | NM_001039396 |
| Genes > 3-fold down in T24 but not TS24 | ||
| NOTCH2 | −3.5 | NM_024408 |
| FCRL4 | −3.3 | NM_031282 |
| HIST1H2BC | −3.2 | NM_003526 |
| TXNIP | −3.0 | NM_006472 |
| Gene . | -Fold change . | RefSeq Transcript ID . |
|---|---|---|
| Genes > 3-fold up in T24 but not TS24 | ||
| CTHRC1 | 7.6 | NM_138455 |
| EMP1 | 7.2 | NM_001423 |
| GLYATL2 | 6.5 | NM_145016 |
| MYLK | 6.1 | NM_053025 |
| PALLD | 5.3 | NM_016081 |
| SERPINB2 | 5.2 | NM_001143818 |
| GBP2 | 5.1 | NM_004120 |
| CD24 | 5.0 | NM_013230 |
| ANXA1 | 4.7 | NM_000700 |
| XCL1 | 4.7 | NM_002995 |
| MUC4 | 4.5 | NM_004532 |
| PLEKHA7 | 4.4 | NM_175058 |
| LY6K | 4.4 | NM_017527 |
| GABRB2 | 4.3 | NM_000813 |
| DHRS9 | 4.3 | NM_001142270 |
| DKK4 | 4.3 | NM_014420 |
| CTNNA2 | 4.3 | NM_004389 |
| KLF5 | 4.2 | NM_001730 |
| FHL5 | 4.2 | NM_020482 |
| ARAP3 | 4.2 | NM_022481 |
| DCHS1 | 4.0 | NM_003737 |
| SFN | 4.0 | NM_006142 |
| DHRS9 | 4.0 | NM_001142270 |
| CD72 | 3.8 | NM_001782 |
| FGFBP2 | 3.7 | NM_031950 |
| SERPIND1 | 3.6 | NM_000185 |
| TNS3 | 3.6 | NM_022748 |
| ANKRD29 | 3.5 | NM_173505 |
| FNIP1 | 3.5 | NM_001008738 |
| GAP43 | 3.5 | NM_001130064 |
| NR4A3 | 3.4 | NM_006981 |
| RAB3B | 3.3 | NM_002867 |
| EVI2A | 3.3 | NM_001003927 |
| RUNDC3B | 3.3 | NM_001134405 |
| CITED2 | 3.3 | NM_006079 |
| IL2 | 3.3 | NM_000586 |
| IL4 | 3.2 | NM_000589 |
| DNAH12 | 3.2 | NM_178504 |
| CCDC88A | 3.1 | NM_001135597 |
| MYADM | 3.1 | NM_001020818 |
| TEX9 | 3.1 | NM_198524 |
| ENDOD1 | 3.1 | NM_015036 |
| MPEG1 | 3.0 | NM_001039396 |
| Genes > 3-fold down in T24 but not TS24 | ||
| NOTCH2 | −3.5 | NM_024408 |
| FCRL4 | −3.3 | NM_031282 |
| HIST1H2BC | −3.2 | NM_003526 |
| TXNIP | −3.0 | NM_006472 |
Individual sets of genes regulated at each time point under each treatment condition, together with their J5 values, are shown in supplemental Tables 1-6.
Discussion
Our data indicate that the induction of ER stress by TG is sufficient to trigger high-copy (> 400 copies/cell) LCLs into lytic replication, including activation of EBV lytic genes, surface gp350 expression, and release of virus. A 3-hour exposure to TG pulse is sufficient for this effect, resulting in a transcriptional and lytic permissive state that occurs as long as 5 days after the initial application of ER stress. A low-copy-number (< 50 copies) LCL did not show activation of EBV lytic promoters in response to TG. These differences in ER stress effects on low- and high-copy-number LCLs may be related to epigenetic differences in LCLs derived from different patient sources.21 Our results indicate that ER stress is an important in vitro trigger for lytic replication, and provide a chemical model for studying the cellular pathways involved. Our approach is validated by the fact that many of the same genes induced by lytic replication secondary to BCR cross-linking (eg, NR4A2 and EGR-1) are also up-regulated by ER-stress–induced lytic replication in our studies.
TG is a chemical that depletes luminal ER calcium. Therefore, it is possible that some of the lytic effects might be due to increases in cellular Ca2+ and PKC activation.18 However, recapitulation of EBV lytic gene activation with TM makes this less likely. As expected, attempts in our laboratory to induce lytic replication with TPA in all LCLs used were unsuccessful. This raises the question of whether ER stress signaling per se (eg, IRE1 signaling) is a cue for lytic replication. More generally, stress-responsive chaperones activated by ER stress and calcium depletion may be critical for initiating the lytic program.
SAL had opposite effects in modulating TG activity, depending on the length of the assay, repressing TG-induced lytic gene activation when cells were harvested 24 hours after TG pulse and synergizing with TG at 72 hours or at 5 days after TG pulse. SAL had no effect on EBV lytic gene activation when used alone in these assays. However, SAL prevented virus release, as measured by accumulation of EBV viral DNA in supernatants, in both unstressed and stressed conditions, while minimally affecting the TG-induced appearance of gp350 on the LCL cell surface. These results strongly suggest that SAL inhibits a key event in viral budding and egress. Further EM experiments should clarify the location of the SAL block to viral assembly and/or exit. In B95-8 cells, SAL prevented lytic replication stimulated by TPA, almost completely inhibiting gp350 expression, a result that differed from the inhibition by SAL of ER-stress induced lytic induction in human LCLs, in which little effect of SAL was observed on gp350 expression. These experiments suggest ER-stress–dependent and -independent mechanisms of SAL antiviral activity.
The production of viral RNA and viral protein are both potent stimuli for initiating the UPR via both PERK and PKR activation. Both kinases phosphorylate eIF2α to arrest m7GTP cap-dependent translation. Interestingly, both TG and SAL resulted in eIF2α phosphorylation in LCLs; however, their effects were uncoupled at the level of EBV lytic gene activation. Whereas TG alone was able to activate lytic gene expression, SAL alone had no effect. Despite equivalent effects on eIF2a phosphorylation, TG resulted in activation of ATF transcription factors, whereas SAL did not. Further, we were unable to reproducibly recapitulate ER stress effects on EBV lytic gene activation by overexpressing a constitutively active mutant of eIF2α (S51D) in transient transfection experiments in LCLs (data not shown). These data indicate that eIF2a is not a control point for lytic replication.
Despite previous reports implicating the ER stress- and plasma cell–specifying transcription factor XBP1 in Z promoter activation, extremely weak XBP1 splicing by TG compared with other cell types was seen (data not shown), suggesting a relative resistance to this arm of the UPR in LCLs. Our previous data indicated that the typical UPR response is blunted in LCLs, with limited XBP1(s) production and ATF6 N-terminal cleavage that increased when cells were stressed in the absence of EBNA3C.22 Despite these effects, TG nevertheless induced lytic replication. However, such a robust induction of the UPR by TG may not reflect physiologic UPR induction, and latent gene expression (including EBNA3C) may be sufficient to limit UPR-induced lytic replication in vivo.
Differentiation to plasma cell phenotype has been implicated in EBV lytic replication; however, the TG-induced changes in the levels of BLIMP1, BCL6, and CD138 mRNAs did not support a role for UPR lytic induction mediated by plasma cell differentiation in our assays. Furthermore, high levels of EBV lytic gene activity were associated with repression rather than activation of plasma cell–associated genes. T24, T72, TS24, and TS72 treatments led to at least a 2-fold down-regulation of multiple probe sets derived from immunoglobulin κ and light chains, as well as 1 gene for the essential plasma cell ER protein 1 (pERp1; supplemental Tables 3 and 4 and data not shown).23
ER stress induction resulted in LMP1 mRNA and protein expression and the mRNA for the chemokine IL-8, which is induced by ER stress in an LMP1-dependent manner in nasopharyngeal carcinoma cell lines.12 Because IL8 mRNA was increased as early as 3 hours after TG exposure, a time point before maximal LMP1 induction, IL-8 regulation is most likely independent of LMP1 in LCLs undergoing ER stress. IL-8 (CXCL8) is a CXC-type cytokine that is mostly produced by macrophages and epithelial and endothelial cells; however, IL-8 receptor expression has been recorded on CD19-positive B cells, resulting in enhanced chemotaxis.24 IL-8 released from B lymphocytes undergoing ER stress in the oropharynx of patients with acute infectious mononucleosis may result in enhanced B-cell chemotaxis to the oropharyngeal epithelium with release of virus.25,26
The type of prolonged ER stress applied in our experiments is likely to have resulted in autophagy, as indicated by the formation of double-membrane vesicles by EM (Figure 6C arrow). Further experiments will investigate the roles of autophagy in EBV lytic replication.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank James Lyons-Weiler and Haiwen Shi from the University of Pittsburgh genomics and proteomics core laboratories for their assistance with Affymetrix interpretation. They also gratefully acknowledge the assistance of Donna Beer-Stolz, University of Pittsburgh Center for Biologic Imaging, who performed the EM studies.
This work was supported by a Physician-Scientist Early Career Grant (57006750) from the Howard Hughes Medical Institute startup and by competitive medical research foundation funding from the University of Pittsburgh (to A.R.).
National Institutes of Health
Authorship
Contribution: G.M.T. and S.K.R. performed the experiments and prepared the figures; A.R. and G.M.T. formulated the hypotheses and devised the experiments; D.T.R. provided helpful suggestions, cell lines, and reagents; R.M.W. performed the EBV DNA quantitation; and A.R. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Adam Rosendorff, Rangos Research Center, 3rd Floor Pathology, 530 45th St, Pittsburgh, PA 15224; e-mail: adam.rosendorff@chp.edu.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal