Abstract
MicroRNAs play a crucial role in chronic lymphocytic leukemia. We investigated whether microRNAs can discriminate patients with a progressive disease from patients with a stable disease. We analyzed microRNA expression on leukemic cells isolated from 358 sequential samples of 114 patients with either stable or progressive disease. We found that during the course of the disease the expression values of miR-181b, the most dysregulated microRNA, decreased in samples of patients with a progressive (P < .001, training and validation sets) but not in samples of patients with a stable disease (P = .3, training set; P = .2, validation set) over time. A drop of ≥ 50% between sequential samples and/or a miR-181b value ≤ 0.005 at the starting time point were significant to differentiate progressive from stable disease (P = .004, training set; P < .001, validation set). These parameters were associated with high risk of requiring treatment (risk ratio, 5.8; 95% confidence interval, 2.5-14.9). We also observed that miR-181b targets Mcl-1 protein and that the decrease of its expression inversely correlated with increased protein levels of MCL1 and BCL2 target genes. We conclude that parameters defined on the basis of the miR-181b expression values specify disease progression in chronic lymphocytic leukemia and are associated with clinical outcome.
Introduction
Patients with chronic lymphocytic leukemia (CLL) have a heterogeneous clinical course, ranging from indolent to highly aggressive.1 The molecular features that have been shown to be reliable for predicting an inferior clinical course in CLL are the immunoglobulin heavy-chain variable-region (IGHV) homology to the germline ≥ 98%, recurrent cytogenetic abnormalities,2 and expression of ζ-associated protein 70 (ZAP-70) in ≥ 20% of the leukemic cells.3,4 Each of these features has been shown to be a predictor of time to treatment on univariate analysis. The clinical staging systems, independently developed by Rai et al5 and Binet et al6 on the basis of biologic and clinical parameters, are also useful for assessing prognosis in patients with CLL. Most patients with CLL have the disease diagnosed while they are asymptomatic and in the early stages; however, these systems are not accurate enough to identify the subgroups of patients whose disease will progress. Over the past few years several studies have shown that microRNAs (miRNAs) play an important role in CLL.7-10 Moreover, the differential leukemia-cell expression of certain miRNAs can help distinguish samples of patients with aggressive versus indolent disease. In this study, we examined whether the CLL-cell expression levels of miRNAs change over time, performing a genomewide miRNA profiling on sequential samples obtained from patients with different rates of disease progression. We then validated our results with the use of samples obtained from patients with a stable disease and in an independent larger cohort of sequential samples from patients with either progressive or stable disease. We also examined the expression of certain proteins encoded by mRNA that were targeted by miRNA found to have the most significant change over time in serial samples of patients with progressive disease.
Methods
Patients and analysis of ZAP-70 and IGHV mutation status
We selected 358 samples obtained from 114 untreated patients with CLL and were enrolled in the CLL Research Consortium on written informed consent in accordance with the Declaration of Helsinki. The study protocols were approved by The Ohio State University Institutional Review Board. The participating cancer research center institutions provided the clinical data associated with each patient at the time sample collection, including the date of the initiation of first therapy. The samples were analyzed to determine both the expression of ZAP-70 and the IGHV mutational status as previously described.4 PBMCs were isolated by density gradient centrifugation with the use of Ficoll-Paque Plus (Amersham Biosciences). The PBMCs obtained from these patients were > 98% leukemic CD5+/CD19+ B cells.
RNA extraction and real-time quantitative PCR
RNA from patients with CLL was extracted with standard Trizol (Invitrogen) methods. The quality of the RNAs has been analyzed for each sample on the Agilent 2100 Bioanalyzer. RNA integrity number was on average 9.2 and on median 9.5, calculated on all samples enrolled in this study. Mature miRNA expression was assayed by Taqman MicroRNA assay (Applied Biosystem) and normalized on RNU44 (P/N: 4373384), according to the manufacturer's protocol. Ten nanograms of total RNA from all samples was reverse transcribed in 2 days (1 day for the training set and 1 day for the validation set) to avoid technical problem. Each sample was analyzed in triplicate on 384-well plates, and all the plates were run on the same machine. The level of miRNA was measured with Ct (threshold-cycle). The amount of target, normalized to an endogenous reference, is given by 2−ΔCt (Comparative Ct method; Applied Biosystem).
Cell culture and transfection
HeLa cell lines (all from American Type Culture Collection) were cultured in RPMI 1640 medium with 10% FBS. Precursor-hsa-miR-181b precursor (Sanger Accession no. MI0000270) and negative control 2 ribo-oligonucleotide were purchased from Applied Biosystems/Ambion. The 2′-O-Me-antisense oligonucleotides (AMOs) against miR-181b and against the green fluorescent protein gene (AMO negative control) were purchased from Fidelity Systems. Transfection of miRNAs, AMOs, and expression vectors was performed with Lipofectamine 2000 (Invitrogen) in accordance with the procedures of the manufacturer.
Luciferase assays and vectors
The human 3′-untranslated region (UTR) of MCL1 was amplified by PCR with the use of the primers MCL1_1212F (5′-CCGctcgagTAACCAACCACCACCACCAC-3′) and MCL1_3899R (5′-ATAAGAATgcggccgcCGTTGGTCCTAACCCTTCCTG-3′) and cloned downstream of the firefly luciferase gene psiCHECK-2 Vector (Promega). Substitutions and deletions into the miR-181b binding sites of the MCL1 3′UTR gene was introduced with the use of the Quick-Change site-directed mutagenesis kit, following the instructions of the manufacturer, and with the use of the primers Mcl1-181 mut F2_5′-TAAGATGACTAAGCCAATGGGGAAGAATTGCCCTG-3′ and Mcl1-181 mut R2_5′-CTTCCCCATTGGCTTAGTCATCTTATTCATACC-3′. Transfection was conducted in HeLa cells cultured in 24-well plates, each well was cotransfected with 400 ng of psiCHECK-2 vector and 100nM miR-181b or negative control 2 or AMOs or with methylated control oligonucleotide. Twenty-four hours after transfection, firefly and renilla luciferase activities were measured with the Dual-Luciferase Report Assay (Promega).
Western blot analysis
HeLa cell lines were transfected with 100nM miR-181b, AMOs, and control sequences in 6-well plates. After 48 and 72 hours, cells were collected, lysed in RIPA buffer (Sigma), loaded with Laemmli 5 × buffer, and analyzed by Western blot to assess the expression of MCL1 with the use of monoclonal antibody (antibody sc-819; Santa Cruz Biotechnology). Detection was conducted by chemiluminescent enhanced assay (WesternBreeze Chemiluminescent Kit; Invitrogen). β-actin antibody (β-actin antibody no. 4976; Cell Signaling) has been used to normalize the loaded protein amount. Samples from patients were lysed in RIPA buffer, and the expression of TCL1 and BCL2 was assessed with the use of specific antibodies (sc-32 331, Santa Cruz Biotechnology; M0887, Dako). To quantify Western blot signals, digital images of autoradiographies were acquired with Fluor-S MultiImager, and band signals were quantified in the linear range of the scanner with the use of specific densitometric software (Quantity One).
Bioinformatics analysis
miRNA microarray profiling RNA from 2 sequential samples obtained from 23 patients with progressive disease was performed as previously described.11 Average values of the replicate spots of each miRNA were background subtracted, normalized, and further analyzed. Normalization was performed with the quantile method. We selected the miRNAs measured as present in at least as many samples as the smallest class in the dataset (50%). Absent calls were threshold to 3.3 (log2 scale) before statistical analysis, representing the average minimum intensity level detectable in the system. Greater than 95% of blank probes (ie, negative controls) fell below the threshold value of 3.3. miRNAs that are differentially expressed in 2 groups were identified with the “class comparison among genes” within Biometric Research Branch the (BRB) Array tools Version 3.6.0 developed by Simon et al.12 The criterion for inclusion of a gene in the gene list is a P value < .05. All microarray data are available in the ArrayExpress database under Accession no. E-MTAB-683.
Statistical analysis
Two-sample t tests that compared miR-181b values between groups were made on the log-scale whereby normality and equal variance assumptions did not appear to be violated. Associations between starting and ending points were examined with the Mann-Whitney test. Associations between dichotomous categorical variables, such as the property, IGHV ≥ 98% and ZAP-70 ≥ 20%, were examined with the Fisher exact test. The time from last time point to initial treatment was estimated by the method of Kaplan-Meier and assessed by the log-rank test. The prognostic value of miR-181b values having a starting value < 0.005 or a start to end drop of ≥ 50% (called the property) or both in determining time to treatment was examined with the Cox regression analysis under the proportional hazards model. Here, we also examined the possible independent predictive value of CLL cell expression of ZAP-70 values ≥ 20% or unmutated IGHV values ≥ 98% while controlling for time on study. All P values were 2 sided.
Results
Dysregulated miRNAs in patients with CLL with progressive disease
The characteristics of the patients with progressive and stable disease in the training and the validation sets are shown in Table 1 and supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Progression was defined as a change to a more advanced clinical stage, the need for treatment according to the parameters defined in the international workshop on CLL,13 or both. In the training set the median time on study was 31 months (interquartile range [IQR], 14-41 months) for patients with a progressive disease and 23 months (IQR, 10-42 months) for those with a stable disease. For the latter group we had an additional median follow-up observation period of 41 months after the time at which the last examined sample was collected to verify that patients in this subgroup had indolent disease. In the validation set, the median time was 35 months (IQR, 16-51 months) for patients with progressive disease and, for patients with a stable disease, 27 months (IQR, 15-40 months) plus an additional median time of follow-up of 46 months.
Molecular and clinical features of patients with progressive and stable CLL included in the training and validation sets
| Samples Characteristics . | FTP . | LTP . | No. of patients with therapy needed . | IGHV homology ≥ 98%, no. of patients . | IGHV homology < 98%, no. of patients . | ZAP-70–positive cells ≥ 20%, no. of patients . | ZAP-70–positive cells < 20%, no. of patients . |
|---|---|---|---|---|---|---|---|
| Training set | |||||||
| Progressive CLL (N = 23) | 18 | 10 | 13 | 8 | 15 | ||
| Median β2m level | 2 | 2.7 | |||||
| Median WBC count | 48.7 | 136.5 | |||||
| Stable CLL (N = 13) | 0 | 3 | 10 | 3 | 10 | ||
| Median β2m level | 1.8 | 1.6 | |||||
| Median WBC count | 26.4 | 30.9 | |||||
| Validation set | |||||||
| Progressive CLL (N = 30) | 27 | 18 | 12 | 17 | 13 | ||
| Median β2m level | 1.7 | 2.5 | |||||
| Median WBC count | 25.6 | 48 | |||||
| Stable CLL (N = 48) | 0 | 6 | 42 | 12 | 36 | ||
| Median β2m level | 1.8 | 1.9 | |||||
| Median WBC count | 19.4 | 20.2 |
| Samples Characteristics . | FTP . | LTP . | No. of patients with therapy needed . | IGHV homology ≥ 98%, no. of patients . | IGHV homology < 98%, no. of patients . | ZAP-70–positive cells ≥ 20%, no. of patients . | ZAP-70–positive cells < 20%, no. of patients . |
|---|---|---|---|---|---|---|---|
| Training set | |||||||
| Progressive CLL (N = 23) | 18 | 10 | 13 | 8 | 15 | ||
| Median β2m level | 2 | 2.7 | |||||
| Median WBC count | 48.7 | 136.5 | |||||
| Stable CLL (N = 13) | 0 | 3 | 10 | 3 | 10 | ||
| Median β2m level | 1.8 | 1.6 | |||||
| Median WBC count | 26.4 | 30.9 | |||||
| Validation set | |||||||
| Progressive CLL (N = 30) | 27 | 18 | 12 | 17 | 13 | ||
| Median β2m level | 1.7 | 2.5 | |||||
| Median WBC count | 25.6 | 48 | |||||
| Stable CLL (N = 48) | 0 | 6 | 42 | 12 | 36 | ||
| Median β2m level | 1.8 | 1.9 | |||||
| Median WBC count | 19.4 | 20.2 |
FTP indicates first time point; LTP, last time point; β2m, β2 microglobulin; and WBC, white blood cell.
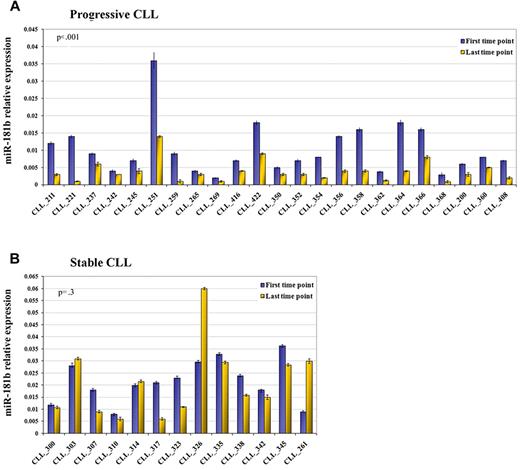
We compared miRNA profiles of 2 sequential samples obtained from 23 patients with a progressive disease in the training cohort. The last time point is a more aggressive form compared with its previous counterpart (parameters defined according to Hallek et al13 ). With the use of the class comparison within BRB Array tools, we identified 15 miRNAs of 474 human mature miRNAs with a P < .05 but only one with a false discovery rate filter < 20% (Table 2). MiR-181b was down-regulated in the more aggressive form of the disease compared with its previous counterpart. This result was confirmed by real-time quantitative PCR (qPCR) on the total cohort (P < .001; Figure 1A). To evaluate the specificity of this finding, we analyzed the expression of miR-181b in 2 sequential samples obtained from 13 patients with the stable course of the disease. We observed that miR-181b values did not undergo significant alteration (P = .3) between the 2 time points (Figure 1B). To validate this data we selected a cohort of 78 patients who had either progressive or stable disease in which the criteria for defining the form of the disease matched those of the training set with the only exception being the white blood cell count. Within the subset of patients who had progressive disease were patients who did not have increasing lymphocytosis, but rather progressive lymphadenopathy, allowing us to assess whether changes in expression levels of miR-181b occurred independent of increases in white blood cell counts. We also noted decreases in expression levels of miR-181b in sequential samples obtained from such patients, as well as in patients with progressive disease in this validation cohort, but not in serial samples from patients with stable disease (supplemental Figure 1).
miRNAs deregulated between 2 sequential time points from patients with CLL with a progressive disease
| P‡ . | FDR . | Fold change† . | miRNA . |
|---|---|---|---|
| .0006 | .18 | 0.32 | miR-181b |
| .0015 | .21 | 0.56 | miR-130b |
| .0032 | .29 | 0.35 | miR-126* |
| .0066 | .46 | 0.54 | miR-296-3p |
| .0092 | .47 | 0.65 | miR-223 |
| .0125 | .47 | 0.61 | miR-130a |
| .0128 | .47 | 1.44 | miR-125a-3p |
| .0134 | .47 | 1.22 | miR-668 |
| .0152 | .47 | 0.64 | miR-299-3p |
| .0258 | .64 | 1.46 | miR-499-3p |
| .0274 | .64 | 1.21 | miR-412 |
| .0275 | .64 | 0.63 | miR-192 |
| .0357 | .72 | 1.24 | miR-193b |
| .0358 | .72 | 0.56 | miR-340 |
| .0427 | .80 | 1.36 | miR-93* |
| P‡ . | FDR . | Fold change† . | miRNA . |
|---|---|---|---|
| .0006 | .18 | 0.32 | miR-181b |
| .0015 | .21 | 0.56 | miR-130b |
| .0032 | .29 | 0.35 | miR-126* |
| .0066 | .46 | 0.54 | miR-296-3p |
| .0092 | .47 | 0.65 | miR-223 |
| .0125 | .47 | 0.61 | miR-130a |
| .0128 | .47 | 1.44 | miR-125a-3p |
| .0134 | .47 | 1.22 | miR-668 |
| .0152 | .47 | 0.64 | miR-299-3p |
| .0258 | .64 | 1.46 | miR-499-3p |
| .0274 | .64 | 1.21 | miR-412 |
| .0275 | .64 | 0.63 | miR-192 |
| .0357 | .72 | 1.24 | miR-193b |
| .0358 | .72 | 0.56 | miR-340 |
| .0427 | .80 | 1.36 | miR-93* |
FDR indicates false discovery rate.
Fold change is the fold ratio of the geometric means of miRNA expressions of paired samples with progressive disease. It is calculated with the use of the class comparison within the BRB Array tool.
Reported are the results of the class comparison analysis with the use of the approach last time point versus first time point. The analysis was performed by using Biometric Research Branch (BRB) Array tool Version 3.6.0.
miR-181b expression values significantly decrease in progressive but not in stable CLL over time (training set). Relative expression of the mature miR-181b in the first time point (violet blocks) and last time point (yellow blocks) from sequential samples of patients with CLL with either progressive (A) or stable (B) disease. The expression has been determined by stem-loop real-time qPCR. Each sample of data was normalized to the endogenous reference RNU44 with the use of the 2−Δct method. P value is the result of the paired t test on log10-transformed values. Error bars indicate SD (mean ± SD), n = 3.
miR-181b expression values significantly decrease in progressive but not in stable CLL over time (training set). Relative expression of the mature miR-181b in the first time point (violet blocks) and last time point (yellow blocks) from sequential samples of patients with CLL with either progressive (A) or stable (B) disease. The expression has been determined by stem-loop real-time qPCR. Each sample of data was normalized to the endogenous reference RNU44 with the use of the 2−Δct method. P value is the result of the paired t test on log10-transformed values. Error bars indicate SD (mean ± SD), n = 3.
MiR-181b expression values specifically decrease during the progression of CLL
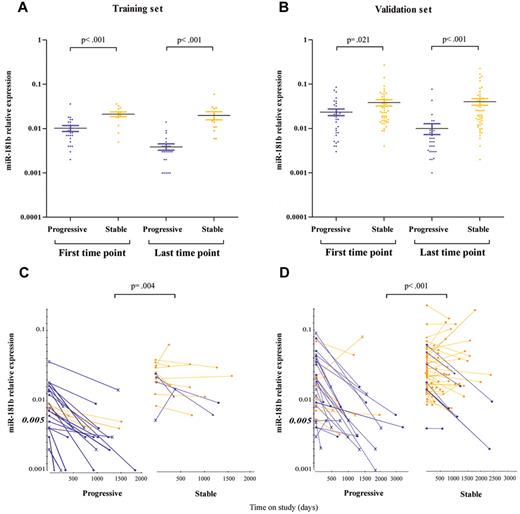
To assess whether the overall expression values of the miR-181b are lower in samples obtained from patients with progressive rather than stable disease, we compared starting and ending values in the 2 subgroups. In the training set, patients who eventually progressed had starting values on average 56% (95% confidence interval [CI], 33%-72%; P < .001 Mann-Whitney test) lower than patients whose disease remained stable, whereas the difference between the ending values increased on average to 81% (95% CI, 69%-89%; P < .001; Figure 2A). With the validation set we confirmed the previous finding in that starting values of samples from patients with progressive CLL are lower than starting values from patients with a stable disease, 39% average (95% CI, 7%-60%; P = .021), whereas between the ending values the difference reached the average of 78% (95% CI, 67%-86%; P < .001; Figure 2B). Moreover, to identify parameters that best distinguish progressive from stable disease, we determined the miR-181b expression at all time points. A decrease of ≥ 50% between the first and the last time point and a miR-181b value at the starting time point ≤ 0.005 significantly differentiated the 2 CLL subgroups (P = .004, training set; P < .001, validation set; Fisher exact test; Figure 2C-D). We observed that 83% and 73% of the progressive patients, in the training and validation sets, respectively, have ≥ 1 of these properties, whereas they are present in only 30% and 15%, respectively, of patients with stable disease. A decrease of ≥ 50% was considered because miR-181b expression undergoes small fluctuation over time, and the value ≤ 0.005 was chosen as cutoff on the basis of the training set in which fewer stable (1 of 13) had such low miR-181b expression at the starting time point compared with samples from patients with progressive disease (6 of 23). In this analysis the decrease between the first and the last time point was considered. This is a conservative approach because our results would appear stronger by looking at all time points of additional patients with progressive disease (CLL_245, training set: CLL_363v, CLL_436, validation set; supplemental Table 1) with a decrease of 50% in miR-181b expression between sequential samples. We further analyzed how well the expression of miR-181b could discriminate a progressive from a stable disease compared with the IGHV mutational status and ZAP-70 expression. We found that miR-181b was the most significant biomarker of progressive disease (Table 3).
MiR-181b expression values discriminate samples of patients with stable versus progressive disease. Relative expression of the mature miR-181b in serial time points from patients with CLL with a progressive (violet dots) or stable (yellow dots) disease in the training (A) and validation (B) set. The bars indicate the mean values and the errors. P value is the result of the Mann-Whitney test. Patients with (violet lines) or without (yellow lines) the properties, defined on the base of miR-181b expression values, are indicated in the training (C) or validation (D) set. The ending points are showed with either a circle for patients with IGHV ≤ 98% or a cross for patients with IGHV ≥ 98%. Squares denote patients with unknown values. P value is the result of the Fisher exact test. MiR-181b relative expression is determined by stem-loop real-time qPCR. Each sample of data was normalized to the endogenous reference RNU44 with the use of 2−Δct method.
MiR-181b expression values discriminate samples of patients with stable versus progressive disease. Relative expression of the mature miR-181b in serial time points from patients with CLL with a progressive (violet dots) or stable (yellow dots) disease in the training (A) and validation (B) set. The bars indicate the mean values and the errors. P value is the result of the Mann-Whitney test. Patients with (violet lines) or without (yellow lines) the properties, defined on the base of miR-181b expression values, are indicated in the training (C) or validation (D) set. The ending points are showed with either a circle for patients with IGHV ≤ 98% or a cross for patients with IGHV ≥ 98%. Squares denote patients with unknown values. P value is the result of the Fisher exact test. MiR-181b relative expression is determined by stem-loop real-time qPCR. Each sample of data was normalized to the endogenous reference RNU44 with the use of 2−Δct method.
Distribution of the subgroup defined on the basis of the properties (drop of the miR-181b expression values of ≥ 50% between the first and the last point and/or value ≤ 0.005 at the starting point), IGHV mutational status and ZAP-70 expression in progressive and stable CLL
| Subgroup . | Training set . | P* . | Validation set . | P* . | ||
|---|---|---|---|---|---|---|
| Progressive (N = 23) . | Stable (N = 13) . | Progressive (N = 30) . | Stable (N = 48) . | |||
| With properties | 19 | 4 | .004 | 22 | 7 | < .001 |
| IGHV unmutated | 10 | 3 | .3 | 18 | 6 | < .001 |
| ZAP-positive cells ≥ 20% | 9 | 3 | .5 | 17 | 12 | .008 |
| Subgroup . | Training set . | P* . | Validation set . | P* . | ||
|---|---|---|---|---|---|---|
| Progressive (N = 23) . | Stable (N = 13) . | Progressive (N = 30) . | Stable (N = 48) . | |||
| With properties | 19 | 4 | .004 | 22 | 7 | < .001 |
| IGHV unmutated | 10 | 3 | .3 | 18 | 6 | < .001 |
| ZAP-positive cells ≥ 20% | 9 | 3 | .5 | 17 | 12 | .008 |
P values were calculated by the Fisher exact test.
Association between miR-181b expression values and clinical outcome in CLL
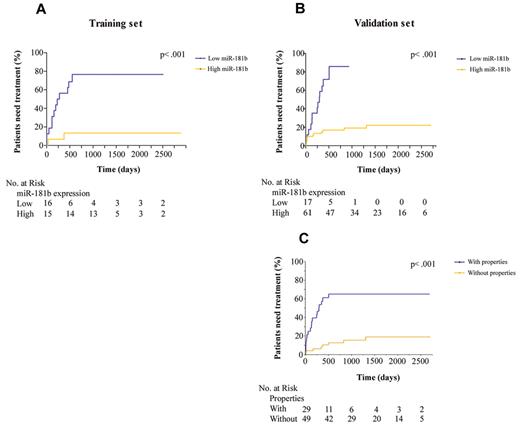
We examined the role of miR-181b expression values as a prognostic indicator of time to first treatment. Kaplan-Meier curves (Figure 3A-B) showed that patients with low expression of miR-181b had a greater risk of needing therapy than patients with high levels; the risk was ∼ 7 times higher (95% CI, 2.2-19.4; P < .001 log-rank test) in the training set and 19 times higher (95% CI: 6.6-55.8; P < .001) in the validation set. With the use of the validation cohort we developed a multivariate proportional hazard model to compare the effect of the IGHV mutational status, the ZAP-70 expression, and the property previously defined according to the miR-181b expression values on the relative risk of requiring treatment. We found a significantly higher hazard of requiring treatment in the group with the property (hazard ratio [HR], 5.8; 95% CI, 6.6-55.8; P < .001; Figure 3C) and for subjects with IGHV ≥ 98% (P ∼ .008). After adjusting for these 2 variables, CLL cell expression of ZAP-70 did not discriminate between groups of patients having different rates of requiring treatment over this observation period. Moreover, among 78 patients in the validation cohort the median time to treatment was 301 days for the 28 patients with the properties, compared with the 50 patients lacking these properties in whom the median was not reached but was already > 4 years.
Relationship between miR-181b expression values and time to treatment. Kaplan-Meier curves depict the clinical outcome of patients with CLL in which the 2 groups were separated on the basis of either a miRNA value dichotomized to ≤ 0.005 (A-B) or the properties defined on the basis of miR-181 expression values (C). The patient numbers were measured at the time intervals of 0, 500, 1000, 1500, 2000, and 2500 days. Log-rank P values are from Kaplan-Meier analysis.
Relationship between miR-181b expression values and time to treatment. Kaplan-Meier curves depict the clinical outcome of patients with CLL in which the 2 groups were separated on the basis of either a miRNA value dichotomized to ≤ 0.005 (A-B) or the properties defined on the basis of miR-181 expression values (C). The patient numbers were measured at the time intervals of 0, 500, 1000, 1500, 2000, and 2500 days. Log-rank P values are from Kaplan-Meier analysis.
Biologic effect of miR-181b in CLL
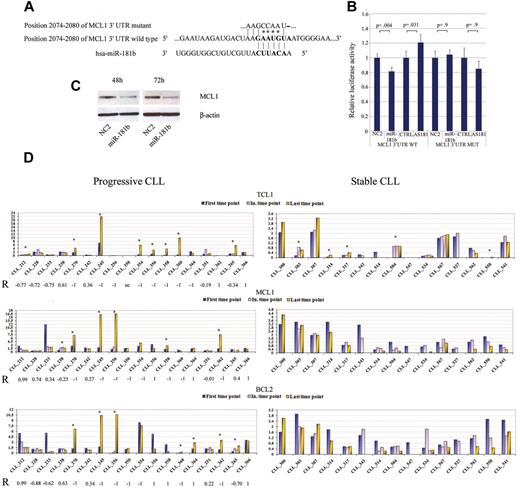
MiR-181b targets TCL114 and BCL215 genes, which are overexpressed in CLL.16-18 Because progression entails an accumulation of leukemic cells as a result of defects in apoptosis and rapid proliferation,19 we explored the possibility that miR-181b regulates the antiapoptotic factor myeloid cell leukemia sequence 1 (MCL1), which is a predicted target (http://www.targetscan.org). Overexpression of this miRNA by oligonucleotide transfection clearly decreased MCL1 protein levels in HeLa cells and inhibited the expression of a reporter vector carrying the MCL1 3′-UTR. Mutation of the predicted miRNA binding sites in the reporter vector abrogated this effect, indicating that miR-181b directly interacts with MCL1 3′-UTR. Contrarily, transfection of AMOs against miR-181b increased the expression of the reporter vector containing MCL1 3′-UTR but not that of the vector carrying the mutated sequence (Figure 4A-C). The protein levels of the 3 targets were then analyzed in samples at different time points obtained from patients with either progressive or stable disease. Considering a minimum fold-change of 2 times, protein expression levels of TCL1, MCL1, and BCL2 increased, respectively, in 5, 0, and 0 of 15 patients with a stable disease, and in 8, 6, and 7 of 18 patients with progressive disease over time, respectively (P = .7 for TCL1; P = .02 for MCL1; and P = .009 for BCL2 Fisher exact test; Figure 4D; supplemental Figure 2). However, in 14 patients with progressive disease, ≥ 1 of the 3 targets increased its expression over time. In several samples of patients with progressive disease, we also observed an inverse correlation between miR-181b and proteins expression (Figure 4D; R = correlation factor).
MiR-181b targets related CLL genes. (A) Putative binding site of miR-181b in MCL1 3′-UTRs (TargetScan5.1 Database). Asterisks indicate the nucleotides substituted in miR-181b–predicted target site to perform luciferase assay. (B) MCL1 3′-UTRs regulated by luciferase activity dependent on miR-181b in HeLa cell lines (WT, wild-type; MUT, mutant; P value t test). Error bars indicate SD (mean ± SD), n = 3. Firefly luciferase activity was normalized on Renilla luciferase activity of the gene included in the same vector. (C) Western blot analysis of MCL1 after either scrambled sequence or precursor-miR-181b transfection in HeLa cell line; β-actin has been used as loading control. Cells were collected after 48 and 72 hours of miRNA transfection. (D) Densitometric display of the TCL1, MCL1, and BCL2 Western blot analyses (supplemental Figure 2) normalized on β-actin on samples from patients with CLL with either progressive or stable disease. Asterisks indicate patients in which the protein expression of the miR-181b target genes increase. R indicates correlation factor by Pearson test; and In. time point, intermediate time point.
MiR-181b targets related CLL genes. (A) Putative binding site of miR-181b in MCL1 3′-UTRs (TargetScan5.1 Database). Asterisks indicate the nucleotides substituted in miR-181b–predicted target site to perform luciferase assay. (B) MCL1 3′-UTRs regulated by luciferase activity dependent on miR-181b in HeLa cell lines (WT, wild-type; MUT, mutant; P value t test). Error bars indicate SD (mean ± SD), n = 3. Firefly luciferase activity was normalized on Renilla luciferase activity of the gene included in the same vector. (C) Western blot analysis of MCL1 after either scrambled sequence or precursor-miR-181b transfection in HeLa cell line; β-actin has been used as loading control. Cells were collected after 48 and 72 hours of miRNA transfection. (D) Densitometric display of the TCL1, MCL1, and BCL2 Western blot analyses (supplemental Figure 2) normalized on β-actin on samples from patients with CLL with either progressive or stable disease. Asterisks indicate patients in which the protein expression of the miR-181b target genes increase. R indicates correlation factor by Pearson test; and In. time point, intermediate time point.
Discussion
The course of CLL is highly variable, and to date it is not possible to identify patients with a disease that will progress. In this study we investigated whether miRNAs play a role in CLL progression. We evaluated 2 independent cohorts of patients over time and found that the miR-181b expression values significantly decreased in samples obtained from patients with progressive disease but not in samples obtained from patients with stable disease. We also observed that several patients with CLL, mostly with progressive disease, had miR-181b expression values ≤ 0.005 at the initial sample. We measured the expression of miR-181b in 16 samples of B-CD19+ and B-CD19+/CD5+ cells obtained from healthy persons. None of these control samples had such low miR-181b expression (data not shown). The miR-181b value ≤ 0.005 was chosen as the low threshold in this study because it was observed in very few of the samples obtained from patients with CLL with stable disease in the training set, and this enabled us to discriminate, at least in part, the 2 manifestations of the disease. Presumably, those patients with low expression levels of miR-181b in the initial sample already had progressive, albeit early-stage, disease that later clinically manifested itself at later time points. Therefore, considering a decrease in the miR-181b expression values of ≥ 50% between sequential samples or a miR-181b value ≤ 0.005 at the starting point or both, we observed that the proportion of progressive patients who have these properties was significantly higher than the proportion for patients with stable disease, indicating that the miR-181b is a biomarker for the disease progression in CLL. To further corroborate our findings, we demonstrated a strong correlation between the low expression of miR-181b at the last time point and the risk of requiring treatment in both the training and the validation cohorts, which is consistent with our previous study on CLL with 17p deletion.9 Last time point was chosen for this analysis because we have shown a big difference at the ending than at the starting time points between the 2 subgroups with either progressive or stable disease. However, the drop of 50% occurs in many progressive cases, before the detection of the classic clinical parameters, which are represented by the last time point (Table S1). To date, the prediction of the risk of requiring treatment on the basis of only miR-181b expression values is not reliable in the laboratory practice. To address this point, we correlated samples with the previously defined properties and time to first treatment in a Cox regression analysis. We found that these patients have a risk of requiring treatment that is 5.8 times that of patients without such properties. This was the case in both validation and training sets, even though in the test cohort the result was not significant because of the small number of samples and 5 samples diagnosed as progressive and having the property were untreated at the last follow-up.
MiR-181b targets TCL1, whose enhanced expression in murine models leads to the aggressive form of CLL,20 and the antiapoptotic genes BCL2 and MCL1, that are up-modulated in CLL cells of patients with progressive disease. In this study we found that, although the MCL1 and BCL2 expression increased in samples from patients with progressive disease but not in samples from patients with stable disease over time, TCL1 increased in both sets, indicating a predominant role for this gene in the early phase of CLL. Overall, in 14 of 18 patients with a progressive disease, ≥ 1 of the 3 targets increased during disease progression, suggesting a functional role of this miRNA.
A larger prospective study is necessary to understand the effect of this miRNA on these genes, and the discovery of new targets may help to clarify how the lack of miR-181b contributes to the progression of CLL. We have shown that miR-181b also targets activation-induced cytidine deaminase (AID).21 Thus, loss of miR-181b will lead to overexpression of AID, leading to genomic instability and cancer progression.
Several markers have been identified in the past years to predict the course of the disease in CLL. In this study we only associated IGHV mutational status and ZAP-70 expression to miR-181b dysregulation; however, cytogenetics may also play a role in this process of miR-181b–related leukemia progression, but we do not have sufficient data in this particular cohort of patients to investigate this correlation.
In conclusion the novel finding of this research is that miR-181b is a unique biomarker for CLL because its expression can be monitored throughout the disease course of a patient and this change in the leukemic cells correlate with the overexpression of 4 genes with great significance in CLL and other cancers (ie, MCL1, TCL1, BCL2, and AID). Collectively, this information together with the analysis of stable prognostic markers (eg, ZAP-70 and IGHV mutation status) could help enable physicians to accurately stratify patients who have variable risks of requiring imminent therapy. To date, although treatment of patients in early stage finds no survival advantage and “wait and watch” is the standard care for patients with CLL,22 the development of more active therapies has rekindled interest in studying the benefit of early treatment for selected high-risk patients with CLL. Hence, in this new context the identification of new biomarkers predicting disease progression should be extremely useful.
In addition, given the relevance of the miR-181b in CLL and the promising features of microRNAs as therapeutic tools, once the molecular mechanism responsible for its action is identified, the restoration of the activity of miR-181b in CLL cells might have therapeutic potential for patients with progressive disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Teresa Monnett (The Ohio State University) for her excellent graphical assistance.
This work was supported by a grant from the National Cancer Institute (C.M.C.) and partially by the National Institutes of Health (grant PO1-CA81534 of the CLL Research Consortium).
National Institutes of Health
Authorship
Contribution: R.V. designed and performed the research, analyzed data, and wrote the paper; L.Z.R. contributed patient samples and data; A.V. performed the research and analyzed the data; C.T., S.V., and D.K.P. performed bioinformatics and statistical analysis; V.B. performed the research and analyzed the data; M.A. performed the research; T.J.K. contributed patient samples and data and helped design the research; and C.M.C. designed the research and wrote the paper. All authors critically reviewed and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Carlo M. Croce, Department of Molecular Virology, Immunology and Medical Genetics and Comprehensive Cancer Center, The Ohio State University, 460 W 12th Ave, Columbus, OH 43210; e-mail: carlo.croce@osumc.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal