To the editor:
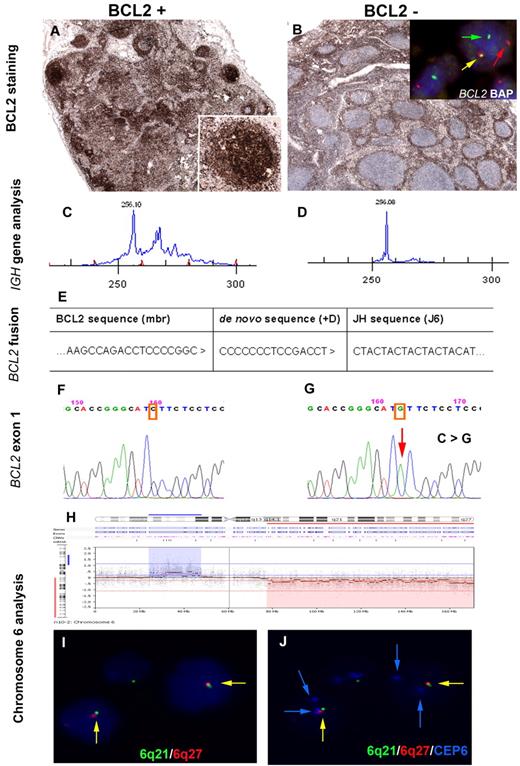
Follicular lymphoma in situ (FLIS) is characterized by the presence of a B-cell population with immunophenotypic and genotypic features of follicular lymphoma (FL) but exclusively localized to germinal centers (GCs) in morphologically reactive lymph nodes.1 FLIS lesions are monoclonal and carry the t(14;18)(q32;q21) translocation, which juxtaposes the BCL2 gene to the immunoglobulin heavy chain (IGH) locus, causing constitutive expression of the antiapoptotic protein BCL2. Most of the cases described showed no evidence of manifest FL (mFL) at diagnosis, however, some individuals had synchronous evidence of mFL at another site or developed mFL during follow-up.2,3 This latter finding suggests that FLIS might represent a very early stage in the development of mFL. However, the genetic events responsible for progression of FLIS to mFL are largely unknown. Here we report a case of FLIS with BCL2 expression synchronously presenting with a BCL2 negative mFL, both lesions clonally related and carrying the t(14;18)(q32;q21) translocation. The case occurred in a 78-year-old man with stage III disease and FLIPI intermediate risk. The diagnostic biopsy was composed of 2 lymph nodes. The larger one showed the typical morphology of FL grade 1/II with the characteristic phenotype (CD20+, CD10+, BCL6+), except for lack of BCL2 expression. The small lymph node showed normal architecture, but with strong BCL2 expression in some morphologically reactive GCs diagnostic of FLIS (Figure 1A-B). FISH analysis using a BCL2 break-apart probe (LSI BCL2 BAP, Vysis) confirmed a BCL2 breakpoint in both areas suggestive of the t(14;18)(q32;21) translocation (Figure 1B; insert). IGH PCR clonality analysis performed in microdissected tissue showed identical monoclonal peaks in both lesions (Figure 1C-D). Sequence analysis of the BCL2 breakpoint region confirmed the clonal identity of both lesions (Figure 1E). To explain the discordant BCL2 reactivity, exon 1 of the BCL2 gene, where the epitope of the BCL2 antibody resides, was amplified and sequenced. A point mutation resulting in amino acid substitution (c.144 C > G; p.I48M ATC–ATG) was found in the mFL, whereas the FLIS showed a wild type sequence (Figure 1F-G). Array CGH (244K platform, Agilent Technologies) analysis was performed on DNA from microdissected tissue of both FLIS and mFL.4 mFL showed gains in 6p22.2-p12.3 and losses in 6q14.1-qter and 8pter-p23.1. Subsequent FISH using probes for 6q21 and 6q27 confirmed 6q deletions in 80% of the mFL cells (Figure 1H-J). In contrast, the FLIS counterpart showed no pathogenetic alterations by array CGH. Nevertheless, 32 benign polymorphisms (CNPs) were observed, of which 16 (50%) were also detected in the mFL confirming the same patient origin.
Immunophenotypic, molecular, and molecular cytogenetic findings in a synchronous, clonally related in situ and manifest follicular lymphoma. Immunohistochemical analysis of BCL2. (A) BCL2 expression in some germinal centers of a morphologically reactive lymph node. Inset: higher magnification of a BCL2 positive germinal center. (Labvision, BCL2 clone 100/D5). (B) Lymph node biopsy with the manifest follicular lymphoma, BCL2 negative. Insert: Interphase FISH analysis using double-color BCL2 break-apart probe (Abbott, Vysis; LSI BCL2 BAP) shows one allele with a normal co-localized signal (yellow arrow) and the second allele with a split of the red and green signal (arrows red and green), indicating a BCL2 break. (C-D) Clonality analysis of the IGH framework 2 (FR2) region shows an identical monoclonal peak of 256 base pairs in both lesions. (E) Sequence analysis of the major BCL2 breakpoint region showed an identical de novo sequence (+D) in both lesions. (F-G) Sequence analysis of BCL2 gene, exon 1 shows a wild-type sequence in the FLIS lesion (left), whereas the mFL shows a point mutation in residue 48 resulting in amino acid substitution (c.144 C > G, p.I48M ATC-ATG). (H) Array-CGH profile of chromosome 6 ideogram reveals loss in 6q14-qter (red label) and gain in 6p22.2-p12.3 (blue label) in the mFL. (I-J) Interphase FISH analysis using FISH probes for 6q21 (RP3–515A4 Spectrum green), 6q27 (RP3–450D5 Spectrum red), and a centromeric probe for chromosome 6 (CEP6 Spectrum aqua; Abbott, Vysis) False color display of the same area in panels I and J. Blue channel in panel I is DAPI counterstain whereas in panel J is Spectrum Aqua (CEP6). Nuclei contain only 1 6q21 and 6q27 signal (green, red) but 2 blue signals (CEP6) indicating deletion in 6q21 and q27 in line with the array CGH. FISH images were acquired with a 100×/1.40 oil-immersion objective in a Zeiss Axioskop fluorescence microscope (Zeiss) equipped with the appropriate filter sets and were documented and processed using the ISIS imaging system (MetaSystems). PCR analysis for IGH gene rearrangements was performed with BIOMED-2 primers for FR2 and FR3 using Phusion hot start DNA polymerase (Finnzymes) with adequate amplification conditions and D4-fluorescent dye primer modification (Sigma-Aldrich). The products were separated by capillary electrophoresis on the GenomeLab GeXP Genetic Analysis System and analyzed with GenomeLab GeXP 10.2 software (Beckman Coulter). Genomic DNA from residue 29-96 of the BCL2 gene was amplified using AmpliTaq Gold DNA Polymerase. PCR products were cloned into the pGEM-T easy vector and transfected into JM109 competent cells. Bacterial plasmids were processed according to standard procedures, and mFL and FLIS clones were subjected to dye terminator cycle sequencing (DTCS-Quick Start Master Mix) using M13 primers and capillary electrophoresis on the GenomeLab GeXP Genetic Analysis System 10.2 software. Array CGH of mFL and FLIS samples was performed on a Human Genome CGH Microarray 244K platform (Agilent Technologies) using Bio Prime Array-CGH Genomic Labeling Kit and hybridized using the manufacturer's protocol (Invitrogen). Slides were scanned with a GenePix 4000B microarray reader (Molecular Devices Corporation). Signal intensities from the generated images were measured and evaluated with Feature Extraction 9.5.3 and DNA Analytics v4.0.81 software packages, respectively (Agilent Technologies). Array CGH gains and losses were defined by use of a trial version of Nexus 6.0 β Discovery Edition.
Immunophenotypic, molecular, and molecular cytogenetic findings in a synchronous, clonally related in situ and manifest follicular lymphoma. Immunohistochemical analysis of BCL2. (A) BCL2 expression in some germinal centers of a morphologically reactive lymph node. Inset: higher magnification of a BCL2 positive germinal center. (Labvision, BCL2 clone 100/D5). (B) Lymph node biopsy with the manifest follicular lymphoma, BCL2 negative. Insert: Interphase FISH analysis using double-color BCL2 break-apart probe (Abbott, Vysis; LSI BCL2 BAP) shows one allele with a normal co-localized signal (yellow arrow) and the second allele with a split of the red and green signal (arrows red and green), indicating a BCL2 break. (C-D) Clonality analysis of the IGH framework 2 (FR2) region shows an identical monoclonal peak of 256 base pairs in both lesions. (E) Sequence analysis of the major BCL2 breakpoint region showed an identical de novo sequence (+D) in both lesions. (F-G) Sequence analysis of BCL2 gene, exon 1 shows a wild-type sequence in the FLIS lesion (left), whereas the mFL shows a point mutation in residue 48 resulting in amino acid substitution (c.144 C > G, p.I48M ATC-ATG). (H) Array-CGH profile of chromosome 6 ideogram reveals loss in 6q14-qter (red label) and gain in 6p22.2-p12.3 (blue label) in the mFL. (I-J) Interphase FISH analysis using FISH probes for 6q21 (RP3–515A4 Spectrum green), 6q27 (RP3–450D5 Spectrum red), and a centromeric probe for chromosome 6 (CEP6 Spectrum aqua; Abbott, Vysis) False color display of the same area in panels I and J. Blue channel in panel I is DAPI counterstain whereas in panel J is Spectrum Aqua (CEP6). Nuclei contain only 1 6q21 and 6q27 signal (green, red) but 2 blue signals (CEP6) indicating deletion in 6q21 and q27 in line with the array CGH. FISH images were acquired with a 100×/1.40 oil-immersion objective in a Zeiss Axioskop fluorescence microscope (Zeiss) equipped with the appropriate filter sets and were documented and processed using the ISIS imaging system (MetaSystems). PCR analysis for IGH gene rearrangements was performed with BIOMED-2 primers for FR2 and FR3 using Phusion hot start DNA polymerase (Finnzymes) with adequate amplification conditions and D4-fluorescent dye primer modification (Sigma-Aldrich). The products were separated by capillary electrophoresis on the GenomeLab GeXP Genetic Analysis System and analyzed with GenomeLab GeXP 10.2 software (Beckman Coulter). Genomic DNA from residue 29-96 of the BCL2 gene was amplified using AmpliTaq Gold DNA Polymerase. PCR products were cloned into the pGEM-T easy vector and transfected into JM109 competent cells. Bacterial plasmids were processed according to standard procedures, and mFL and FLIS clones were subjected to dye terminator cycle sequencing (DTCS-Quick Start Master Mix) using M13 primers and capillary electrophoresis on the GenomeLab GeXP Genetic Analysis System 10.2 software. Array CGH of mFL and FLIS samples was performed on a Human Genome CGH Microarray 244K platform (Agilent Technologies) using Bio Prime Array-CGH Genomic Labeling Kit and hybridized using the manufacturer's protocol (Invitrogen). Slides were scanned with a GenePix 4000B microarray reader (Molecular Devices Corporation). Signal intensities from the generated images were measured and evaluated with Feature Extraction 9.5.3 and DNA Analytics v4.0.81 software packages, respectively (Agilent Technologies). Array CGH gains and losses were defined by use of a trial version of Nexus 6.0 β Discovery Edition.
This is the first description of a FLIS clonally related to the mFL where secondary genetic alterations are demonstrated, probably representing clonal evolution as sign of disease progression. Our findings provide evidence that FLIS is in fact a very early lesion carrying the t(14;18)(q32;q21) translocation without additional secondary genetic alterations.5,6 In this case, acquisition of numeric chromosomal aberrations and BCL2 mutation indicates that the synchronous FLIS represents cells of the founder clone rather than early colonization of reactive GCs by mFL.
Authorship
Acknowledgments: The authors thank the technical staff of the laboratories in Tübingen and in Kiel for expert technical assistance. The work of R.S. on clonal evolution of t(14;18)-positive lymphomas is supported from Bundesministerium für Bildung und Forschung through the network “HämatoSys” The work of L.Q.-M., F.F. and I.B. is supported by the SFB 685.
Contribution: I.B., I.S. and A.H. performed research and analyzed data; G.G. contributed with vital patient information; P.A. analyzed data; and R.S., F.F. and L.Q.-M. designed the research, analyzed data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Leticia Quintanilla-Martinez, Institute of Pathology, University Hospital Tuebingen, Liebermeisterstrasse 8, 72076 Tuebingen, Germany; e-mail: leticia.quintanilla-fend@med.uni-tuebingen.de.
References
Author notes
I.B. and I.S. contributed equally to this article.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal