Abstract
Studying human antigen-specific memory B cells has been challenging because of low frequencies in peripheral blood, slow proliferation, and lack of antibody secretion. Therefore, most studies have relied on conversion of memory B cells into antibody-secreting cells by in vitro culture. To facilitate direct ex vivo isolation, we generated fluorescent antigen tetramers for characterization of memory B cells by using tetanus toxoid as a model antigen. Brightly labeled memory B cells were identified even 4 years after last immunization, despite low frequencies ranging from 0.01% to 0.11% of class-switched memory B cells. A direct comparison of monomeric to tetrameric antigen labeling demonstrated that a substantial fraction of the B-cell repertoire can be missed when monomeric antigens are used. The specificity of the method was confirmed by antibody reconstruction from single-cell sorted tetramer+ B cells with single-cell RT-PCR of the B-cell receptor. All antibodies bound to tetanus antigen with high affinity, ranging from 0.23 to 2.2 nM. Furthermore, sequence analysis identified related memory B cell and plasmablast clones isolated more than a year apart. Therefore, antigen tetramers enable specific and sensitive ex vivo characterization of rare memory B cells as well as the production of fully human antibodies.
Introduction
The development of vaccines to cancer antigens and problem pathogens requires the ability to monitor induced T-cell and B-cell responses. Although antibodies secreted by plasma cells can be readily measured, there is a need for methods that enable quantitative ex vivo analysis of human memory B cells with defined antigen specificity. The analysis of antigen-specific human memory B cells has been challenging because they circulate at very low frequencies in peripheral blood, do not secrete antibodies, and proliferate only at a very slow rate under steady-state conditions.1,2 Therefore, most investigators have relied on expansion and conversion of memory B cells into antibody-secreting cells by in vitro culture by the use of several different stimuli, including Toll-like receptor ligands (CpG oligonucleotide 2006 or R848), pokeweed mitogen, cytokines or cytokine cocktails (IL-2, IL-6, IL-15, IL-10, IL-21), CD40 ligation, and BCR crosslinking.3-6 After in vitro culture for up to 1 week, antibodies can be measured in culture supernatants and the frequencies of antibody secreting cells determined by the use of ELISPOT assays. However, depending on the stimulation condition used, different antibody secretion rates and different frequencies of antibody-secreting cells can be observed, which makes comparisons across studies difficult.7,8 Furthermore, in vitro culture not only induces antibody secretion but also alters the memory B-cell phenotype to resemble plasmablasts with distinct functional properties.9-11
As an alternative, fluorescently labeled antigen has been used to identify B cells with particular BCR specificities by flow cytometry. This approach has been used to label plasmablasts, which are present at a relatively high frequency in peripheral blood after vaccination.12,13 However, plasmablasts only circulate in the blood for a few days in transit to the BM.14 Fluorescently labeled HIV surface proteins have been used to isolate HIV-specific B cells for the generation of neutralizing antibodies from HIV elite controllers who have high frequencies of HIV-specific B cells.15,16 However, the frequency of most memory B-cell specificities is very low because the antigen has been cleared, making detection of these cells very challenging. Furthermore, the signal generated by fluorescently labeled antigens is typically not bright and tends to overlap with the unlabeled cell population, making frequency analysis difficult.
Here we describe a sensitive method that enables reliable detection of memory B cells with defined specificity from small quantities of peripheral blood, despite very low frequencies. Antigen tetramers were used to increase the avidity of BCR labeling and the brightness of staining, and sorting procedures were optimized to minimize background as much as possible. This approach enabled visualization and isolation of memory B cells months to years after antigen had been cleared. The specificity of the approach was validated by single-cell sorting of tetramer-labeled B cells and reconstruction of antibodies, all of which bound to their target antigen with high affinity.
Methods
Protein expression
Tetanus toxin C-fragment (TTCF) was cloned into the pET-15b expression vector (Novagen) containing an N-terminal, thrombin-cleavable 6x histidine-tag followed by a BirA site (amino acids: GLNDIFEAQKIEWHE) and a short, flexible linker sequence. Protein expression was induced in BL21(DE3) Escherichia coli cells with 1mM isopropyl β-D-1-thiogalactopyranoside for 4 hours at 28°C. Cells were washed and lysed by sonication in 50mM tris(hydroxymethyl)aminomethane pH 8/500mM NaCl buffer. Supernatant was collected by ultracentrifugation, and TTCF was purified by the use of a HIS-Select affinity column (Sigma-Aldrich) according to the manufacturer's instructions. The histidine-tag was removed by cleavage with 10 U thrombin (Novagen) per 1 mg of TTCF for 2 hours at room temperature. TTCF was subsequently purified by the use of MonoQ anion exchange chromatography (GE Healthcare). For tetramer formation TTCF was mono-biotinylated by incubation with BirA enzyme at a 1:20 molar ratio for 4 hours at 30°C in a buffer containing 10mM tris(hydroxymethyl)aminomethane, pH 8; 0.1mM biotin; 10mM adenosine-5′-triphosphate; 10mM magnesium acetate; and 50mM bicine, pH8.3. Excess biotin was removed via Superose 12 gel filtration chromatography (GE Healthcare).
Murine CD80 membrane proximal domain (amino acids 145-239) was cloned into the pET-22b expression vector containing an N-terminal BirA site and a short linker sequence. Protein production was induced in BL21(DE3) E coli cells with 1mM isopropyl β-D-1-thiogalactopyranoside for 4 hours at 37°C. Inclusion bodies were dissolved in 6M guanidine hydrochloride and reduced with 65mM dithiothreitol. CD80 was refolded at 10°C for 2 days in buffer containing 100mM tris(hydroxymethyl)aminomethane, pH 8; 400mM L-arginine; 2mM ethylenediaminetetraacetic acid; 5mM reduced glutathione; 0.5mM oxidized glutathione; and 0.1M PMSF. After refolding, protein was first dialyzed against 20mM tris(hydroxymethyl)aminomethane, pH 7.4, and 100mM urea followed by dialysis against 10mM tris(hydroxymethyl)aminomethane, pH 8. CD80 was purified by anion-exchange chromatography (MonoQ column) and biotinylated with BirA enzyme as described above. For TTCF monomer versus tetramer experiments, Alexa-488 dye molecules (Molecular Probes) were linked to primary amines on biotinylated CD80 or TTCF.
Sample preparation and cell staining
This study was approved by the Dana Farber/Harvard Cancer Center Institutional Review Board, and written informed consent was obtained from healthy participants in accordance with the Declaration of Helsinki. Blood samples were donated by healthy volunteers, and the date of the last tetanus vaccination was recorded when available. Two healthy volunteers were immunized with a combination vaccine (tetanus, diphtheria, and pertussis) for analysis of tetanus-specific plasmablasts. Leukocytes were isolated by the use of density gradient centrifugation with Ficoll-Paque PLUS (GE Healthcare). Cells were washed, counted, and resuspended in PBS/5% FCS. For memory B-cell labeling, cells were enriched with the use of EasySep Human B Cell Enrichment Kit (StemCell Technologies Inc). After enrichment, cells were adjusted to a cell density of 5 × 106 cells/mL and stained with 0.125 μg/mL of TTCF or CD80 control tetramer and incubated on ice for 30 minutes with intermittent gentle vortexing. Cells were costained with CD19-Pacific-Blue (AbD Serotec), CD27-fluorescein, IgM-allophycocyanin, CD3-PerCP, CD14-PerCP, 7AAD (BD Biosciences), and CD16 PerCP (BioLegend) for an additional 20 minutes on ice. For monomer versus tetramer experiments, CD27-fluorescein isothiocyanate was replaced with CD27-phycoerythrin (BD Biosciences). Cells were washed at least twice with 5 mL of phosphate-buffered saline/5% fetal calf serum and filtered through a 35-μm nylon mesh before single cell sorting. The gating strategies are depicted in supplemental Figures 1 and 3 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Tetramer preparation
All tetramers were prepared freshly for each experiment. Biotinylated TTCF or CD80 were incubated with premium-grade phycoerythrin-labeled streptavidin (Molecular Probes) for at least 20 minutes on ice at a molar ratio of 4:1. Before cell staining, tetramer preparations were centrifuged for 10 minutes at maximum speed to remove aggregates. For experiments investigating TTCF monomer versus tetramer staining, tetramers were formed with Alexa-fluor-488–tagged antigens and nonfluorescent streptavidin (Thermo Scientific) at a 4:1 molar ratio.
Single-cell sorting
Cells were sorted on a BD fluorescence-activated cell sorter Aria II. Proper alignment of the laser was checked before each experiment by sorting of single Flouresbrite microspheres (Polysciences Inc) into 8-tube PCR strips (USA Scientific) and visual inspection under a fluorescent microscope to ensure that beads were found at the bottom center of each tube. After alignment, PCR strips containing 3 μL of mRNA extraction buffer (Epicentre Biotechnologies) were set up on a cooled 96-well metal block (Stratagene) to keep the buffer cold during sorting. After cells were collected, PCR strips were briefly centrifuged, frozen immediately on dry ice and stored at −80°C until further processing. As a positive control for mRNA amplification and PCR processing, 100 CD19+ CD27+ IgM− B cells were sorted into a single tube and processed in the same manner as single cells.
mRNA preamplification
mRNA amplification was performed in a designated laminar flow hood to minimize contamination. Workspace and equipment were routinely cleaned with RNaseZAP wipes (Ambion). All mRNA and PCR steps were carried out using a C1000 96-well Thermal Cycler (BioRad). The mRNA amplification protocol was adapted from MessageBOOSTER cDNA Synthesis From Cell Lysates kit (Epicentre Biotechnologies) with the following modifications. T7-oligo(dT) primer was replaced with BCR gene-specific primers tagged with a single stranded T7 polymerase recognition site (underlined): IgG-T7 TAATACGACTCACTATAGGTTCGGGGAAGTAGTCCTTGACCAGG; Kappa-T7 TAATACGACTCACTATAGGGATAGAAGTTATTCAGCAGGCACAC; and Lambda-T7 TAATACGACTCACTATAGGCGTCAGGCTCAGRTAGCTGCTGGCCGC. All 3 primers were mixed together at a concentration of 50 ng/μL each. MMLV reverse transcriptase provided with the kit was replaced with Superscript3 (Invitrogen), and RT reactions were carried out at 50°C for 1 hour. In vitro transcription was carried out for 4 hours at 42°C. Before final complementary DNA (cDNA) synthesis, mRNA was concentrated by the use of RNA clean/concentrator-5 columns (Zymo Research), eluted in 8 μL of water, and transferred into 8-tube PCR strips for cDNA synthesis. Final cDNA was suspended in a volume of approximately 12 μL and provided sufficient template for up to 6 separate PCR reactions.
Nested RT-PCR
Primers for nested PCR were used as described previously, but restriction enzyme sequences were omitted from second-round primers.17 cDNA (2 μL) generated via mRNA amplification was used as a template for first-round PCR, with the following cycling conditions: 3 cycles of preamplification (94°C/45 seconds, 45°C/45 seconds, 72°C/105 seconds), followed by 30 cycles of amplification (94°C/45 seconds, 50°C/45 seconds, 72°C/105 seconds), and 10 minutes of final extension at 72°C. The first-round PCR (3 μL) product served as a template for the second round of nested PCR. The same cycling conditions were used for the first round of PCR, but the 3 cycles of preamplification were omitted. Both PCR steps were performed by the use of cloned Pfu polymerase AD (Agilent Technologies). PCR products were separated on 1% agarose gels and products of 300-400 nucleotides in size isolated with the use of Zymoclean DNA gel recovery kit (Zymo Research). Sequencing was performed by the use of forward and reverse primers used for the second-round nested PCR.
Antibody production and purification
Isolated heavy- and light-chain variable-domain DNA was cloned into separate pcDNA3.3 expression vectors containing the bovine prolactin signal peptide sequence as well as full-length IgG1 heavy- or κ light-chain constant domains. Antibodies were expressed by Chinese hamster ovary (CHO)-S cells (Invitrogen) cultured in serum free FreeStyle CHO-S media (Invitrogen) supplemented with 8mM Glutamax (Gibco) in 100-mL spinner flasks (Bellco) at 37°C with 8% CO2. One day before transfection, CHO-S cells were split to 6 × 105 cells/mL. On the day of transfection, cells were adjusted to 1 × 106 when necessary. Heavy- and light-chain plasmid DNA (25 μg of each) were cotransfected with the use of MAX transfection reagent (Invitrogen) according to the manufacturer's instructions. Cells were cultured after transfection for 6-8 days. Protein G Sepharose beads (GE Healthcare) were added to filtered supernatant and rotated overnight at 4°C. Beads were collected and antibody was eluted with 100mM glycine, pH 2.5, and separated from beads by the use of Spin-X centrifuge tube filters (Corning). Eluted protein was quickly neutralized by addition of 1M tris(hydroxymethyl)aminomethane, pH 9. Purified antibody was buffer exchanged into PBS by the use of Micro Bio-spin columns (BioRad). Protein concentration was determined by measurement of absorbance at 280 nm with a NanoDrop (Thermo Scientific) instrument.
Saturation binding assay
Nonbiotinylated, MonoQ-purified TTCF was labeled with europium according to the manufacturer's instructions (PerkinElmer). Free europium was removed by Superose 200 gel filtration chromatography (GE Healthcare). Then, 96-well flat bottom plates (PerkinElmer) were coated overnight at 4°C with 20 ng of antibody per well in 100mM NaHCO3 buffer, pH 9.6. Plates were blocked with assay buffer containing BSA and bovine γ-globulins (PerkinElmer) at room temperature for 3 hours. TTCF-europium was diluted in assay buffer (100nM to 4pM), and 200 μL was added per well in triplicates. Plates were incubated for 2 hours at 37°C and washed 3 times with 200 μL of buffer [50mM tris(hydroxymethyl)aminomethane, pH 8; 150mM NaCl; 20μM ethylenediaminetetraacetic acid; 0.05% Tween]. Then, 100 μL of enhancement solution (PerkinElmer) was added to each well and fluorescence counts measured with a Victor3 plate reader (PerkinElmer) at a wavelength of 615 nm. When necessary, fluorescence counts were normalized to compare different experiments. To control for nonspecific binding, plates were coated with a control antibody (clone 8.18.C5) generated in the same manner as TTCF-specific Abs.
Analysis
Heavy- and light-chain variable domain sequences were analyzed with IMGT/V-Quest and JOINSOLVER software. Flow cytometry data were evaluated with FlowJo analysis software. Statistical analysis was conducted with GraphPad Prism 5 software by the use of unpaired t tests. To determine antibody KD values, saturation binding data were fitted with GraphPad Prism 5 software by the use of nonlinear regression analysis.
Results
Generation of tetanus toxoid tetramer
Our approach enabling isolation of memory B cells by flow cytometry using antigen tetramers is illustrated in Figure 1. We chose the 52 kDa, nontoxic, carboxyl-terminal fragment of tetanus toxoid (TTCF) as a model antigen because most members of the population have been vaccinated with tetanus toxoid and because persistent IgG antibody titers are induced by this vaccine.18 TTCF was expressed in E coli, and a BirA site was attached to the N-terminus for site-specific mono-biotinylation by BirA enzyme. A flexible linker was placed between the protein and this biotinylation site to prevent steric hindrance of antibody binding. TTCF was purified by anion-exchange chromatography, biotinylated with BirA, and separated from free biotin and BirA by gel filtration chromatography. TTCF tetramers were generated by incubation of fluorescently tagged streptavidin with biotinylated TTCF antigen at a molar ratio of 1:4. These tetramers were then used along with a panel of mAbs for the identification of tetanus toxoid-specific memory B cells.
Overview of the technique. (A) Soluble antigen or antigen domain is expressed with a BirA tag for site-specific biotinylatation and tetramerization with fluorescently labeled streptavidin. (B) B cells are stained with tetramer and a panel of monoclonal antibodies. Tetramer-positive class-switched memory B cells (CD19+ CD27+ IgM−) are single-cell sorted into PCR strips. (C) mRNA preamplification is performed with T7 RNA polymerase. Single-stranded cDNA is synthesized by the use of a primer with a single-stranded T7 RNA polymerase site. Conversion to double-stranded cDNA enables an in vitro transcription reaction with T7 RNA polymerase, which provides sufficient amounts of RNA for RT-PCR from resting, recirculating memory B cells. (D) Sequencing of PCR products is carried out directly from 300- to 400-bp PCR products by the use of second-round forward and reverse primers. (E) Overlap PCR is used for construction of full-length IgG1 heavy chain and κ light sequences, which are cloned into separate vectors. These vectors are transiently transfected into CHO-S cells for expression of fully human recombinant antibodies.
Overview of the technique. (A) Soluble antigen or antigen domain is expressed with a BirA tag for site-specific biotinylatation and tetramerization with fluorescently labeled streptavidin. (B) B cells are stained with tetramer and a panel of monoclonal antibodies. Tetramer-positive class-switched memory B cells (CD19+ CD27+ IgM−) are single-cell sorted into PCR strips. (C) mRNA preamplification is performed with T7 RNA polymerase. Single-stranded cDNA is synthesized by the use of a primer with a single-stranded T7 RNA polymerase site. Conversion to double-stranded cDNA enables an in vitro transcription reaction with T7 RNA polymerase, which provides sufficient amounts of RNA for RT-PCR from resting, recirculating memory B cells. (D) Sequencing of PCR products is carried out directly from 300- to 400-bp PCR products by the use of second-round forward and reverse primers. (E) Overlap PCR is used for construction of full-length IgG1 heavy chain and κ light sequences, which are cloned into separate vectors. These vectors are transiently transfected into CHO-S cells for expression of fully human recombinant antibodies.
Tetramer-based detection of antigen-specific plasmablasts
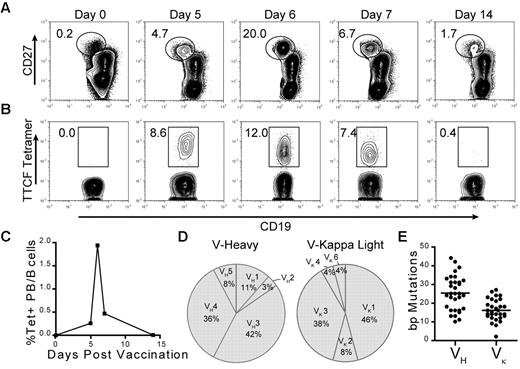
After a booster vaccination, a burst of plasmablasts containing a fraction of antigen-specific cells can be detected in peripheral blood that can reach levels up to 100-fold greater than baseline.19 Therefore, we first tested the technique by using blood samples from 2 donors who had received a tetanus booster shot (tetanus and diphtheria toxoids, pertussis vaccine). Consistent with previous studies, we detected an expanded population of plasmablasts, characterized by high level expression of CD27 and moderately decreased expression of CD19.13 The peak frequency was detected on day 6 after vaccination when plasmablasts constituted 20% and 12% of total CD19+ B cells in donors 1 and 2, respectively (Figure 2A; supplemental Figure 1B). Furthermore, as shown in donor 1, the plasmablast population contracted quickly in the blood within 24 hours (day 6-7). The TTCF tetramer brightly labeled a substantial subpopulation of plasmablasts on days 5, 6, and 7, when 8.6%, 12.0%, and 7.4% of all plasmablasts were tetramer-positive (Figure 2B). Bright tetramer labeling of plasmablasts was also observed in donor 2 on day 6 when 10.6% of plasmablasts were tetramer-positive (supplemental Figure 1C). TTCF-specific plasmablasts represented approximately 1%-2% of total peripheral blood B cells in the 2 donors on day 6 (Figure 2C; supplemental Figure 1D). However, on day 9 (donor 2) or day 14 (donor 1) TTCF tetramer-positive populations could no longer be detected within the plasmablast compartment. Interestingly, the intensity of tetramer labeling decreased from day 5 to day 7, suggesting that surface BCR levels were down-regulated as plasmablasts differentiated toward plasma cells, which no longer express surface BCR (Figure 2B).20
Detection of tetanus toxoid specific plasmablasts after vaccination. (A) Plasmablasts were identified on the basis of intermediate CD19 and high CD27 expression. Fluorescence-activated cell sorter plots were gated on total CD19+ B cells, which were negative for a panel of exclusion markers (CD3, CD14, CD16, and 7AAD). Numbers adjacent to the gate represent the percentage of plasmablasts within the CD19+ B-cell population. (B) Identification of TTCF tetramer-positive cells within the plasmablast population (gated in panel A). Numbers adjacent to gate represent the percentage of TTCF tetramer-positive within the plasmablast population (identified in panel A). (C) Frequency of tetramer-positive plasmablasts within the total CD19+ B-cell population during the first 2 weeks after vaccination. (D) Variable gene segment usage by unique B-cell clones for heavy (n = 26) and light (n = 25) chains from single cell sorted tetramer-positive plasmablasts isolated from days 6 and 7. (E) Scatter plot of somatic mutations detected in variable gene segments of heavy and light chains with mean values (VH = 25.3 and VK = 16.2) indicated by vertical solid lines.
Detection of tetanus toxoid specific plasmablasts after vaccination. (A) Plasmablasts were identified on the basis of intermediate CD19 and high CD27 expression. Fluorescence-activated cell sorter plots were gated on total CD19+ B cells, which were negative for a panel of exclusion markers (CD3, CD14, CD16, and 7AAD). Numbers adjacent to the gate represent the percentage of plasmablasts within the CD19+ B-cell population. (B) Identification of TTCF tetramer-positive cells within the plasmablast population (gated in panel A). Numbers adjacent to gate represent the percentage of TTCF tetramer-positive within the plasmablast population (identified in panel A). (C) Frequency of tetramer-positive plasmablasts within the total CD19+ B-cell population during the first 2 weeks after vaccination. (D) Variable gene segment usage by unique B-cell clones for heavy (n = 26) and light (n = 25) chains from single cell sorted tetramer-positive plasmablasts isolated from days 6 and 7. (E) Scatter plot of somatic mutations detected in variable gene segments of heavy and light chains with mean values (VH = 25.3 and VK = 16.2) indicated by vertical solid lines.
To analyze the diversity BCR sequences and generate recombinant antibodies, tetramer-positive cells from donor 1 (days 6 and 7) were single-cell sorted (Table 1). We first gated on CD19+ cells that were negative for a panel of exclusion markers (CD3, CD14, CD16, 7AAD), then gated on plasmablasts, identified by high levels of CD27 and an intermediate level of CD19 expression, and finally on tetramer+ CD19+ cells (supplemental Figure 1A). Single cells were directly sorted into PCR strips containing lysis buffer and immediately frozen. It has been previously shown that the efficiency of single-cell PCR can be substantially improved by mRNA preamplification, which increases the amount of mRNA more than 80-fold.21 Gene-specific reverse primers tagged with a single-stranded T7 RNA polymerase promoter sequence were annealed to the constant regions of heavy and light chains approximately 80 base pairs 3′ to the variable domains. Double-stranded cDNA was used for an in vitro transcription reaction with T7 RNA polymerase, and the resulting mRNA served as the template for cDNA synthesis. A previously reported nested PCR protocol enabled amplification of heavy- and light-chain variable segments,17 and negative controls were included to carefully monitor for contamination. From a total of 35 single cells, 32 heavy and 30 light chain segments were amplified and directly sequenced from gel-purified PCR products, corresponding to an overall PCR efficiency of 89%. Sequence analysis revealed that TTCF tetramer-positive cells used a variety of different VH-D-JH gene segments, without dominance of one particular gene segment (Figure 2D). Interestingly, 19% of unique B-cell clones used IGHV4-31, indicating a potential selection preference for this particular heavy-chain variable-gene segment (Table 1). All light chains presented κ locus sequences, with primary use of IGKV1 and IGKV3 segments. A large number of somatic mutations were present in all analyzed heavy and light chains, indicating that cells had undergone extensive somatic hypermutation (Figure 2E). Five sets of related sequences were observed (4 pairs and one cluster composed of 3 related clones) that shared use of the same gene segments and a subset of mutations (Table 1). This pattern is characteristic of clones whose daughter cells have diversified by somatic hypermutation.
BCR variable-domain analysis of tetramer-positive plasmablasts isolated from donor 1
| ID . | Day . | Heavy chain . | Kappa light chain . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V-D-J junction . | V . | D . | J . | V-Mut nt/aa* . | CDRs† . | V-J junction . | V . | J . | V-Mut nt/aa* . | CDRs† . | ||
| PB01 | D6 | CARGRRYTFGYKSIPHDFW | V1-8 | D5-5 | J4 | 37/19 | 8.8.17 | CMQSSQIPHTF | V2-24 | J5 | 18/12 | 11.3.9 |
| PB02 | D7 | CARDRRQYSGHADLDYW | V1-18 | D5-12 | J4 | 11/5 | 8.8.15 | CQQTYSTLYTF | V1-39 | J2 | 14/4 | 6.3.9 |
| PB03 | D7 | CVRDSPYSSGSIPSAFDIW | V1-18 | D6-19 | J3 | 24/11 | 8.8.17 | CLQHFTYPYTF | V1-17 | J1 | 16/9 | 6.3.9 |
| PB04 | D7 | CARSVRGVVPFDYW | V2-5 | D3-10 | J4 | 18/13 | 10.7.12 | CQQYNTYSPWTF | V1-5 | J1 | 15/10 | 6.3.10 |
| PB05 | D6 | CSRDGGSRRGFAYW | V3-11 | D3-16 | J4 | 44/17 | 8.7.12 | CQHRSKWPPMVTF | V3-11 | J2 | 13/8 | 6.3.11 |
| PB06 | D6 | CGRSVIGTVDNW | V3-11 | D6-19 | J4 | 34/18 | 8.8.10 | CQQSKSWPQLTF | V3-15 | J4 | 14/9 | 6.3.10 |
| PB07 | D6 | CARATLPAAIGYYGMDVW | V3-21 | D2-2 | J6 | 10/4 | 8.8.16 | |||||
| PB08 | D6 | CAKDLQLWLLGAFDIR | V3-23 | D5-5 | J3 | 16/10 | 8.8.14 | CLQNCNCHRF | V1-6 | J1 | 24/18 | 6.3.8 |
| PB09‡ | D6 | CVKREYYNDNSGYHVWGDYW | V3-23 | D3-22 | J5 | 32/18 | 8.8.18 | CQEYDNWPRFTF | V3D-15 | J3 | 14/7 | 6.3.10 |
| PB10‡ | D7 | CVKREYYNDNSGYHVWGDYW | V3-23 | D3-22 | J5 | 33/21 | 8.8.18 | CQNYNNWPRFSF | V3-15 | J3 | 14/4 | 6.3.10 |
| PB11 | D7 | CARGQIWSNLPTLFEYW | V3-30 | D5-5 | J4 | 22/11 | 8.8.15 | CQHYNSYPLTF | V1-16 | J4 | 18/11 | 6.3.9 |
| PB12‡ | D6 | CARERGALLTRGHFDYW | V3-30-3 | D2-2 | J4 | 16/13 | 8.8.15 | CQHRSKWPPMVTF | V3-11 | J2 | 14/9 | 6.3.11 |
| PB13‡ | D7 | CARERGAMVTRGHFDYW | V3-30-3 | D5-5 | J4 | 25/16 | 8.8.15 | CQLRTTWPSMYTF | V3-11 | J2 | 22/12 | 6.3.11 |
| PB14‡ | D6 | CARDVFRVSGPLVAVGGFDYW | V3-30-3 | D6-19 | J4 | 29/12 | 8.8.19 | CQQYENWPPLTF | V3-15 | J4 | 12/9 | 6.3.10 |
| PB15‡ | D7 | CARDVFRVSGPLVAVGGFDYW | V3-30-3 | D6-19 | J4 | 31/12 | 8.8.19 | CQQYEKWPPLTF | V3-15 | J4 | 21/13 | 6.3.10 |
| PB16 | D7 | CARGLDHNSYVYYFDHW | V3-33 | D1-1 | J4 | 23/12 | 8.8.15 | CQQYNTWPPFTF | V3-15 | J3 | 12/7 | 6.3.10 |
| PB17 | D6 | CARDRDYSGSGIPPSFDPW | V3-33 | D3-10 | J5 | 17/11 | 8.8.17 | CQQSYIPPPAF | V1-39 | J4 | 17/12 | 6.3.9 |
| PB18 | D6 | CVREDIVLRVFAIFDYW | V3-74 | D2-8 | J4 | 25/12 | 8.8.15 | CQQSHTTPRTF | V1-39 or V1D-39 | J1 | 26/14 | 6.3.9 |
| PB19 | D7 | CARSGGSSSGSFYPGALYYNGMDVW | V4-4 | D3-10 | J6 | 16/9 | 8.7.23 | CQQSYSATWTF | V1-39 or V1D-39 | J1 | 18/9 | 6.3.9 |
| PB20 | D6 | CARAGPVGGSWHSVWFDLW | V4-31 | D2-15 | J5 | 24/13 | 10.7.17 | CQQRHNLITF | V3-11 | J5 | 16/7 | 6.3.8 |
| PB21 | D6 | CVRGYRFCSSSSCNEWYFDLW | V4-31 | D2-2 | J2 | 30/17 | 10.7.19 | CHQSRSLPHTF | V6-21 or V6D-21 | J2 | 8/5 | 6.3.9 |
| PB22 | D7 | CATEPGLHCGPDCPDAFEIW | V4-31 | D2-21 | J3 | 25/14 | 10.7.18 | CQHHHTF | V1-9 | J2 | 18/11 | 6.3.5 |
| PB23‡ | D6 | CAREFINTLGGVVDHYGIDVW | V4-31 | D3-16 | J6 | 13/8 | 10.7.19 | |||||
| PB24‡ | D6 | CARGFMNTLGGVIDTEGLDVW | V4-31 | D3-16 | J6 | 42/23 | 10.7.19 | |||||
| PB25 | D6 | CARVTTHYDPPFDHW | V4-31 | D4-4 | J4 | 39/19 | 10.7.13 | CQQYYSTPWTF | V4-1 | J1 | 10/4 | 12.3.9 |
| PB26 | D6 | CARNVVITLGGVIESNYYFDYW | V4-39 | D3-16 | J4 | 23/13 | 10.7.20 | CMQGIQTPTF | V2-28 | J1 | 11/4 | 11.3.8 |
| PB27‡ | D7 | CGLMADNWFDPW | V4-39 | D2-8 | J5 | 28/13 | 10.7.10 | CQQYNNWPPITF | V3-15 | J5 | 2/1 | 6.3.10 |
| PB28‡ | D7 | CALQTDNWFDPW | V4-39 | D4-4 | J5 | 34/17 | 10.7.10 | CQQYYQWPPYTF | V3D-15 | J2 | 22/10 | 6.3.10 |
| PB29‡ | D7 | CARQADNWCDPW | V4-39 | D6-19 | J5 | 31/16 | 10.7.10 | CQQYYNWPPYTF | V3D-15 | J2 | 9/8 | 6.3.10 |
| PB30 | D7 | CARGWFGYLDYW | V4-59 | D3-10 | J4 | 13/8 | 8.7.10 | |||||
| PB31 | D7 | CARYCSSPTCSVVYGMDVW | V5-51 | D2-2 | J6 | 15/8 | 8.8.17 | CQQTNSFPFNF | V1D-12 | J4 | 27/13 | 6.3.9 |
| PB32 | D6 | CWSSVYQSDGGGYYYVDYW | V5-51 | D3-22 | J4 | 31/14 | 8.8.17 | CQETYTTPPLTF | V1-39 | J4 | 24/11 | 6.3.10 |
| PB33 | D6 | CQHRSKWPPMVTF | V3-11 | J2 | 13/8 | 6.3.11 | ||||||
| PB34 | D6 | CQQYSSYPLTF | V1-16 | J4 | 19/10 | 6.3.9 | ||||||
| ID . | Day . | Heavy chain . | Kappa light chain . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V-D-J junction . | V . | D . | J . | V-Mut nt/aa* . | CDRs† . | V-J junction . | V . | J . | V-Mut nt/aa* . | CDRs† . | ||
| PB01 | D6 | CARGRRYTFGYKSIPHDFW | V1-8 | D5-5 | J4 | 37/19 | 8.8.17 | CMQSSQIPHTF | V2-24 | J5 | 18/12 | 11.3.9 |
| PB02 | D7 | CARDRRQYSGHADLDYW | V1-18 | D5-12 | J4 | 11/5 | 8.8.15 | CQQTYSTLYTF | V1-39 | J2 | 14/4 | 6.3.9 |
| PB03 | D7 | CVRDSPYSSGSIPSAFDIW | V1-18 | D6-19 | J3 | 24/11 | 8.8.17 | CLQHFTYPYTF | V1-17 | J1 | 16/9 | 6.3.9 |
| PB04 | D7 | CARSVRGVVPFDYW | V2-5 | D3-10 | J4 | 18/13 | 10.7.12 | CQQYNTYSPWTF | V1-5 | J1 | 15/10 | 6.3.10 |
| PB05 | D6 | CSRDGGSRRGFAYW | V3-11 | D3-16 | J4 | 44/17 | 8.7.12 | CQHRSKWPPMVTF | V3-11 | J2 | 13/8 | 6.3.11 |
| PB06 | D6 | CGRSVIGTVDNW | V3-11 | D6-19 | J4 | 34/18 | 8.8.10 | CQQSKSWPQLTF | V3-15 | J4 | 14/9 | 6.3.10 |
| PB07 | D6 | CARATLPAAIGYYGMDVW | V3-21 | D2-2 | J6 | 10/4 | 8.8.16 | |||||
| PB08 | D6 | CAKDLQLWLLGAFDIR | V3-23 | D5-5 | J3 | 16/10 | 8.8.14 | CLQNCNCHRF | V1-6 | J1 | 24/18 | 6.3.8 |
| PB09‡ | D6 | CVKREYYNDNSGYHVWGDYW | V3-23 | D3-22 | J5 | 32/18 | 8.8.18 | CQEYDNWPRFTF | V3D-15 | J3 | 14/7 | 6.3.10 |
| PB10‡ | D7 | CVKREYYNDNSGYHVWGDYW | V3-23 | D3-22 | J5 | 33/21 | 8.8.18 | CQNYNNWPRFSF | V3-15 | J3 | 14/4 | 6.3.10 |
| PB11 | D7 | CARGQIWSNLPTLFEYW | V3-30 | D5-5 | J4 | 22/11 | 8.8.15 | CQHYNSYPLTF | V1-16 | J4 | 18/11 | 6.3.9 |
| PB12‡ | D6 | CARERGALLTRGHFDYW | V3-30-3 | D2-2 | J4 | 16/13 | 8.8.15 | CQHRSKWPPMVTF | V3-11 | J2 | 14/9 | 6.3.11 |
| PB13‡ | D7 | CARERGAMVTRGHFDYW | V3-30-3 | D5-5 | J4 | 25/16 | 8.8.15 | CQLRTTWPSMYTF | V3-11 | J2 | 22/12 | 6.3.11 |
| PB14‡ | D6 | CARDVFRVSGPLVAVGGFDYW | V3-30-3 | D6-19 | J4 | 29/12 | 8.8.19 | CQQYENWPPLTF | V3-15 | J4 | 12/9 | 6.3.10 |
| PB15‡ | D7 | CARDVFRVSGPLVAVGGFDYW | V3-30-3 | D6-19 | J4 | 31/12 | 8.8.19 | CQQYEKWPPLTF | V3-15 | J4 | 21/13 | 6.3.10 |
| PB16 | D7 | CARGLDHNSYVYYFDHW | V3-33 | D1-1 | J4 | 23/12 | 8.8.15 | CQQYNTWPPFTF | V3-15 | J3 | 12/7 | 6.3.10 |
| PB17 | D6 | CARDRDYSGSGIPPSFDPW | V3-33 | D3-10 | J5 | 17/11 | 8.8.17 | CQQSYIPPPAF | V1-39 | J4 | 17/12 | 6.3.9 |
| PB18 | D6 | CVREDIVLRVFAIFDYW | V3-74 | D2-8 | J4 | 25/12 | 8.8.15 | CQQSHTTPRTF | V1-39 or V1D-39 | J1 | 26/14 | 6.3.9 |
| PB19 | D7 | CARSGGSSSGSFYPGALYYNGMDVW | V4-4 | D3-10 | J6 | 16/9 | 8.7.23 | CQQSYSATWTF | V1-39 or V1D-39 | J1 | 18/9 | 6.3.9 |
| PB20 | D6 | CARAGPVGGSWHSVWFDLW | V4-31 | D2-15 | J5 | 24/13 | 10.7.17 | CQQRHNLITF | V3-11 | J5 | 16/7 | 6.3.8 |
| PB21 | D6 | CVRGYRFCSSSSCNEWYFDLW | V4-31 | D2-2 | J2 | 30/17 | 10.7.19 | CHQSRSLPHTF | V6-21 or V6D-21 | J2 | 8/5 | 6.3.9 |
| PB22 | D7 | CATEPGLHCGPDCPDAFEIW | V4-31 | D2-21 | J3 | 25/14 | 10.7.18 | CQHHHTF | V1-9 | J2 | 18/11 | 6.3.5 |
| PB23‡ | D6 | CAREFINTLGGVVDHYGIDVW | V4-31 | D3-16 | J6 | 13/8 | 10.7.19 | |||||
| PB24‡ | D6 | CARGFMNTLGGVIDTEGLDVW | V4-31 | D3-16 | J6 | 42/23 | 10.7.19 | |||||
| PB25 | D6 | CARVTTHYDPPFDHW | V4-31 | D4-4 | J4 | 39/19 | 10.7.13 | CQQYYSTPWTF | V4-1 | J1 | 10/4 | 12.3.9 |
| PB26 | D6 | CARNVVITLGGVIESNYYFDYW | V4-39 | D3-16 | J4 | 23/13 | 10.7.20 | CMQGIQTPTF | V2-28 | J1 | 11/4 | 11.3.8 |
| PB27‡ | D7 | CGLMADNWFDPW | V4-39 | D2-8 | J5 | 28/13 | 10.7.10 | CQQYNNWPPITF | V3-15 | J5 | 2/1 | 6.3.10 |
| PB28‡ | D7 | CALQTDNWFDPW | V4-39 | D4-4 | J5 | 34/17 | 10.7.10 | CQQYYQWPPYTF | V3D-15 | J2 | 22/10 | 6.3.10 |
| PB29‡ | D7 | CARQADNWCDPW | V4-39 | D6-19 | J5 | 31/16 | 10.7.10 | CQQYYNWPPYTF | V3D-15 | J2 | 9/8 | 6.3.10 |
| PB30 | D7 | CARGWFGYLDYW | V4-59 | D3-10 | J4 | 13/8 | 8.7.10 | |||||
| PB31 | D7 | CARYCSSPTCSVVYGMDVW | V5-51 | D2-2 | J6 | 15/8 | 8.8.17 | CQQTNSFPFNF | V1D-12 | J4 | 27/13 | 6.3.9 |
| PB32 | D6 | CWSSVYQSDGGGYYYVDYW | V5-51 | D3-22 | J4 | 31/14 | 8.8.17 | CQETYTTPPLTF | V1-39 | J4 | 24/11 | 6.3.10 |
| PB33 | D6 | CQHRSKWPPMVTF | V3-11 | J2 | 13/8 | 6.3.11 | ||||||
| PB34 | D6 | CQQYSSYPLTF | V1-16 | J4 | 19/10 | 6.3.9 | ||||||
Bolded sequence pairs represent clonally related B cells.
CDR indicates complementarity determining region.
Number of mutations detected in V-gene segment (nucleotides/amino acids).
CDR1, 2, and 3 lengths in amino acids.
Five sets of related sequences were observed (4 pairs and one cluster composed of 3 related clones) that shared use of the same gene segments and a subset of mutations.
Tetramer-based detection of memory B cells with defined specificity
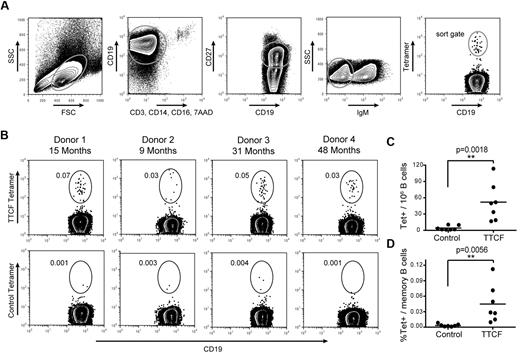
The plasmablast population containing TTCF-specific B cells disappeared rapidly from the blood, and there was only a short window of opportunity for the isolation of these cells. In contrast, memory B cells continue to circulate for many years after infection or vaccination, but their frequency is very low. Memory B cells from patients who spontaneously recovered from an infectious disease are of significant interest for the generation of human mAbs. We therefore asked whether antigen tetramers could enable ex vivo isolation of these rare cells without any previous in vitro expansion. Because of the low frequency of memory B cells, it was necessary to carefully reduce background as much as possible. B cells were first enriched by negative selection (cocktail of antibodies to CD2, CD3, CD14, CD16, CD56, and glycophorin A) to remove most cells that could nonspecifically bind the tetramer. Enriched cells were split evenly and stained with TTCF or a control tetramer followed by labeling with CD19, CD27, and IgM to specifically select class-switched memory B cells. The gating strategy considered expression of CD19, lack of labeling with a panel of exclusion markers (CD3, CD14, CD16, 7AAD), expression of the memory marker CD27, and lack of IgM expression as evidence of class switching (Figure 3A). The control tetramer contained the membrane proximal domain of CD80 (which has no known interaction partner). A population of TTCF tetramer-positive B cells was identified in all analyzed donors, whereas background staining with the control tetramer was very low (Figure 3B). TTCF tetramer-positive B cells were brightly labeled, with an intensity of more than 1 log above background for the majority of cells. TTCF tetramer-positive B cells represented 0.01%-0.11% of class-switched memory B cells, with frequencies ranging from 16-113 TTCF-specific B cells per 106 CD19+ B cells (Figure 3C-D). Therefore, the average number of TTCF-specific B cells expected per 106 PBMCs would range from 0.52-3.64 cells depending on total B-cell frequencies. The control tetramer labeled significantly fewer cells, ranging from 0.0005% to 0.008% of class-switched memory B cells (corresponding to 1-10 cells per 106 CD19+ B cells). Binding by this control tetramer may be nonspecific or represent BCR cross-reactivity to a component of the tetramer.
Detection of tetanus toxoid specific memory B cells. (A) Gating scheme for detection of TTCF tetramer-positive B cells among class-switched memory B cells. (B) Identification of TTCF tetramer-positive class-switched memory B cells from 4 donors. The elapsed time since the last tetanus boost is indicated (top). Labeling with a control tetramer (membrane-proximal domain of CD80) is shown for each donor (bottom). Frequencies represent tetramer-positive cells among CD19+ CD27+ IgM− B cells. (C) Scatter plot of tetramer+ cells detected per 1 × 106 B cells, with the mean indicated by a solid vertical line. The mean frequency was 52.0 cells per 1 × 106 B cells with the TTCF tetramer, and 4.3 cells per 1 × 106 B cells with the control tetramer. (D) Scatter plot showing percentage of tetramer-positive cells within the memory B-cell population. The mean frequency of TTCF tetramer-positive was 0.0447% (0.0032% for the control tetramer).
Detection of tetanus toxoid specific memory B cells. (A) Gating scheme for detection of TTCF tetramer-positive B cells among class-switched memory B cells. (B) Identification of TTCF tetramer-positive class-switched memory B cells from 4 donors. The elapsed time since the last tetanus boost is indicated (top). Labeling with a control tetramer (membrane-proximal domain of CD80) is shown for each donor (bottom). Frequencies represent tetramer-positive cells among CD19+ CD27+ IgM− B cells. (C) Scatter plot of tetramer+ cells detected per 1 × 106 B cells, with the mean indicated by a solid vertical line. The mean frequency was 52.0 cells per 1 × 106 B cells with the TTCF tetramer, and 4.3 cells per 1 × 106 B cells with the control tetramer. (D) Scatter plot showing percentage of tetramer-positive cells within the memory B-cell population. The mean frequency of TTCF tetramer-positive was 0.0447% (0.0032% for the control tetramer).
Tetramers are more sensitive than monomers
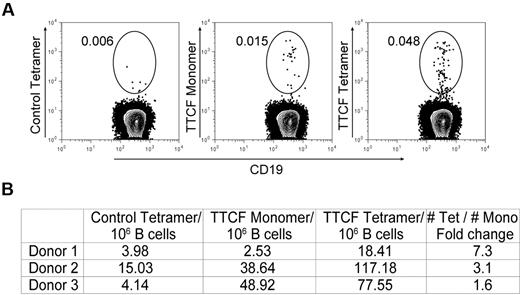
We also examined whether multimerization of TTCF is important for the isolation of rare memory B cells by using an approach that allowed direct side-by-side comparison of tetrameric and monomeric antigen. TTCF was fluorescently labeled with Alexa-488 and then either used in this monomeric form or converted to a tetramer by the use of unlabeled streptavidin. Enriched B cells were incubated with tetrameric or monomeric TTCF-Alexa-488 at the same concentration. The control protein (CD80 membrane proximal domain) was labeled in the same manner and also used as a tetramer. TTCF monomer labeled some memory B cells, but the frequencies identified with the tetramer were substantially larger (1.6- to 7.3-fold) when we used cells from 3 donors (Figure 4). In 1 of the 3 donors, TTCF-specific memory B cells could only be detected above background with the tetramer but not with the monomer (Figure 4B, donor 1). These data show that labeling with an antigen monomer does not capture the entire repertoire of memory B cells specific for TTCF.
Comparison of monomeric and tetrameric antigen for identification of memory B cells. (A) Mono-biotinylated TTCF or CD80 antigens were directly labeled with the Alexa-488 fluorophore; tetramers were generated with unlabeled streptavidin. Enriched B cells from each donor were split into 3 fractions and stained with control CD80 tetramer, TTCF monomer, or TTCF tetramer at the same total antigen concentration of 0.125 μg/mL. Fluorescence-activated cell sorter plots depict CD19+ CD27+ IgM− class-switched memory B cells; numbers adjacent to the gate represent the percentage of the parental gate. (B) Frequencies of tetramer+ memory B cells detected in 3 different donors. Numbers are calculated as tetramer-positive cells per 1 × 106 CD19+ memory B cells.
Comparison of monomeric and tetrameric antigen for identification of memory B cells. (A) Mono-biotinylated TTCF or CD80 antigens were directly labeled with the Alexa-488 fluorophore; tetramers were generated with unlabeled streptavidin. Enriched B cells from each donor were split into 3 fractions and stained with control CD80 tetramer, TTCF monomer, or TTCF tetramer at the same total antigen concentration of 0.125 μg/mL. Fluorescence-activated cell sorter plots depict CD19+ CD27+ IgM− class-switched memory B cells; numbers adjacent to the gate represent the percentage of the parental gate. (B) Frequencies of tetramer+ memory B cells detected in 3 different donors. Numbers are calculated as tetramer-positive cells per 1 × 106 CD19+ memory B cells.
Sequence analysis of TTCF tetramer-positive memory B cells
To analyze the BCR sequence diversity among TTCF-positive cells, we single-cell sorted 20 memory cells from donor 1 (Table 2), gating cells that were at least 1 log brighter than the negative population. BCR mRNA amplification and nested PCR were performed in the same manner as described previously for plasmablasts. PCR amplification yielded 9 of 20 heavy chains and 11 of 20 light chains (overall PCR efficiency of 50%). The reduced PCR efficiency compared with the plasmablast population (89%) may be attributable to lower quantities of BCR mRNA in resting B cells compared with recently activated plasmablasts.22,23 Two of the memory B-cell clones (M1 and M2) had identical VH-D-JH and Vκ-Jκ gene rearrangements in both heavy and light chains, respectively (Table 2). At the nucleotide level, they shared 99% sequence homology as well as all silent and nonsilent mutations in the VH and Vκ gene segments (Figure 5), which is indicative of close clonal relatedness.
BCR variable-domain analysis of tetramer-positive memory B cells isolated from donor 1
| ID . | Heavy chain . | Kappa light chain . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| V-D-J junction . | V . | D . | J . | V-Mut nt/aa* . | CDRs† . | V-J junction . | V . | J . | V-Mut nt/aa* . | CDRs† . | |
| M1‡ | CARVRAVVRMQHYYYGMDLW | V1-2 | D3-10 | J6 | 18/13 | 8.8.18 | CMQALQTPPTF | V2-28 or V2D-28 | J1 | 5/3 | 11.3.9 |
| M2‡ | CARVRAVVRTQHYYYGMDLW | V1-2 | D3-10 | J6 | 18/13 | 8.8.18 | CMQALQTPPTF | V2-28 or V2D-28 | J1 | 5/3 | 11.3.9 |
| M3 | CAGGGYTNPLIIG | V3-23 | D4-4 | J4 | 42/22 | 8.8.11 | CQQYANSPRTF | V3-20 | J1 | 14/6 | 7.3.9 |
| M4 | CARDGAPYTRKDRFDPW | V3-33 | D1-7 | J5 | 12/9 | 8.8.15 | CQQYNTYSPWTF | V1-5 | J1 | 5/2 | 6.3.10 |
| M5 | CARSGGSASGSFYPGALYYNGMDVW | V4-4 | D3-10 | J6 | 25/11 | 8.7.23 | CQQSYSATWTF | V1-39 or V1D-39 | J1 | 23/11 | 6.3.9 |
| M6 | CATEPGLHCGPDCPDAFEIW | V4-31 | D2-21 | J3 | 20/13 | 10.7.18 | CQHHHTF | V1-9 | J2 | 17/10 | 6.3.5 |
| M7 | CARGFMVTLGGVIEHNGLHIW | V4-31 | D3-16 | J3 | 21/13 | 10.7.19 | CQQRSNWPPVPTF | V3-11 | J3 | 7/6 | 6.3.11 |
| M8 | CGRLWGRSYSYYSGSYYENFDYW | V5-51 | D3-10 | J4 | 21/14 | 8.8.21 | CHQYGSLPPTF | V3-20 | J1 | 35/20 | 7.3.9 |
| M9 | CARPSNYDFWSGYYETGYAFDMW | V5-51 | D3-3 | J3 | 8/4 | 8.8.21 | CQQYGSLPRTF | V3-20 | J1 | 9/3 | 7.3.9 |
| M10 | CQQSHNTPLTF | V1-39 or V1D-39 | J4 | 17/11 | 6.3.9 | ||||||
| M11 | CQQSFSTLYTF | V1-39 or V1D-39 | J2 | 6/3 | 6.3.9 | ||||||
| ID . | Heavy chain . | Kappa light chain . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| V-D-J junction . | V . | D . | J . | V-Mut nt/aa* . | CDRs† . | V-J junction . | V . | J . | V-Mut nt/aa* . | CDRs† . | |
| M1‡ | CARVRAVVRMQHYYYGMDLW | V1-2 | D3-10 | J6 | 18/13 | 8.8.18 | CMQALQTPPTF | V2-28 or V2D-28 | J1 | 5/3 | 11.3.9 |
| M2‡ | CARVRAVVRTQHYYYGMDLW | V1-2 | D3-10 | J6 | 18/13 | 8.8.18 | CMQALQTPPTF | V2-28 or V2D-28 | J1 | 5/3 | 11.3.9 |
| M3 | CAGGGYTNPLIIG | V3-23 | D4-4 | J4 | 42/22 | 8.8.11 | CQQYANSPRTF | V3-20 | J1 | 14/6 | 7.3.9 |
| M4 | CARDGAPYTRKDRFDPW | V3-33 | D1-7 | J5 | 12/9 | 8.8.15 | CQQYNTYSPWTF | V1-5 | J1 | 5/2 | 6.3.10 |
| M5 | CARSGGSASGSFYPGALYYNGMDVW | V4-4 | D3-10 | J6 | 25/11 | 8.7.23 | CQQSYSATWTF | V1-39 or V1D-39 | J1 | 23/11 | 6.3.9 |
| M6 | CATEPGLHCGPDCPDAFEIW | V4-31 | D2-21 | J3 | 20/13 | 10.7.18 | CQHHHTF | V1-9 | J2 | 17/10 | 6.3.5 |
| M7 | CARGFMVTLGGVIEHNGLHIW | V4-31 | D3-16 | J3 | 21/13 | 10.7.19 | CQQRSNWPPVPTF | V3-11 | J3 | 7/6 | 6.3.11 |
| M8 | CGRLWGRSYSYYSGSYYENFDYW | V5-51 | D3-10 | J4 | 21/14 | 8.8.21 | CHQYGSLPPTF | V3-20 | J1 | 35/20 | 7.3.9 |
| M9 | CARPSNYDFWSGYYETGYAFDMW | V5-51 | D3-3 | J3 | 8/4 | 8.8.21 | CQQYGSLPRTF | V3-20 | J1 | 9/3 | 7.3.9 |
| M10 | CQQSHNTPLTF | V1-39 or V1D-39 | J4 | 17/11 | 6.3.9 | ||||||
| M11 | CQQSFSTLYTF | V1-39 or V1D-39 | J2 | 6/3 | 6.3.9 | ||||||
Bolded sequence pair represents clonally related B cells.
CDR indicates Ca2+-dependent regulator.
Number of mutations detected in V-gene segment (nucleotides/amino acids).
CDR1, 2, and 3 lengths in amino acids.
Identical VH-D-JH and Vκ-Jκ gene rearrangements in both heavy and light chains, respectively.
Clonally related B cells detected in both memory and plasmablast populations. (A) Alignment of CDR3 protein sequences as well as VH-D-JH and Vκ-Jκ gene segment use of clonally related memory B cells and plasmablasts. Amino acid differences are shaded in gray. (B) Variable gene segments were aligned at the nucleotide level for clonally related memory B cells and plasmablasts. Solid vertical lines represent coding/replacement mutations, and dashed lines silent mutations per codon, compared with the most homologous germline segment. Asterisks denote coding mutations that occurred at the same codon positions but resulted in different amino acids between the aligned sequences.
Clonally related B cells detected in both memory and plasmablast populations. (A) Alignment of CDR3 protein sequences as well as VH-D-JH and Vκ-Jκ gene segment use of clonally related memory B cells and plasmablasts. Amino acid differences are shaded in gray. (B) Variable gene segments were aligned at the nucleotide level for clonally related memory B cells and plasmablasts. Solid vertical lines represent coding/replacement mutations, and dashed lines silent mutations per codon, compared with the most homologous germline segment. Asterisks denote coding mutations that occurred at the same codon positions but resulted in different amino acids between the aligned sequences.
In addition, 3 of the memory B cells were clonally related to plasmablasts isolated 15 months previously from the same donor (Figure 5). Comparison of variable gene segments of memory cells to plasmablasts and their closest matching germline sequences identified a pair of cells (M6 and PB22) with high sequence homology and identical germline mutations in the VH and Vκ gene segments, except for one amino acid difference in the heavy chains and 2 differences among the light chains (Figure 5B). The remaining cell pairs (M5 and PB19; M7 and PB24), showed a larger number of amino acid differences, indicating distant clonal relatedness to a parental clone (Figure 5).
Generation of fully human antibodies
To conclusively demonstrate the specificity of TTCF tetramer staining, we generated 5 fully human recombinant antibodies that were randomly chosen from 2 plasmablasts and 3 memory B cells from 3 different donors (Table 3). We joined the constant regions of IgG1 heavy and κ chains to the isolated variable segments via overlap PCR. Antibodies were then expressed in a transient, serum-free mammalian expression system (CHO-S cells) over the course of 6-8 days and were purified by the use of protein G and gel filtration chromatography.
TTCF antibodies generated from plasmablasts and memory B cells
| B-cell subset . | TTCF Ab ID (donor) . | KD, nM . | Heavy chain . | Kappa light chain . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V-D-J junction . | V . | D . | J . | V-Mut nt/aa* . | CDRs† . | V-J junction . | V . | J . | V-Mut nt/aa* . | CDRs† . | |||
| Plasmablast | Ab 1 (D1) | 2.2 | CVKREYYNDNSGYHVWGDYW | V3-23 | D3-22 | J5 | 32/18 | 8.8.18 | CQEYDNWPRFTF | V3D-15 | J3 | 14/7 | 6.3.10 |
| Ab 2 (D1) | 0.32 | CGRSVIGTVDNW | V3-11 | D6-19 | J4 | 34/18 | 8.8.10 | CQQSKSWPQLTF | V3-15 | J4 | 14/9 | 6.3.10 | |
| Memory | Ab 3 (D4) | 0.38 | CAKDTGSSELLLDNW | V3-30 | D1-26 | J4 | 23/14 | 8.8.13 | CQQNSIWPPLTF | V3-11 | J4 | 15/10 | 6.3.10 |
| Ab 4 (D2) | 0.23 | CARDPTTVTGNSLLDMW | V1-18 | D4-17 | J3 | 28/14 | 8.8.15 | CQQSYSTPRVTF | V1-39 or V1D-39 | J3 | 25/12 | 6.3.10 | |
| Ab 5 (D1) | 1.4 | CATEPGLHCGPDCPDAFEIW | V4-31 | D2-21 | J3 | 20/13 | 10.7.18 | CQHHHTF | V1-9 | J2 | 17/10 | 6.3.5 | |
| B-cell subset . | TTCF Ab ID (donor) . | KD, nM . | Heavy chain . | Kappa light chain . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V-D-J junction . | V . | D . | J . | V-Mut nt/aa* . | CDRs† . | V-J junction . | V . | J . | V-Mut nt/aa* . | CDRs† . | |||
| Plasmablast | Ab 1 (D1) | 2.2 | CVKREYYNDNSGYHVWGDYW | V3-23 | D3-22 | J5 | 32/18 | 8.8.18 | CQEYDNWPRFTF | V3D-15 | J3 | 14/7 | 6.3.10 |
| Ab 2 (D1) | 0.32 | CGRSVIGTVDNW | V3-11 | D6-19 | J4 | 34/18 | 8.8.10 | CQQSKSWPQLTF | V3-15 | J4 | 14/9 | 6.3.10 | |
| Memory | Ab 3 (D4) | 0.38 | CAKDTGSSELLLDNW | V3-30 | D1-26 | J4 | 23/14 | 8.8.13 | CQQNSIWPPLTF | V3-11 | J4 | 15/10 | 6.3.10 |
| Ab 4 (D2) | 0.23 | CARDPTTVTGNSLLDMW | V1-18 | D4-17 | J3 | 28/14 | 8.8.15 | CQQSYSTPRVTF | V1-39 or V1D-39 | J3 | 25/12 | 6.3.10 | |
| Ab 5 (D1) | 1.4 | CATEPGLHCGPDCPDAFEIW | V4-31 | D2-21 | J3 | 20/13 | 10.7.18 | CQHHHTF | V1-9 | J2 | 17/10 | 6.3.5 | |
CDR indicates complementary determining region.
Number of mutations detected in V-gene segment (nucleotides/amino acids).
CDR1, 2, and 3 lengths in amino acids.
The binding specificity and affinity of these antibodies was measured in a 96-well–based saturation binding assay. Nonbiotinylated TTCF was labeled with europium and unbound europium was removed by gel filtration chromatography. TTCF antibodies were captured overnight in 96-well plates, and after a blocking step, wells were incubated for 2 hours at 37°C with TTCF-europium at concentrations ranging from 100nM to 4pM. Unbound TTCF was quickly washed away, europium was released from the bottom of the wells by incubation with an acidic chelating solution, and the fluorescence signal was measured on a plate reader. As a negative control, wells were coated with a control antibody (clone 8.18.C5) generated with identical heavy- and light-chain constant regions in the same CHO-S expression system. As seen in Figure 6A, both antibodies generated from plasmablasts showed high binding affinities to TTCF antigen, with a KD of 2.2nM (antibody 1) and 323pM (antibody 2). The 3 antibodies generated from memory B cells also exhibited very high binding affinities, with a KD of 382pM, 228pM, and 1.4nM for antibodies 3, 4, and 5, respectively (Figure 6B). The binding of all generated recombinant antibodies to TTCF demonstrates the specificity of the technique reported here.
High-affinity binding of TTCF by antibodies generated from plasmablasts and memory B cells. Saturation binding experiments were performed to determine the affinities of recombinant antibodies. TTCF antigen was labeled with europium, which emits a strong fluorescent signal at 615 nm on incubation with a chelating reagent. Antibodies were immobilized in a 96-well plate and incubated with TTCF-europium (100nM to 4pM) for 2 hours at 37°C. Fluorescence counts at 615 nm were recorded and KD calculated by the use of nonlinear regression analysis. A control antibody (clone 8.18.C5) that was also produced in CHO-S cells was included in all experiments. (A) Recombinant TTCF Abs 1 and 2 were generated from TTCF tetramer-positive plasmablasts (donor 1). (B) TTCF Abs 3, 4, and 5 originated from TTCF tetramer-positive memory B cells of 3 different donors.
High-affinity binding of TTCF by antibodies generated from plasmablasts and memory B cells. Saturation binding experiments were performed to determine the affinities of recombinant antibodies. TTCF antigen was labeled with europium, which emits a strong fluorescent signal at 615 nm on incubation with a chelating reagent. Antibodies were immobilized in a 96-well plate and incubated with TTCF-europium (100nM to 4pM) for 2 hours at 37°C. Fluorescence counts at 615 nm were recorded and KD calculated by the use of nonlinear regression analysis. A control antibody (clone 8.18.C5) that was also produced in CHO-S cells was included in all experiments. (A) Recombinant TTCF Abs 1 and 2 were generated from TTCF tetramer-positive plasmablasts (donor 1). (B) TTCF Abs 3, 4, and 5 originated from TTCF tetramer-positive memory B cells of 3 different donors.
Discussion
The goal of this project was to develop an approach for direct ex vivo isolation of human memory B cells with defined antigen specificity. Such cells are present at very low frequencies in peripheral blood, and their isolation by flow cytometry thus requires a ligand for the BCR that brightly labels these cells on the basis of their antigen specificity. We reasoned that tetrameric forms of antigens could offer several distinct advantages compared with antigen monomers. Bivalent antibodies bind with significant greater avidity to their target antigen than monomeric Fab fragments, and this increase in avidity is even greater for pentameric IgM molecules with their 10 antigen binding sites.24,25 This principle has been exploited to develop peptide-major histocompatibility complex tetramers, which label T cells on the basis of the specificity of their TCR.26 Another significant advantage is that tetrameric antigen complexes can be generated with brightly labeled streptavidin molecules; this obviates the need to chemically modify the antigen with a fluorophore, a procedure that can destroy epitopes. We show that such antigen tetramers enable sensitive identification and isolation of memory B cells specific for defined antigens by flow cytometry, despite their low frequency. This approach thereby enables efficient isolation of these rare cells for the production of fully human monoclonal antibodies. When antibodies with very high affinities are desired, this sorting procedure can focus on the brightest cells in the gate.
By using tetanus toxoid as a model antigen, we demonstrated that antigen tetramers enabled sensitive detection of memory B cells on the basis of the antigen specificity of their BCR, even though these cells were very rare in peripheral blood. Class-switched memory B cells specific for TTCF were brightly labeled by the appropriate antigen tetramer, whereas background labeling with a control tetramer was consistently low. The specificity of this technique was unambiguously demonstrated by the construction of 5 recombinant antibodies from 3 different donors, all of which bound TTCF with high affinity. The technique also enabled sequence analysis of a panel of tetramer-positive plasmablasts and memory B cells isolated more than a year apart. Consistent with previous reports, tetanus-specific plasmablasts induced by booster vaccination were oligoclonally expanded and had highly mutated BCR variable domains, indicative of affinity maturation.27 Interestingly, several memory B cells were clonally related to plasmablasts isolated previously from the same donor. This observation is consistent with previous studies showing clonal relatedness among plasmablasts and memory B-cell populations specific for the same antigen.28 These data demonstrate that antigen tetramers enable sensitive detection of memory B cells long after clearance of the antigen.
We anticipate that this technique can be widely applied for the study of human memory B cells, as long as the antigen or antigen domain can be expressed in a soluble form. B cells have previously been stained with labeled antigen,13,29 but a direct comparison of monomeric versus tetrameric labeled antigen showed that a substantial fraction of memory B cells can be missed with antigen monomers. In contrast, we observed very bright labeling with antigen tetramers that increased both the sensitivity and the specificity of the technique. The increased sensitivity of this approach may be valuable when limited numbers of cells from clinical specimens are available.
We envision several applications of this technique. First, this approach can be used for in depth characterization of human memory B cells after infection or vaccination in terms of phenotype, intracellular protein expression, or gene expression. The mRNA amplification step may be adapted to other target mRNAs and generate sufficient cDNA template for investigation of cytokine, chemokine, or transcription factor gene expression. Sequencing of BCR light and heavy chains can be used to track clonal evolution of memory B cells in humans over time and related to the affinity of reconstructed antibodies. Second, this approach will enable the production of fully human antibodies from memory B cells rather than plasmablasts, which only transiently circulate in the blood after infection or vaccination. Of particular interest will be the isolation of memory B cells from subjects who have survived infection by a major pathogen, including an emerging infectious disease, so that neutralizing antibodies for passive immunotherapy can be generated. Third, the technique may enable the isolation of tumor-specific B cells from patients who responded favorably to cancer vaccines or other therapies that amplify a tumor-directed immune response. In such patients, the frequency of B cells of interest and the avidity of their BCR may be considerably lower than those studied here because of immune tolerance to self-antigens. The gain in the brightness of labeling resulting from the use of tetrameric antigen may be essential in such studies to enable isolation of the very rare cells of interest.
B cell antigen tetramers could also enhance the sensitivity and specificity of existing techniques for the generation of human antibodies.19,30,31 For example, this method could be combined with approaches targeting plasmablasts by enabling sorting of the cells with the greatest avidity for the antigen of interest. It could also be incorporated into a method in which peripheral blood B cells are immortalized with Epstein-Barr virus because immortalized B cells with the desired specificity could be isolated with a relevant antigen tetramer, rather than by limiting dilution cloning.32 The methodology reported in this article may thereby enhance our understanding of human memory B cells and facilitate the development of human antibody therapeutics.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Suzan Lazo-Kallanian for outstanding technical assistance, Dr Shiv Pillai for reading the manuscript, and Drs Kevin O'Connor and David A. Hafler for advice on the CHO-S expression system.
This study was supported by a grant from the Melanoma Research Alliance (to G.D. and K.W.), a grant from the Research Foundation for the Treatment of Ovarian Cancer (to G.D.), and the National Institutes of Health (PO1 AI045757 to K.W.). B.F. is a PhD candidate at Heidelberg University, Germany and this work is submitted in partial fulfillment of the requirement for the PhD.
National Institutes of Health
Authorship
Contribution: B.F. designed and performed research, analyzed data, and wrote the paper; K.F.M. performed research and edited manuscript; G.D. designed research and edited manuscript; and K.W. conceived and supervised the study, designed research, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kai Wucherpfennig, Dana-Farber Cancer Institute, Dana 1410, 450 Brookline Ave, Boston, MA 02215-5450; e-mail: kai_wucherpfennig@dfci.harvard.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal