Abstract
Genomewide association meta-analysis studies have identified > 100 independent genetic loci associated with blood cell indices, including volume and count of platelets and erythrocytes. Although several of these loci encode known regulators of hematopoiesis, the mechanism by which most sequence variants exert their effect on blood cell formation remains elusive. An example is the Rho guanine nucleotide exchange factor, ARHGEF3, which was previously implicated by genomewide association meta-analysis studies in bone cell biology. Here, we report on the unexpected role of ARHGEF3 in regulation of iron uptake and erythroid cell maturation. Although early erythroid differentiation progressed normally, silencing of arhgef3 in Danio rerio resulted in microcytic and hypochromic anemia. This was rescued by intracellular supplementation of iron, showing that arhgef3-depleted erythroid cells are fully capable of hemoglobinization. Disruption of the arhgef3 target, RhoA, also produced severe anemia, which was, again, corrected by iron injection. Moreover, silencing of ARHGEF3 in erythromyeloblastoid cells K562 showed that the uptake of transferrin was severely impaired. Taken together, this is the first study to provide evidence for ARHGEF3 being a regulator of transferrin uptake in erythroid cells, through activation of RHOA.
Introduction
The formation of blood cells from pluripotent hematopoietic stem cells has been used extensively to systematically catalog all key regulators in stem cell biology. So far only a fraction have been identified by forward and reverse genetics studies in model organisms and by studies of the genetic and molecular basis of hematologic cancers.1 In an individual person, the mass (number of cells times their volume) of both erythrocytes and platelets are highly heritable and tightly regulated within narrow ranges,2,3 but there is a wide variation of the number and volume of both cells in the population. This has provided the foundation for recent successful genomewide association studies (GWASs) as a novel approach to discover important regulators of stem cell fate determination, blood cell mass, and their half-life.4-7 We reported the results of our initial GWAS meta-analysis for platelets and erythrocytes4,5 and complemented this with a recently completed meta-analysis in ∼ 60 000 persons, leading to a catalog with > 100 loci that are associated with the 2 aforementioned traits of both cells (Gieger et al, manuscript submitted, August 2011). Interestingly, and in sharp contrast with the results of GWASs in common diseases, > 80% of associated single nucleotide polymorphisms (SNPs) are localized in or within a 10-Kb window of genes, providing a sound argument to infer biologically relevant candidate genes (Gieger et al, manuscript submitted, August 2011). For two-thirds of association signals the mechanisms by which the corresponding genes exert their effect on hematopoiesis are not understood. In a first step to address this lack of knowledge we selected 6 genes and determined their effect on hematopoiesis in zebrafish embryos by morpholino (MO) knockdown (Gieger et al, manuscript submitted, August 2011). For all but one we observed a variable decrease in the number of mature erythrocytes and a complete abrogation of thrombocyte formation (Gieger et al, manuscript submitted, August 2011). Silencing of arhgef3 resulted in the most profound phenotype, prompting further analysis, which is the subject of this study.
In humans ARHGEF3 belongs to the family of Rho guanine nucleotide exchange factors (RhoGEFs), which activate Rho GTPases by catalyzing the exchange of GDP for GTP.8 Of the ∼ 70 known members of the RhoGEF family, a few have been extensively studied,9 and several have been reported to have a function in normal and malignant hematopoiesis. For instance, mutations in DOCK2,10 DOCK8,11 ARHGEF6,12 and VAV 1 to 313 were associated with impaired lymphocyte number or function and aberrant regulation of several GEF genes by somatic events are causative of leukemias.14 Finally, knockdown of Lsc in mice has been implicated in neutrophil chemotaxis and leukocyte homeostasis.15 Considering the main regulatory function of GEFs in controlling Rho proteins, this represents a major gap in the present understanding of this signaling pathway.9 ARHGEF3 is to date a unique example of a RhoGEF with selectivity within the Rho family, because it has been shown to activate RhoA and RhoB but not RhoC.16 In humans, its transcript is present widely, in the brain, skeletal muscle, heart, kidney,16 and all main blood cell types, including megakaryocytes and erythroblasts.17 It has been shown by a GWAS that SNP rs7646054 in ARHGEF3 locus confers risk for postmenopausal osteoporosis, but the cellular mechanism of this association has not yet been elucidated.18 SNP rs12485738 in ARHGEF3, which is not in linkage disequilibrium with the former SNP, exerts an effect on hematopoiesis. Here, we provide functional evidence on the importance of this GEF in hematopoiesis with the use of zebrafish and the K562 human erythromyeloblastoid leukemia cells. Our detailed studies clearly show a quintessential role for ARHGEF3 in vasculogenesis and erythropoiesis. The role in the latter is in part explained by a major effect, most probably by RhoA, on transferrin (Tf) uptake and iron homeostasis.
Methods
Zebrafish maintenance and strains
General maintenance, collection, and staging of the wild-type (Tübingen Long Fin) and transgenic lines (gata1:DsRed,19 gata1:EGFP,20 fli1a:EGFP21 ) were performed as previously described.22,23 To prevent melanization, embryos were treated with 0.002% phenylthiourea from Sigma-Aldrich at 24 hours post fertilization (hpf).
Whole-mount in situ hybridization
Whole-mount RNA in situ hybridization for arhgef3, scl, gata2, gata1, alas2, hbbe3, l-plastin, and mpx was performed with digoxigenin-labeled antisense riboprobes as previously described.24 Photomicrographs were taken with a Zeiss camera AxioCam HRC attached to a LeicaMZ16 FA dissecting microscope with Planapo objective 1.0× (Leica Microsystems) using the AxioVision 4.5 software (Carl Zeiss).
Embryo injection experiments
MO antisense oligonucleotides were obtained from GeneTools LLC. The oligos were resuspended in sterile water, and ∼ 1 nL was injected in zebrafish embryos, at the 1- to 2-cell stage. MO names and sequences are listed in supplemental Figure 4A (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Full-length zebrafish arhgef3 cDNA was obtained by PCR with the use of gene-specific primers (forward primer, 5′-GCCAGGATCGATATGGTTGCGAAGGACTAC-3′; reverse primer, 5′-AAGCTAGAATTCGCTCTCTCACAGGGCTGAC-3′) with added ClaI/EcoRI restriction enzyme sites (underlined) for cloning into pCS2-EGFP vector. Arhgef3-EGFP sense mRNA was synthesized with mMESSAGE mMACHINE kit (Ambion), according to the manufacturer's protocol and injected (250 pg) immediately after the injection of arhgef3 MO or control MO. For the iron rescue experiments, iron-dextran (2.5 ng; Sigma-Aldrich) was coinjected with MO into 1-cell stage embryos as previously described.25
O-Dianisidine staining
Staining of hemoglobin by o-Dianisidine was performed as previously described.26 To obtain blood cells, live embryos were anesthetized with a solution of PBS containing 0.02% tricaine. Blood collected by cardiac puncture was immediately smeared onto poly-L-lysine–coated glass slide and stained with o-Dianisidine solution for 5 minutes. Photomicrographs of blood smears were taken with a Zeiss camera AxioCam HRC attached to a Zeiss Axio Imager M1 compound microscope (40×/0.75 NA dry Plan Neofluar objective) with the use of the AxioVision 4.5 software (Carl Zeiss). To determine cell size and hemoglobin content, images of blood smears stained with o-Dianisidine were analyzed with ImageJ software (National Institutes of Health). Cell area was used as a measure of cell size. To quantify hemoglobin content of the cells, the images were first converted to grayscale, and cell mean gray value (MGV) was subsequently measured for individual cells. Gray values range from 0 for black to 255 for white; therefore, cells more abundant in hemoglobin had lower MGVs, whereas paler, more hypochromic cells had higher MGVs. Obtained values were normalized against the respective control value.
TUNEL assay
TUNEL assay was performed as previously described.26
Confocal microscopy
For in vivo imaging, embryos were anesthetized with 0.02% tricaine. Images of live embryos with 0.02% tricaine were captured with the use of a Leica TCS SP5 confocal microscope with the Leica LAS AF software (Leica Microsystems). Images of TUNEL-stained embryos were captured using a 10×/0.3 NA dry objective.
Reverse Transcription-PCR
RT-PCR was used to determine the efficiency of the knockdown mediated by splice-blocking MOs. Total RNA was isolated from control- and MO-injected embryos at 30 hpf with the use of the RNeasy Mini Kit (QIAGEN) and subjected to cDNA synthesis with the use of Supersript III Reverse Transcriptase (Invitrogen), according to the manufacturer's protocol. PCR reactions were performed with gene-specific primers (listed in supplemental Figure 4A) and KOD Hot Start DNA Polymerase (Novagen). Resulting PCR products were analyzed on 1% agarose gel.
Lentiviral construction and production
Plasmids were constructed as described with some modifications because the shRNAs were cloned 3′ of the eGFP gene instead of 5′ to improve knockdown efficiency.27,28 Briefly, synthesized 97-nucleotide PCR templates (Invitrogen) containing gene-specific RNAi sequences targeting human ARHGEF3 mRNA (5′-CAGCAGTGGCTTAACTGTA-3′) or a mismatch (mm)RNAi sequence against human WDR66 (5′-CAGAGGTACGTTCTCAAAGC-3′) as a negative control were each flanked by artificial sequences containing various restriction sites.27,28 The PCR products were cloned into peGFP-N1 (Clontech) and confirmed by sequencing. A second hairpin was cloned in a different cloning site of the same mono hairpin peGFP-N1 vector, and the double hairpin shRNA construct was further cloned into SpeI and SmaI sites 3′ of the GFP gene of the lentiviral pCMV-GFP vector to generate the lenti-GFP/RNAi-ARHGEF3 and lenti-GFP/mmRNAi-WDR66 vectors. Lentiviral vectors were produced as described,29 and concentrated vector particles (1 × 107 pg/mL of normalized p24 antigen) were stored at −80°C.
Lentiviral transduction and transferrin uptake and binding
Human K562 cells (1.25 × 105) were seeded in 6-well plates in RPMI 1640 medium supplemented with 10% FBS and transduced with the lentiviral vectors with a MOI of 10 for 24 hours. K562 cells were washed and cultured for 5 days in fresh medium. Transduced cells were washed and then incubated for 20 minutes in serum-free medium (SFM) to enhance the uptake of fluorescently labeled transferrin (Alexa Flour 633 Tf [Tf-633]; Invitrogen). For RhoA inhibition experiments, cells were incubated with or without 1 μg/mL cell-permeable C3 transferase (Cytoskeleton; Tebu-Bio) for 2 hours in SFM before Tf uptake measurements. The maximum Tf intracellular uptake was quantified after the addition of 5 μg/mL Tf-633 for 30 minutes at 37°C. Cells were immediately fixed in 500 μL of CellFIX (BD Biosciences), and images were acquired with a Leica TCS SP5 laser scanning confocal microscope (40×/1.25 NA oil objective. For measuring fluid-phase endocytosis 1 mg/mL Texas red dextran (molecular weight = 40 000; Invitrogen) was applied to transduced cells and incubated for 60 minutes at 37°C. Cells were washed and fixed in 500 μL of CellFIX, and mean fluorescence intensity (MFI) was measured by flow cytometry. The binding of Tf-633 to the cellular membrane before uptake was determined by flow cytometry after incubation of transduced cells with 20 μg/mL Tf-633 at 4°C for 30 minutes in ice-cold SFM. Cells were immediately fixed in 500 μL of CellFIX and analyzed on the FACSCalibur, and results were analyzed with FACSDiva software (BD Biosciences).
RhoA activity assay
RhoA activity was measured 5 days after transduction in K562 cells with and without the addition of 1 μg/mL C3 transferase with the use of a commercially available assay as described by the manufacturer (G-LISA RhoA activation; Cytoskeleton).
Cell sorting and real-time qRT-PCR
Whole transgenic embryos (gata1:EGFP or fli1a:EGFP) were dissociated at 22 hpf and processed for flow cytometry as previously described.30 Propidium iodide (Sigma-Aldrich) was added at a final concentration of 1 μg/mL to exclude dead cells. Flow cytometric analysis and sorting were based on propidium iodide exclusion and green fluorescent protein (GFP) fluorescence. Data analysis was performed with FlowJo flow cytometric analysis software (TreeStar). For the real-time quantitative RT-PCR (qRT-PCR) analysis, total RNA was extracted from the sorted gata1+ or fli1+ cells (3 × 105 and 6 × 105 cells, respectively) with the use of TRIzol reagent (Invitrogen), according to the manufacturer's instructions. First-strand cDNA was synthesized with Superscript III Reverse Transcriptase (Invitrogen), according to the manufacturer's protocol. Arhgef3 expression was measured with FAM-labeled Taqman probe (assay Dr03092365_mH; Applied Biosystems). Each sample was tested in triplicate, and β-actin was used as an endogenous control to normalize the samples. Transduced K562 cells were sorted on GFP by flow cytometry (FACSAria Sorter; BD Biosciences) to isolate GFP+ cells. Total RNA was extracted from 1 × 106 GFP+ K562 cells with the use of the RNeasy mini kit (QIAGEN). Total RNA (300 ng) was used for cDNA synthesis with M-MLV reverse transcriptase. ARHGEF3 expression was measured with a FAM-labeled Taqman probe (assay Hs00219609_m1) in combination with the VIC-labeled β-actin as housekeeping gene (both from Applied Biosystems). Reactions were analyzed with the ABI 7000 real-time PCR machine (Applied Biosystems).
Results
ARHGEF3 ablation induces severe cytopenia
Zebrafish arhgef3 gene encodes a protein of 506 amino acids and shares 76% identity with human ARHGEF3 (supplemental Figure 2A-B). Whole-mount arhgef3 in situ hybridization at 24 hpf showed expression in eyes, brain, and intermediate cell mass (ICM) of developing embryos (Figure 1A-B). By 48 hpf it was mostly expressed in the trunk, and no expression was detectable at 72 hpf (data not shown). To further increase the sensitivity of arhgef3 detection and to enrich for the cells of interest, with the use of FACS, we sorted GFP+ cells from Tg(gata1:EGFP) and Tg(fli1:EGFP) transgenic lines at 22 hpf (supplemental Figure 1). qPCR of reverse-transcribed RNA from sorted gata1- and fli1-positive cells confirmed the presence of the arhgef3 transcript (Figure 1C).
Arhgef3 is essential for late stages of erythroid differentiation. (A-B) Whole-mount in situ hybridization showing arhgef3 expression in Danio rerio embryos at 24 hpf. (A) Arhgef3 was expressed in eyes, brain, and ICM of the developing embryos. (B) Higher magnification view of the boxed area in (A) shows the expression of arhgef3 in the ICM (nc indicates notochord; pd, pronephric duct). (C) Quantitative real-time PCR analysis of arhgef3 expression in gata1+ and fli1+ cells sorted by FACS. Arhgef3 mRNA expression was normalized to β-actin expression. Results are presented as quotient of Ct values for arhgef3 and β-actin ± SD. (D-E) O-Dianisidine staining of hemoglobin was used to assess the number of mature erythrocytes in arhgef3 MO-injected embryos. At 30 hpf, hemoglobin-positive cells were present on the yolk and in circulation of control embryos (D and D″ black arrows); however, arhgef3 MO-injected embryos completely lacked such cells at the same time point (E,E″). At 48 (G,G′) and 72 (I,I′) hpf, the number of o-Dianisidine–positive cells in arhgef3 MO-injected embryos was severely reduced compared with time-matched controls (F,F′ and H,H′, respectively). (D′-E″) TUNEL assay in control- and arhgef3 MO-injected embryos at 24 hpf. There was no change in the number of apoptotic cells in the ICM in the arhgef3 MO-injected (E′) compared with control embryos (D′). Insets in (D′) and (E′) show 1 TUNEL-positive cell under higher magnification.
Arhgef3 is essential for late stages of erythroid differentiation. (A-B) Whole-mount in situ hybridization showing arhgef3 expression in Danio rerio embryos at 24 hpf. (A) Arhgef3 was expressed in eyes, brain, and ICM of the developing embryos. (B) Higher magnification view of the boxed area in (A) shows the expression of arhgef3 in the ICM (nc indicates notochord; pd, pronephric duct). (C) Quantitative real-time PCR analysis of arhgef3 expression in gata1+ and fli1+ cells sorted by FACS. Arhgef3 mRNA expression was normalized to β-actin expression. Results are presented as quotient of Ct values for arhgef3 and β-actin ± SD. (D-E) O-Dianisidine staining of hemoglobin was used to assess the number of mature erythrocytes in arhgef3 MO-injected embryos. At 30 hpf, hemoglobin-positive cells were present on the yolk and in circulation of control embryos (D and D″ black arrows); however, arhgef3 MO-injected embryos completely lacked such cells at the same time point (E,E″). At 48 (G,G′) and 72 (I,I′) hpf, the number of o-Dianisidine–positive cells in arhgef3 MO-injected embryos was severely reduced compared with time-matched controls (F,F′ and H,H′, respectively). (D′-E″) TUNEL assay in control- and arhgef3 MO-injected embryos at 24 hpf. There was no change in the number of apoptotic cells in the ICM in the arhgef3 MO-injected (E′) compared with control embryos (D′). Insets in (D′) and (E′) show 1 TUNEL-positive cell under higher magnification.
To better understand the in vivo requirement of this GEF we have transiently knocked down arhgef3 with the use of translation blocking (atg) MO antisense oligonucleotides. The ability of atg MOs to block translation of arhgef3 was validated by coinjecting arhgef3-EGFP reporter construct with arhgef3 atg MO into 1-cell stage embryos. This showed robust GFP expression in control MOs but not in arhgef3 atg MOs injected embryos (supplemental Figure 2C).
In contrast to the relatively normal morphologic development of the arhgef3 MO-injected embryos (supplemental Figure 2D), striking defects in hematopoiesis were observed. Whereas control MO-injected embryos showed vigorous circulation at 30 hpf, light microscopy showed no circulating blood cells in arhgef3 MO-injected embryos, although they displayed a normal heart beat. This blood-forming defect was confirmed by o-Dianisidine staining, which stains hemoglobin in mature erythrocytes (nctrl = 31, nMO = 42; Figure 1D,D″ and E,E″). As development proceeded, embryos continued to show reduced numbers of circulating erythrocytes (Figure 1F-F′, G-G′) with < 50% of wild-type cell numbers at 72 hpf (nctrl = 122, nMO = 103; Figure 1H-H′, I-I′). To understand the underlying process responsible for this cytopenia, we considered the possibility of enhanced apoptosis of progenitor cells in the ICM. Whole-mount TUNEL staining of arhgef3 MO-injected embryos 24 hpf was not indicative of such an increase compared with control MO-injected ones (nctrl = 32, nMO = 35; Figure 1D′,E′), suggesting that arhgef3 may be required for erythroid differentiation rather than survival of early progenitor cells.
Arhgef3 is essential in erythrocyte formation and vascular development but is expandable for primitive myelopoiesis
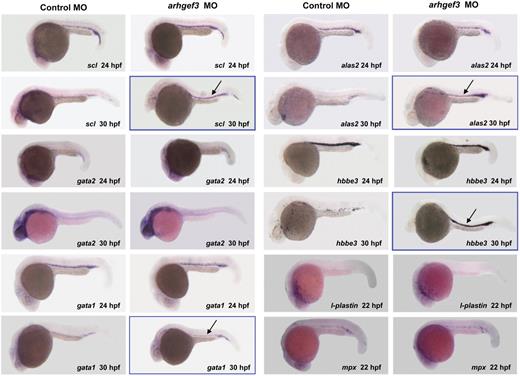
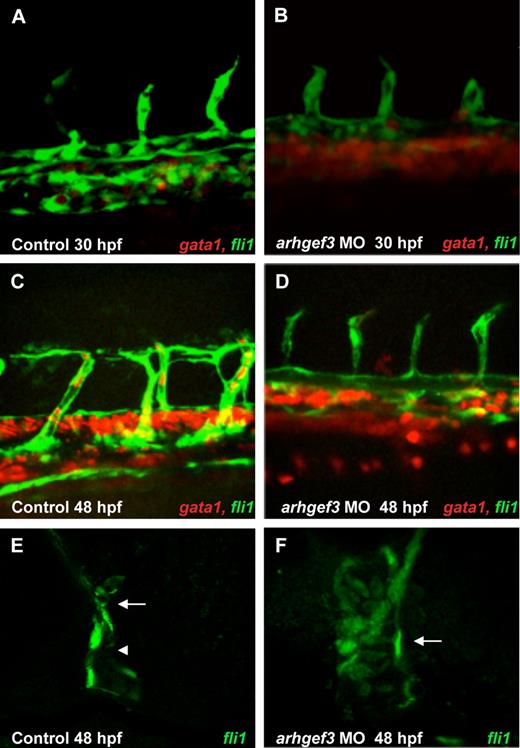
Hematopoietic progenitors undergo numerous fate-determining changes to reach the final functional stage, which concludes with the generation of thrombocytes and mature erythrocytes. We observed a complete ablation of the formation of thrombocytes (Gieger et al, manuscript submitted, August 2011) and erythrocytes (as reported here and in Gieger et al, manuscript submitted, August 2011) in arhgef3 MO-injected zebrafish embryos. To assess the exact stage at which arhgef3 is required in erythropoiesis and to determine its hierarchical position in the regulatory cascade, we performed in situ hybridization of several early and late hematopoietic markers, namely scl, gata2, gata1, alas2, and hbbe3 (Figure 2). All the tested markers appeared indistinguishable compared with control embryos at 24 hpf, suggesting that hematopoietic commitment and early erythroid differentiation progressed normally in arhgef3-depleted embryos (Figure 2). However, at 30 hpf we observed a dramatic increase in the number of scl-, gata1-, alas2-, and hbbe3-positive cells in the ICM (Figure 2) with the concomitant absence of o-Dianisidine–positive cells (Figure 1D-E,D″,E″). These results further show 2 findings. First, that arhgef3 functions at a late stage in erythroid differentiation. Second, that arhgef3 depletion transiently alters the ability of immature erythroid cells to enter into circulation, resulting in their accumulation in the ICM. We reasoned that the latter could indicate defects in vascular development. Thus, we examined the expression of the endothelial- and erythroid-specific transcription factors fli1a and gata1 in double transgenic embryos Tg(fli1a:EGFP) Tg(gata1:DsRed) at 30 and 48 hpf (Figure 3A-F). Although fli1a was expressed in arhgef3 MO-injected embryos at levels comparable with those of control MO-injected embryos, we noticed a poor segregation of artery and vein at 30 hpf (Figure 3A-B) and 48 hpf (Figure 3C-D) in arhgef3-depleted embryos. Detailed analysis of the double transgenic embryos showed abnormal accumulation of gata1-positive cells in the subaortic region of arhgef3 MO-injected embryos because of the lack of lumen formation at 30 hpf (Figure 3B). To better understand the observed phenotype, we next examined transverse sections of arhgef3 MO-injected and control embryos at 48 hpf (Figure 3E-F). Unlike in control embryos, in arhgef3 MO-injected ones we observed rare stretches of normal and physically separated axial vessels, alternating with regions in which the lumen formation was perturbed (Figure 3F). Altogether, our observations suggest that arhgef3 is essential for normal vascular development.
Whole-mount in situ hybridization analysis of hematopoietic markers in control- and arhgef3 MO-injected embryos. Expression analysis of the hematopoietic-specific markers scl, gata2, gata1, alas2, and hbbe3 in arhgef3 MO-injected embryos showed that all 5 genes were normally expressed at 24 hpf compared with control embryos. However, whereas in control embryos scl, gata1, alas2, and hbbe3 expression diminished in the ICM by 30 hpf and persisted only in its posterior region, arhgef3 MO-injected embryos exhibited a persistently strong expression of all 4 markers in the ICM (denoted by framed figures and black arrows). In situ hybridization for the late myelomonocytic markers at 22 hpf showed that there is no alteration in the expression of l-plastin or mpx in arhgef3 MO-injected embryos compared with the controls.
Whole-mount in situ hybridization analysis of hematopoietic markers in control- and arhgef3 MO-injected embryos. Expression analysis of the hematopoietic-specific markers scl, gata2, gata1, alas2, and hbbe3 in arhgef3 MO-injected embryos showed that all 5 genes were normally expressed at 24 hpf compared with control embryos. However, whereas in control embryos scl, gata1, alas2, and hbbe3 expression diminished in the ICM by 30 hpf and persisted only in its posterior region, arhgef3 MO-injected embryos exhibited a persistently strong expression of all 4 markers in the ICM (denoted by framed figures and black arrows). In situ hybridization for the late myelomonocytic markers at 22 hpf showed that there is no alteration in the expression of l-plastin or mpx in arhgef3 MO-injected embryos compared with the controls.
Expression analysis of endothelial marker fli1a and erythroid marker gata1 in double transgenic embryos Tg(fli1a:EGFP) Tg(gata1:DsRed) shows that vascular development is impaired in arhgef3-depleted embryos. At 30 and 48 hpf arhgef3 MO-injected embryos (B,D) showed a poor segregation of artery and vein compared with control embryos (A,C). Furthermore, arhgef3-depleted embryos exhibited an accumulation of gata1+ cells in the subaortic region (B,D). By 48 hpf, all gata1+ cells from the subaortic space of control embryos have entered the circulation (C), whereas in arhgef3 MO-injected ones a reduced number of gata1+ cells were observed in the circulation (D). (E-F) Transverse agarose sections (200-μm thick) of Tg(fli1a:EGFP) control embryos at 48 hpf show 2 distinct lumenized vessels, artery (E arrow) and vein (E arrowhead), whereas lumen formation of arhgef3 MO-injected ones is perturbed and axial vessels form a single tube (F arrow).
Expression analysis of endothelial marker fli1a and erythroid marker gata1 in double transgenic embryos Tg(fli1a:EGFP) Tg(gata1:DsRed) shows that vascular development is impaired in arhgef3-depleted embryos. At 30 and 48 hpf arhgef3 MO-injected embryos (B,D) showed a poor segregation of artery and vein compared with control embryos (A,C). Furthermore, arhgef3-depleted embryos exhibited an accumulation of gata1+ cells in the subaortic region (B,D). By 48 hpf, all gata1+ cells from the subaortic space of control embryos have entered the circulation (C), whereas in arhgef3 MO-injected ones a reduced number of gata1+ cells were observed in the circulation (D). (E-F) Transverse agarose sections (200-μm thick) of Tg(fli1a:EGFP) control embryos at 48 hpf show 2 distinct lumenized vessels, artery (E arrow) and vein (E arrowhead), whereas lumen formation of arhgef3 MO-injected ones is perturbed and axial vessels form a single tube (F arrow).
The profound effect of arhgef3 silencing on erythropoiesis prompted us to examine myeloid lineage development. No alteration was detectable in the early pu.1 (data not shown) and late, mpx and l-plastin, myelomonocytic lineage markers at 22 hpf (Figure 2). These results imply that arhgef3 is dispensable for specification and differentiation of premacrophages and neutrophils from cephalic mesoderm and point toward its specific role in both thrombopoiesis and primitive erythropoiesis.
Knockdown of rhoad and rhoae phenocopies anemia seen in arhgef3 knockdown embryos
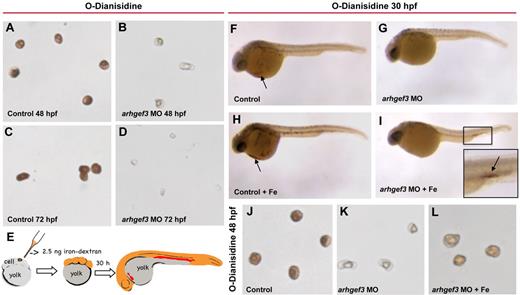
Morphologic features of erythrocytes may further show the underlying mechanism of inadequate erythropoiesis. Indeed, examination of peripheral blood obtained by cardiac puncture from 48 and 72 hpf in arhgef3 MO-injected embryos showed hypochromic and microcytic erythrocytes (Figure 4A-D) that are highly indicative of iron deficiency anemia.31 Lack of iron availability affects the late stages of erythropoiesis at several levels of regulation.32 To investigate whether the anemia in arhgef3 MO-injected embryos was caused by a lack of iron accessibility, rescue experiments were performed using an approach used in a study of the chianti zebrafish, which are anemic because of a mutation in the Tf receptor 1 (tfr1a).25 We reasoned that by overloading the embryo cytoplasm with iron dextran at the 1-cell stage, we would deposit a supply of bioavailable iron to be distributed during subsequent cell divisions, so that the cytoplasm of all cells, including erythroid ones, would directly receive iron (Figure 4E). Indeed, the provision of usable iron to the arhgef3-depleted embryos restored hemoglobin production as shown with o-Dianisidine staining in the posterior blood island (Figure 4I). At 30 hpf, hemoglobin-positive cells were detected in 92.7% ± 2.5% of the control embryos (nembryos = 79), compared with only 8.9% ± 1.6% of the arhgef3 MO-injected embryos (nembryos = 78). Coinjection of arhgef3 MO with iron dextran resulted in a significant increase in the number of embryos exhibiting staining for hemoglobin compared with the arhegf3-depleted embryos (Figure 4F-I; supplemental Figure 3A). Furthermore, injection of iron dextran partially normalized the hemoglobin levels in arhgef3-depleted erythrocytes and wild-type morphology of these cells at 48 hpf (ncells = 50; Figure 4J-L; supplemental Figure 3B-C). This suggests that arhgef3-depleted erythroid cells are fully capable of hemoglobinization, but only after iron supplementation, and that the hypochromia is most probably caused by inadequate iron uptake.
Injection of iron dextran rescues the anemia in arhgef3 MO-injected embryos. (A-D) O-Dianisidine staining of blood collected by cardiac puncture from control and arhgef3 MO-injected embryos at 48 and 72 hpf. Erythrocytes from the arhgef3 MO-injected embryos (B,D) have less hemoglobin and are smaller compared with erythrocytes from control embryos (A,C). (E) Injection of iron dextran (Fe) at the 1-cell stage deposits a supply of usable iron, which is distributed to all cells of the developing embryos during subsequent cell divisions, including erythroid cells. (F-L) Whereas no difference in hemoglobin staining (indicated by black arrows) was detected in the control embryos injected with Fe (H) compared with noninjected controls (F), arhgef3 MO-injected embryos supplemented with Fe (I) showed a significant recovery in hemoglobin levels compared with the ones without supplementation (G) at 30 hpf. (I) Inset shows higher magnification view of the boxed area (black arrow shows accumulated erythrocytes stained with o-Dianisidine). Erythrocytes from arhgef3-depleted embryos supplemented with Fe showed wild-type structure and restored hemoglobin levels at 48 hpf (L) compared with arhgef3 MO-injected embryos (K).
Injection of iron dextran rescues the anemia in arhgef3 MO-injected embryos. (A-D) O-Dianisidine staining of blood collected by cardiac puncture from control and arhgef3 MO-injected embryos at 48 and 72 hpf. Erythrocytes from the arhgef3 MO-injected embryos (B,D) have less hemoglobin and are smaller compared with erythrocytes from control embryos (A,C). (E) Injection of iron dextran (Fe) at the 1-cell stage deposits a supply of usable iron, which is distributed to all cells of the developing embryos during subsequent cell divisions, including erythroid cells. (F-L) Whereas no difference in hemoglobin staining (indicated by black arrows) was detected in the control embryos injected with Fe (H) compared with noninjected controls (F), arhgef3 MO-injected embryos supplemented with Fe (I) showed a significant recovery in hemoglobin levels compared with the ones without supplementation (G) at 30 hpf. (I) Inset shows higher magnification view of the boxed area (black arrow shows accumulated erythrocytes stained with o-Dianisidine). Erythrocytes from arhgef3-depleted embryos supplemented with Fe showed wild-type structure and restored hemoglobin levels at 48 hpf (L) compared with arhgef3 MO-injected embryos (K).
To further dissect the signaling pathway of arhgef3, we examined the role of its downstream target, RhoA, in erythrocyte maturation. We reasoned that, if RhoA is the key arhgef3 target to support its function in iron uptake, then MO knockdown of RhoA would mimic the functional loss of arhegf3 and tfr1a. Because there are 5 rhoa genes in zebrafish33 with > 90% amino acid identity, there is a potential for redundancy of function, so we injected 1-cell stage embryos with translation blocking MOs designed to specifically knock down rhoaa and rhoac (rhoaa/ac) or rhoad and rhoae (rhoad/ae) and a splice-blocking MO to knock down rhoab (supplemental Figure 4A-B). Whereas knockdown of rhoab resulted in severe developmental defects (n = 40), rhoaa/ac and rhoad/ae MO-injected embryos showed no gross structural abnormalities (data not shown). However, rhoad/ae (nctrl = 48, nMO = 52) but not rhoaa/ac MO-injected embryos (nctrl = 48, nMO = 57) had total abrogation of the formation of mature erythrocytes at 48 hpf (Figure 5A-C). Subsequent selective as well as double knockdown of rhoad (nctrl = 35, nMO = 38) and rhoae (nctrl = 35, nMO = 42) by 2 nonoverlapping splice-blocking MOs confirmed their additive effect in erythrocyte maturation (Figure 5D-F; supplemental Figure 4C). As in arhgef3-depleted embryos, severe anemia in rhoad/ae MO-injected embryos was again partially rescued by intracellular iron dextran deposition (nctrl = 96% ± 0.9%, nMO = 39% ± 6.6%, nMO+Fe = 64% ± 3.2%; Figure 5G-J). Altogether, these data indicate that arhgef3 in zebrafish exerts its function in iron uptake through activation of rhoad and rhoae.
Arhgef3 regulates iron uptake through activation of rhoad and rhoae. (A-C) Knockdown of rhoad/rhoae (C), but not rhoaa/rhoac (B), results in severe anemia compared with control embryos (A) as shown by o-Dianisidine staining at 48 hpf. (D-F) O-Dianisidine staining of rhoad and rhoae MO-injected embryos at 48 hpf shows a reduction in the number of hemoglobin-positive cells, which is more profound in rhoae-depleted embryos. (G-J) Provision of intracellular iron rescues the anemia of rhoad/rhoae-depleted embryos. Note o-Dianisidine–positive cells (indicated with black arrow) in the posterior blood island of embryos coinjected with rhoad/rhoae MO and iron dextran (Fe; J). (G) Graph to illustrate the difference (on the y-axis) between the percentages of embryos with hemoglobin positive (Hb+) erythrocytes. At 30 hpf, Hb+ cells were detected in 96% ± 0.9% of the control embryos (nembryos = 53; G,H), whereas only 39% ± 6.6% of rhoad/rhoae MO-injected ones (nembryos = 52) had Hb+ cells (G,I). Coinjection of rhoad/rhoae MO with Fe resulted in a significant increase in the number of embryos showing Hb staining (64% ± 3.2%; nembryos = 64; G,J) compared with the rhoad/rhoae-depleted embryos (*P < .05 in graph). Error bars represent the SEM. (J) Inset shows higher magnification view of the boxed area; black arrow shows accumulated erythrocytes stained with o-Dianisidine.
Arhgef3 regulates iron uptake through activation of rhoad and rhoae. (A-C) Knockdown of rhoad/rhoae (C), but not rhoaa/rhoac (B), results in severe anemia compared with control embryos (A) as shown by o-Dianisidine staining at 48 hpf. (D-F) O-Dianisidine staining of rhoad and rhoae MO-injected embryos at 48 hpf shows a reduction in the number of hemoglobin-positive cells, which is more profound in rhoae-depleted embryos. (G-J) Provision of intracellular iron rescues the anemia of rhoad/rhoae-depleted embryos. Note o-Dianisidine–positive cells (indicated with black arrow) in the posterior blood island of embryos coinjected with rhoad/rhoae MO and iron dextran (Fe; J). (G) Graph to illustrate the difference (on the y-axis) between the percentages of embryos with hemoglobin positive (Hb+) erythrocytes. At 30 hpf, Hb+ cells were detected in 96% ± 0.9% of the control embryos (nembryos = 53; G,H), whereas only 39% ± 6.6% of rhoad/rhoae MO-injected ones (nembryos = 52) had Hb+ cells (G,I). Coinjection of rhoad/rhoae MO with Fe resulted in a significant increase in the number of embryos showing Hb staining (64% ± 3.2%; nembryos = 64; G,J) compared with the rhoad/rhoae-depleted embryos (*P < .05 in graph). Error bars represent the SEM. (J) Inset shows higher magnification view of the boxed area; black arrow shows accumulated erythrocytes stained with o-Dianisidine.
ARHGEF3-depleted K562 cells have impaired transferrin uptake
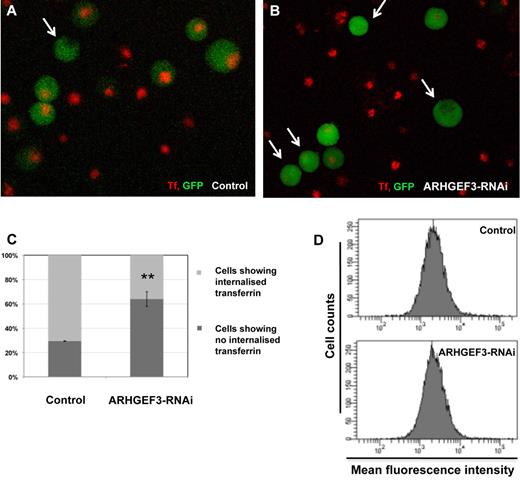
Nonheme iron in the circulation is bound to the plasma protein Tf. All tissues acquire iron by the binding of the iron-loaded Tf to the Tf receptor 1 (TFRC, also named TFR1 or CD71), with this complex then being internalized by receptor-mediated endocytosis.34,35 We postulated that inadequate iron uptake observed in erythroid cells of arhgef3-depleted zebrafish embryos may be a result of ineffective receptor internalization. To test this hypothesis we turned to K562 cells and performed Tf uptake and binding assays. K562 cells, in which the transcription of ARHGEF3 was knocked down by 60% with the use of a lentiviral RNAi construct (supplemental Figure 4D) were allowed to take up Tf-633 for 30 minutes at 37°C. ARHGEF3-depleted cells showed a significant inhibition in Tf uptake, with 64% ± 6.1% of cells (ncells = 104) lacking internalized Tf compared with 29.5% ± 0.3% of control cells (ncells = 105; Figure 6A-C). In contrast, the uptake of an unrelated fluid-phase endocytic marker, Texas red dextran, was not affected by ARHGEF3 depletion, suggesting that ARHGEF3 is not required for all forms of endocytosis (Texas-red MFIcontrol = 8648 ± 344 vs MFIARHGEF3-RNAi = 6792 ± 150; error is SEM). Furthermore, the decrease in Tf internalization attributable to a reduction in TFRC abundance or occupancy was excluded because the amount of membrane-bound Tf-633 to transduced GFP+ K562 cells at strictly 4°C did not differ between ARHGEF3-depleted and control cells (MFI of 2680 ± 114 vs 2618 ± 40, respectively; Figure 6D). The results obtained with the human K562 cells corroborate the phenotype observed in zebrafish and show that silencing of ARHGEF3 does not reduce the amount of Tf that is initially bound to cells, but it does prevent cells to internalize membrane-bound Tf efficiently.
Effect of ARHGEF3 RNAi on Tf uptake in erythroid cells. Tf binding and uptake were assayed in K562 cells that have been transduced with either the lenti-GFP/RNAi-ARHGEF3 construct or lenti-GFP/mmRNAi-WDR66, which was used as a control. (A-B) In arhgef3-depleted K562 cells (B) Tf uptake was dramatically reduced compared with the control (A; white arrows indicate cells showing no Tf). (C) Tf-633 uptake by K562 control cells (n = 105) versus RNAi-ARHGEF3-transduced cells (n = 104), (**P < .01 in the graph). (D) Tf binding to the membrane is unaltered in ARHGEF3 knockdown cells. Control and ARHGEF3-depleted K562 cells were incubated with Tf-633 at 4°C, and excess of unbound Tf was removed by washing. Membrane-bound Tf was measured by flow cytometry, MFI is on the x-axis, and the number of cells assessed is on the y-axis.
Effect of ARHGEF3 RNAi on Tf uptake in erythroid cells. Tf binding and uptake were assayed in K562 cells that have been transduced with either the lenti-GFP/RNAi-ARHGEF3 construct or lenti-GFP/mmRNAi-WDR66, which was used as a control. (A-B) In arhgef3-depleted K562 cells (B) Tf uptake was dramatically reduced compared with the control (A; white arrows indicate cells showing no Tf). (C) Tf-633 uptake by K562 control cells (n = 105) versus RNAi-ARHGEF3-transduced cells (n = 104), (**P < .01 in the graph). (D) Tf binding to the membrane is unaltered in ARHGEF3 knockdown cells. Control and ARHGEF3-depleted K562 cells were incubated with Tf-633 at 4°C, and excess of unbound Tf was removed by washing. Membrane-bound Tf was measured by flow cytometry, MFI is on the x-axis, and the number of cells assessed is on the y-axis.
To investigate whether inhibition of RhoA activity in human K562 cells would mimic the Tf internalization defect seen in ARHGEF3-depleted cells, we examined the effect of exoenzyme C3 transferase. C3 transferase is commonly used to selectively inactivate RhoA, RhoB, and RhoC GTPases. Inhibition of RhoA activity in control cells resulted in the 77.8% increase in the number of cells that were not able to internalize Tf compared with control untreated cells (supplemental Figure 5A-B). The observed reduction of Tf uptake was comparable to the one seen in ARHGEF3-depleted cells. To further test for possible additive effects of ARHGEF3 and RhoA activity, simultaneous knockdown of ARHGEF3 and inactivation of RhoA was performed. Indeed, additional inhibition of RhoA activity in ARHGEF3-depleted cells, by C3 transferase treatment, resulted in 68.8% increase in the number of cells not able to internalize Tf compared with untreated ARHGEF3-depleted cells (supplemental Figure 5A-B). Taken together, our data strongly suggest that ARHGEF3 and RhoA belong to the same essential pathway for Tf internalization and iron uptake.
Discussion
In this study, we used zebrafish and the human erythromyeloblastoid cells K562 for the functional analysis of 1 of 3 GEF genes that were identified in our GWAS on hematologic traits in > 68 000 persons (Gieger et al, manuscript submitted, August 2011). We have functionally validated 5 GWAS association signals in zebrafish and observed that silencing of arhgef3 has profound inhibitory effects on both thrombopoiesis and primitive erythropoiesis (Gieger et al, manuscript submitted, August 2011). Here, we report on the discovery of the mechanism by which arhgef3 exerts its function on the latter and provide further evidence for its additional role in vasculogenesis.
The most striking feature of ARHGEF3-depleted embryos was a profound reduction in the number of the circulating blood cells. Interestingly, this cytopenia was accompanied with a lack of mature, o-Dianisidine–positive erythrocytes. We have demonstrated that the lack of circulating hemoglobin-positive cells is not caused by excessive apoptosis of progenitor blood cells but resulted from a block in the terminal differentiation of erythroid cells and their inability to enter into the circulation. Indeed, the analyses of early and late markers of primitive erythropoiesis (ie, scl, gata2, gata1, alas2, hbbe3) in arhgef3 MO-injected embryos showed no change in their expression at 24 hpf when contrasted with control ones. Interestingly, at 30 hpf we observed an accumulation of cells positive for all markers but gata2 in the ICM in arhgef3-depleted embryos, with the concomitant lack of o-Dianisidine staining. We hypothesized that the accumulation of cells could be partly explained by defects in vessel lumen formation. This was confirmed by extensive confocal imaging in fli1agata1 double transgenic embryos, which showed the accumulation of gata1 cells in the subaortic region of the ICM at 30 hpf in arhgef3 knockdown embryos. The vascular phenotype of arhgef3-depleted embryos is reminiscent of those observed in ephb2 and meis1 knockdown embryos, whereby aberrant vessel lumen formation was also observed.26,36 Further studies are however warranted to determine how arhgef3 exerts its function in vascular development.
Despite severe cytopenia in arhgef3 MO-injected embryos we were able to obtain blood cells by cardiac puncture at 48 hpf and to examine their morphology and hemoglobin content. The smaller size of the isolated erythrocytes and their reduced hemoglobin content strongly resembled the phenotype observed in cia zebrafish mutants25 that lack a type 1 receptor for Tf (tfr1a). Tfr1a is orthologue of human TFRC and is critical for iron uptake in erythroid precursors.25 Mendelian mutations in TFRC leading to a null phenotype have not been identified, and it is assumed that they are incompatible with life, but we have shown that subtle changes in the regulation of transcription of TFRC by common sequence variation exerts an effect on erythrocyte volume and hemoglobin content, confirming its essential role in erythropoiesis.5 It is therefore of interest to search for rare function-modifying variants that may cause microcytosis. Furthermore, Tfrc−/− mice die early in embryogenesis because of anemia.37 The striking similarities between the erythroid phenotypes of cia-mutant and arhgef3-depleted embryos supported the assumption that the GEF may also play a pivotal role in the uptake of Tf by trf1a in erythroid precursors. We reasoned that injection of iron before the onset of cleavage would bypass the postulated defect in internalization of the Tf-tfr1a complex, because during subsequent divisions all cells in the embryo would have been saturated with an excess of usable iron. Although injected iron at high (10 ng) concentration was toxic and caused excessive mortality, at 2.5 ng effective rescue of the anemic phenotype was attained in 41% of arhgef3 MO-injected embryos. Thus, our data strongly suggest that arhgef3-depleted erythroid cells can traffic and metabolize directly delivered intracellular iron but lack the ability to internalize it. However, iron deposition did not restore the circulation at 30 hpf from which we infer that the block in erythrocyte maturation and the vascular defect in arhgef3-depleted embryos are uncoupled events.
As a part of our assessment of arhgef3 function in iron homeostasis, we have tested potential arhgef3 downstream targets. In humans ARHGEF3 specifically activates 2 members of the family of Rho GTPases: RhoA and RhoB.16 Expression analysis of the zebrafish small Rho GTPases during development identified 5 homologs of human RHOA, namely rhoaa, rhoab, rhoac, rhoad, rhoae, and no RHOB homologs.33 By exploiting various MO knockdowns, we have demonstrated that rhoad and rhoae, but not rhoaa and rhoac, are required for hemoglobin production. We were not able to assess the role of rhoab in hematopoiesis because MO-injected embryos died early during development. Clearly, there are several plausible mechanisms that can account for the observed disparity in MO knockdown phenotypes of specific rhoa genes. One possibility is the temporal and/or spatial difference in their expression. Alternatively, arhgef3 might be involved in pathway-specific activation of only rhoad and rhoae but not the other rhoa genes. In any case, the positive results of the rescue experiment with intracellular delivery of usable iron in rhoad- or rhoae-depleted embryos provided further evidence for the functional relatedness of arhgef3 and rhoad or rhoae in iron uptake.
Hemoglobin synthesis is the hallmark of terminal erythropoiesis, and maturing erythroid cells therefore depend on large amounts of bioavailable iron for hem synthesis. Cellular uptake of iron-loaded Tf by erythroid cells is primarily by internalization of the Tf-TFRC complex.34,35 In addition to TFRC, zebrafish and mammals also possess a second receptor for Tf (tfr2 and TFR2, respectively),38 and studies in man and mice strongly suggest its functional interplay with the hereditary hemochromatosis protein. Nonsense mutations in TFR2 in humans cause hemochromatosis and Tfr2−/− mice experience increased intestinal iron absorption.39,40 The recent observation that common sequence variation at the human locus exerts an effect on red cell mass and hemoglobin concentration further confirms the role of TFR2 as critical gatekeeper of iron absorption.4 The 2 Tf receptors (TFRC and TFR2) function at different stages in iron uptake, and Tfr2 cannot compensate for the lack of Tfrc. To determine whether the anemia in arhgef3-depleted erythroid cells was because of ineffective Tf internalization, we performed Tf binding and uptake assays in K562 cells. Notwithstanding that both receptors are present in these cells,38 we observed a strong reduction in Tf uptake in ARHGEF3-depleted cells. This further supports the notion that ARHGEF3 is important in iron uptake, most probably as a regulator of the activity of Rho GTPases in the endocytosis of the Tf-TFRC complex. This result is in concordance with the reported role of RhoA in the regulation of receptor-mediated endocytosis, although the precise mechanism remains to be determined.41 The defect in delivery of Tf attributable to a more rapid exocytoses of the Tf-TFRC complex to the cell surface as seen in hbd-mutant mouse42 was excluded because the amount of membrane-bound Tf-633 to transduced GFP+ K562 cells at strictly 4°C did not differ between ARHGEF3-depleted and control cells.
In summary, iron homeostasis is essential for the formation of adequate numbers of erythroid cells; thus, its intestinal absorption and subsequent internalization into cells must be tightly regulated. Although there have been major advances in our understanding of disorders that perturb iron metabolism, greater knowledge of the interplay between the key factors involved in the regulation of iron metabolism and erythropoiesis will help to develop more effective therapies for inherited and acquired diseases of erythropoiesis. We have provided direct evidence for an important role of ARHGEF3 in Tf uptake and have therefore identified a novel regulator of iron homeostasis. In addition, our study shows the value of pursuing GWAS signals as a new and exciting forward genetics approach in identifying novel regulatory molecules and signaling pathways in hematopoiesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Prof Dorine Swinkels (University of Nijmegen, Nijmegen, The Netherlands) for discussions and advice about iron homeostasis. Transgenic lines gata1:Ds Red and gata1:EGFP were kindly provided by Prof Roger Patient (Univeristy of Oxford, Oxford, United Kingdom).
This work was supported by the Wellcome Trust (grant WT 082597/Z/07/Z, A.C.) and (grants WT 077037/Z/05/Z and WT 077047/Z/05/Z, D.L.S.), the European Union NetSim (training fellowship scheme no. 215820, J.S.-C.), the Research Council of the University of Leuven (grant GOA/2009/13, R.F.), and NIHR (program grant RP-PG-0310-1002 to NHSBT, W.H.O.).
Wellcome Trust
Authorship
Contribution: J.S.-C., A.C., and K.F. designed and performed the experimental work, analyzed the data, and wrote the manuscript; N.S. performed research; D.L.S. planned the project, analyzed the data, and edited the manuscript; and W.H.O. planned the project, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Willem H. Ouwehand, Department of Haematology, University of Cambridge, NHS Blood and Transplant Centre, Long Road, Cambridge CB2 0PT, United Kingdom; e-mail: who1000@cam.ac.uk.
References
Author notes
J.S.-C. and A.C. contributed equally to this study.
W.H.O. and K.F. are shared senior authors.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal