Abstract
Deficiency in Msh2, a component of the mismatch repair (MMR) system, leads to an approximately 10-fold increase in the mutation frequency in most tissues. By contrast, Msh2 deficiency in germinal center (GC) B cells decreases the mutation frequency at the IgH V region as a dU:dG mismatch produced by AID initiates modifications by MMR, resulting in mutations at nearby A:T base pairs. This raises the possibility that GC B cells express a factor that converts MMR into a globally mutagenic pathway. To test this notion, we investigated whether MMR corrects mutations in GC B cells at a gene that is not mutated by AID. Strikingly, we found that GC B cells accumulate 5 times more mutations at a reporter gene than during the development of the mouse. Notably, the mutation frequency at this reporter gene was approximately 10 times greater in Msh2−/− compared with wild-type GC B cells cells. In contrast to the V region, the increased level of mutations at A:T base pairs in GC B cells was not caused by MMR. These results show that in GC B cells, (1) MMR functions normally at an AID-insensitive gene and (2) the frequency of background mutagenesis is greater in GC B cells than in their precursor follicular B cells.
Introduction
Mismatch repair (MMR) is an evolutionary conserved DNA repair pathway that is required to repair mutations that arise through DNA replication or other processes.1,2 MutS homologue 2 (Msh2) dimerizes with either Msh6 or Msh3 to recognize single base-pair mismatches or mismatches caused by insertion/deletions, respectively.2 After binding to the mismatch, Msh2 hydrolyzes ATP, which induces a change in the heterodimer that allows for the recruitment of downstream repair factors such as MutL homologue 1, postmeiotic segregation 2, and exonuclease 1 (Exo1).3,4 Because of Msh2's essential role in repairing DNA mutations, Msh2−/− mice have 5-fold, 11-fold, and 15-fold greater mutation frequencies in the brain, small intestine, and thymus, respectively,5 and are predisposed to cancer.6-8
During an immune response, Ig genes undergo somatic hypermutation (SHM) to produce high-affinity antibodies. SHM is initiated by activation-induced cytidine deaminase (AID),9 which deaminates cytidine molecules within the V region to produce dU:dG mispairs.10-12 The dU:dG mismatch is recognized by the Msh2/Msh6 heterodimer, but instead of repairing this mutation, MMR produces mutations.13-18 Mice deficient in Msh2, Msh6, postmeiotic segregation 2, MutL homologue 1, or Exo1 have an approximately 2-fold decrease in overall mutation frequency at the V region.13,14,16-18 In particular, there is an approximately10-fold decrease in mutations at A:T base pairs at the V region in MSH2-deficient mice, whereas the mutation frequency at G:C base pairs is unaltered.15-19 These results indicate that the MMR pathway is required to produce mutations at A:T base pairs in the V region. MMR proteins have also been implicated in mutagenic processes that cause disease, including trinucleotide-repeat diseases.20
Two sets of recent results suggest the possibility that the MMR system is generally defective in germinal center (GC) B cells. Ouchida et al21 reported that GC B cells have a generally greater mutation rate than other cells, as measured by the frequency of mutations on a lacZ transgene. Liu et al22 found that in Msh2 mutant mice, the mutation frequency of AID-sensitive genes was either lower or only slightly greater than in wild-type (WT) cells; they argued that the Msh2-dependent pathway was not functioning normally in GC cells. Together, these results would fit a simple unified model in which the expression of a GC/B cell–specific factor commandeers MMR and transforms this DNA repair pathway into a mutagenic pathway.
An alternative explanation is that the MMR pathway is only mutagenic at sites mutated by AID, which would argue against a factor that disrupts global MMR function in GC B cells. To explore this issue, we examined the impact of Msh2-deficiency at a lacI transgene that is not mutated by AID in GC B cells. Our results indicate that Msh2 efficiently repairs lacI mutations in all cell populations examined, including GC B cells. Thus, MMR functions normally in GC B cells except at AID-targeted loci.
Methods
Mice
Msh2+/− mice were kindly provided by Tak W. Mak (Ontario Cancer Institute). Big Blue C57BL/6 hemizygous male mice were purchased from Stratagene. Msh2−/− were bred with Big Blue (BB+) mice to generate BB+ Msh2−/− mice. All mice were on the C57BL/6 genetic background. Animals carrying the shuttle vector (BB+) and Msh2 WT, heterozygous, or homozygous for the “null allele” were identified with the use of 5′-ACCACACCTATGGTGTATGCA and 3′-CCTCTGCCGAAGTTGAGTAT primers for the PCR of the transgene (95°C for 3 minutes, and 30 cycles at 95°C for 30 seconds, 50°C for 60 seconds, 72°C for 60 seconds, and a final extension at 72°C for 10 minutes) and Msh2 PCR as previously described.23 Mice were housed in a specific pathogen-free facility. Approximately 3-month-old WT and Msh2−/− mice heterozygous for the lacI-transgene were immunized once intraperitoneally with 150 μL of hapten 4-hydroxy-3-nitrophenyl conjugated to chicken γ globulin (Bioresearch Technologies) and precipitated in 1:1 ratio with alum (Thermo Scientific). All animal study protocols were approved by the University of Toronto's Division of Comparative Medicine Committee.
Cells
BM cells were harvested from ∼ 3-month-old WT and Msh2−/− mice carrying the lacI transgene and differentiated into macrophages by culturing the cells in the presence of GM-CSF as described previously.24 Spleen cells were harvested from BB+ WT and BB+ Msh2−/− mice that were ∼ 3 months of age and heterozygous for the transgene 9-12 days after immunization. Each mouse was studied individually. Cells were treated with 1 mL of RBC lysis buffer (Sigma-Aldrich) for 3 minutes at room temperature. Cells were then blocked in PBS with 5% NMS (Jackson ImmunoResearch Laboratories) and anti-FcγIII/II antibody (2.4G2).
To visualize follicular B cells and GC B cells by flow cytometry, we used the following antibodies: anti–mouse B220 (Southern Biotech), anti–mouse Fas (clone 15A7, eBioscience), and anti–mouse GL7 (clone GL7, Biosciences Pharmingen). Approximately 5 × 106 follicular B cells and 2-5 × 105 GC B cells were sorted from each mouse on a FACSAria II (BD Biosciences) flow cytometer. Follicular B cells were defined as B220+GL7−Fas−, and GC B cells were defined as B220+GL7+Fas+. High-molecular-weight genomic DNA was isolated from these cells by the use of a standard genomic DNA isolation protocol.
Mutation analysis
Transgenic λ phage rescue was performed as described.25 Phage was packaged with the Stratagene Transpack Packaging Extract (Sigma-Aldrich). The dilution assay was performed on 10-cm plates with SCS-8 bacterial lawn, and the next day, the virus was plated on 15-cm plates in presence of X-gal. The number of blue plaques (mutants) over the total number of plaques indicates mutation frequency. Sectored plaques, arising from bacterial processing of DNA damage acquired within the murine host, were not included in the results (ie, only plaques that were at least 50% blue were counted). We confirmed that the mutation frequency was not affected by the amount of DNA in the preparation: We found that the mutation frequency from DNA isolated from 5 million and 0.5 million BB+ spleen cells was 7.5 × 10−5 and 6 × 10−5, respectively, indicating that the number of cells used to isolate DNA does not affect the overall mutation frequency. To sequence the lacI gene, the blue plaques were isolated in 100 μL of SM buffer and stored at 4°C. A total of 1.5 μL of virus was used for the PCR experiment in the section “PCR amplification and sequence analysis.” If the blue plaques were contaminated by colorless plaques, we purified them by plating a small volume of virus in a 10-cm plate and isolating blue plaques that were not contaminated.
PCR amplification and sequence analysis
LacI mutants were chosen at random from indicated cell types derived from Msh2−/− and control animals for DNA sequencing. These blue plaques were amplified by PCR for the lacI region and LacZ promoter with primers previously reported (55D and 1283U).26 PCR products were purified by a MultiScreen PCR96 Filter Plate (Millipore) and sent to Macrogen for sequencing with the use of primers 55D and 1232U.26 Alignment and analysis of unique mutations was performed by SeqMan.
Reverse transcriptase
RNA was extracted from splenic B cells from a 3-month-old BB+ and a WT mouse with TRIzol (Invitrogen) per the manufacturer's instructions. RNA was treated with DNase I (Fermentas) and reverse-transcribed with the use of superscript III reverse transcriptase (Invitrogen) and random primers following the company's protocol. The resulting cDNA was diluted 1 in 5, 1 in 25, and 1 in 125 dilution, and 1.5 μL of each dilution was used for the PCR. Gene expression was examined by the use of appropriate GAPDH-specific primers (forward primer AACTTTGGCATTGTGGAAGG and reverse primer GGAGACAACCTGGTCCTCAG). The lacI gene was amplified by PCR with primers 331D and 803U.26
Data analysis
Data were analyzed with GraphPad Prism Version 5.0, in which we performed 2-tailed Student t tests.
Results
The lacI mutation frequency is elevated in GC B cells from Msh2−/− mice relative to WT mice
AID deficiency leads to significant alterations in the GCs in mice9,27 and might therefore complicate the interpretation of any results. For this reason, we measured the effects of Msh2 deficiency in AID-sufficient mice by using the lacI-containing transgenic mouse mutation detection system (Big Blue, hereafter referred to as BB+). This transgene lacks eukaryotic transcriptional promoters and is expected not to be transcribed and consequently is expected not to be a substrate for AID, which generally requires a high rate of transcription.28-31
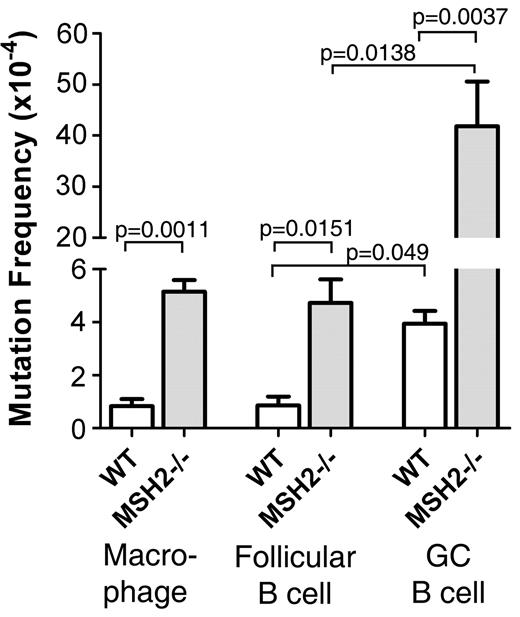
The lacI transgene is part of a shuttle vector that can be recovered as phage particles giving rise to white plaques (unmutated) or blue plaques (lacI mutated), thereby yielding information regarding tissue specific- or cell-specific mutation frequencies. To validate our system, we first examined the mutation frequency in macrophages. Macrophages were generated from the BM of WT and Msh2−/−BB+ mice. We found a 6-fold increase in the mutation frequency in Msh2−/− mice compared with WT controls (Figure 1, P = .0011). This result confirms the important role of MMR in reducing the cellular mutation load.
Frequency of mutations at the lacI gene in macrophages, follicular B cells, and GC B cells from WT and Msh2−/− mice. Figure is plotted with data from supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Statistical analysis involved a 2-tailed t test.
Frequency of mutations at the lacI gene in macrophages, follicular B cells, and GC B cells from WT and Msh2−/− mice. Figure is plotted with data from supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Statistical analysis involved a 2-tailed t test.
To determine whether MMR is mutagenic at the lacI gene in GC B cells, it was necessary to know the mutation frequency at the stage of differentiation before GC B cells to determine whether mutations were inherited from follicular B cells or whether they occurred at the GC B-cell stage. Therefore, we measured the mutation frequency in follicular B cells from WT and Msh2−/−BB+ mice. At 10 days after mice were immunized with hapten 4-hydroxy-3-nitrophenyl conjugated to chicken γ globulin, B220+GL7−Fas− and follicular B cells from spleen were sorted and analyzed for mutations in the lacI gene. We found that Msh2−/− follicular B cells had a significant 5.5-fold increase in the mutation frequency compared with WT follicular B cells (Figure 1 and supplemental Table 1).
From the same immunized WT and Msh2−/− mice, we prepared GC B cells from spleen by sorting for B220+GL7+Fas+ and analyzed the mutation frequency of the lacI gene. If MMR repairs mutation at the lacI gene in GC B cells, we would expect to observe an increase in mutations in Msh2−/− mice. Indeed, we observed a 10-fold increase in the lacI mutation frequency in Msh2−/− GC B cells compared with WT GC B cells (Figure 1 and supplemental Table 1). This result is in striking contrast to the ∼ 2-fold decrease in mutation frequency observed at the V region of the IgH gene in Msh2−/− mice.17 Importantly, there was a 4.6-fold and an 8.8-fold increase in lacI mutation frequencies in GC B cells relative to follicular B cells from WT and Msh2−/− mice, respectively (Figure 1 and supplemental Table 1). This result indicates that > 80% of mutations in GC B cells were not inherited from follicular B cells but occurred at the GC B-cell stage. Collectively, these results show that the MMR system repairs mutations that arise at the lacI gene in GC B cells, indicating that MMR functions normally at this genetic locus in GC B cells.
AID does not mutate the lacI transgene
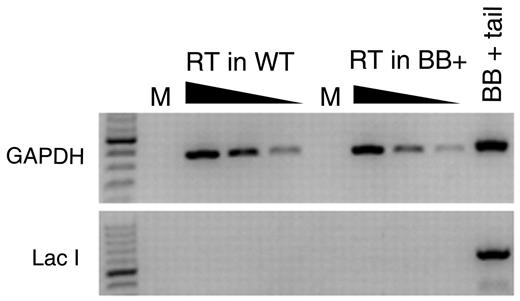
We selected the lacI-containing transgenic BB+ mice because they are expected to be refractory to AID because they lack eukaryotic transcriptional promoters. However, it is possible that the transgene was inserted into a transcriptionally active site in B cells and might be subjected to AID activity. Indeed, the significant increase in mutation frequency from follicular B cells to GC B cells could be because of AID's promiscuous activity.22,32-37 To determine whether the lacI transgene could be a target of AID, we first tested whether the lacI gene is transcribed in B cells. We carried out a reverse-transcriptase PCR by using random hexamers (in case the lacI transcript is not polyadenylated) on RNA isolated from splenocytes from BB− and BB+ WT mice. Although GAPDH was readily amplified from these cDNA preparations, we could not amplify the lacI transcript from either BB− or BB+ splenocytes (Figure 2), indicating that this transgene is not detectably transcribed in splenocytes.
Measurement of lacI mRNA in splenocytes. RNA was extracted from splenocytes harvested from BB+ and BB− mice and used to make cDNA with random hexamers. PCR amplifications for GAPDH and for lacI were performed on 5-fold serial dilutions of cDNA. Genomic DNA from a BB+ mouse was used as a positive control in the GAPDH and lacI PCR. M indicates Mock; and RT, reverse transcription.
Measurement of lacI mRNA in splenocytes. RNA was extracted from splenocytes harvested from BB+ and BB− mice and used to make cDNA with random hexamers. PCR amplifications for GAPDH and for lacI were performed on 5-fold serial dilutions of cDNA. Genomic DNA from a BB+ mouse was used as a positive control in the GAPDH and lacI PCR. M indicates Mock; and RT, reverse transcription.
To more precisely investigate whether the lacI transgene was mutated by AID, we sequenced blue plaques from GC B cells to determine whether mutations occurred at WRCY motifs (W = A or T, R = A or G, Y = T or C). WRCY is a SHM hotspot sequence that is preferentially mutated by AID. A total of 16% and 26% of mutations within the IgH JH2-JH4-region occur within WRCY motifs in WT and Msh2−/− mice, respectively.17 However, the frequency of WRCY mutations within the lacI gene in WT and Msh2−/− GC B cells was only 7% and 4%, respectively (Table 1), although there are more WRCY motifs in the lacI gene than in the IgH JH2-JH4-region (ie, 60.3 WRCY/kb in the lacI gene vs 50.8 WRCY/kb in the IgH JH2-JH4-region). Collectively, these results show that the increase in mutation frequency in GC B cells compared with follicular B cells is not because of AID.
Characteristics of mutations within the lacI gene in wild-type and Msh2−/− mice
| . | BB+MSH2+/+ . | BB+MSH2−/− . | ||||
|---|---|---|---|---|---|---|
| Macrophage (n = 3),* n (%)† . | Follicular B cell (n = 3), n (%) . | GC B cell (n = 4), n (%) . | Macrophage (n = 3), n (%) . | Follicular B cell (n = 3), n (%) . | GC B cell (n = 3), n (%) . | |
| Sequences analyzed | 11 | 2 | 35 | 39 | 31 | 23 |
| Mutated sequences | 9 | 2 | 27 | 28 | 21 | 21 |
| Total mutations‡ | 7 (100) | 2 (100) | 30 (100) | 33 (100) | 20 (100) | 24 (100) |
| Base substitutions | 6 (86) | 2 (100) | 30 (100) | 27 (82) | 19 (95) | 23 (96) |
| Mutations at C:G | 6 (86) | 2 (100) | 17 (57) | 20 (61) | 13 (65) | 11 (46) |
| Mutations at A:T | 0 | 0 | 13 (43) | 7 (21) | 6 (30) | 12 (50) |
| Mutations at CpG§ | 3 (43) | 2 (100) | 9 (30) | 13 (40) | 13 (65) | 6 (25) |
| Mutations at WRCY¶ | 2 (29) | 0 | 2 (7) | 1 (3) | 0 | 1 (4) |
| Mutations at WA | 0 | 0 | 9 (30) | 4 (12) | 5 (25) | 7 (29) |
| Insertions/deletions | 1 (14) | 0 | 0 | 6 (18) | 1 (5) | 1 (4) |
| Insertions | 0 | 0 | 0 | 2 (6) | 0 | 0 |
| Deletions | 1 (14) | 0 | 0 | 4 (12) | 1 (5) | 1 (4) |
| . | BB+MSH2+/+ . | BB+MSH2−/− . | ||||
|---|---|---|---|---|---|---|
| Macrophage (n = 3),* n (%)† . | Follicular B cell (n = 3), n (%) . | GC B cell (n = 4), n (%) . | Macrophage (n = 3), n (%) . | Follicular B cell (n = 3), n (%) . | GC B cell (n = 3), n (%) . | |
| Sequences analyzed | 11 | 2 | 35 | 39 | 31 | 23 |
| Mutated sequences | 9 | 2 | 27 | 28 | 21 | 21 |
| Total mutations‡ | 7 (100) | 2 (100) | 30 (100) | 33 (100) | 20 (100) | 24 (100) |
| Base substitutions | 6 (86) | 2 (100) | 30 (100) | 27 (82) | 19 (95) | 23 (96) |
| Mutations at C:G | 6 (86) | 2 (100) | 17 (57) | 20 (61) | 13 (65) | 11 (46) |
| Mutations at A:T | 0 | 0 | 13 (43) | 7 (21) | 6 (30) | 12 (50) |
| Mutations at CpG§ | 3 (43) | 2 (100) | 9 (30) | 13 (40) | 13 (65) | 6 (25) |
| Mutations at WRCY¶ | 2 (29) | 0 | 2 (7) | 1 (3) | 0 | 1 (4) |
| Mutations at WA | 0 | 0 | 9 (30) | 4 (12) | 5 (25) | 7 (29) |
| Insertions/deletions | 1 (14) | 0 | 0 | 6 (18) | 1 (5) | 1 (4) |
| Insertions | 0 | 0 | 0 | 2 (6) | 0 | 0 |
| Deletions | 1 (14) | 0 | 0 | 4 (12) | 1 (5) | 1 (4) |
GC indicates germinal center.
Denotes the number of animals assayed in each group.
Percent mutations were calculated from total mutations (ie, base substitutions and insertion/deletion mutations).
Only unique mutations were considered.
Mutations at CpG include transition and transversion mutations at dC (on either DNA strand).
Mutations at WRCY occur within the underlined dC nucleotide of this motif on either DNA strand.
The increased mutation frequency at A:T base pairs in GC B cells is not because of Msh2
In a separate approach to investigate whether MMR is inducing mutations at A:T base pairs within the lacI gene as it does at the V region, we characterized the mutations at the lacI gene by sequencing blue plaques from macrophages, follicular B cells, and GC B cells. We found that the majority of lacI mutations in macrophages and follicular B cells occurred at G:C base pairs (Figures 3–4) and that ∼ 50% of all mutations occurred within CpG dinucleotides in both WT and Msh2−/− mice (Table 1). CpG dinucleotides frequently undergo spontaneous deamination,38 and thus our data suggest that this is a dominant type of mutation during hematopoiesis or the development of the mouse, as previously shown.39,40
The mutation spectrum at the lacI gene in macrophages, follicular B cells, and GC B cells from WT and Msh2−/− mice. (A) The mutation spectrum in the indicated cell types and genotypes showing the original nucleotide (left) and the mutation (top). Most of the mutations in macrophages and follicular B cells are G:C to A:T transition mutations, whereas in GC B cells, there is a high proportion of mutations at A:T base pairs. (B) The percentage of G:C and A:T mutations are plotted for the indicated cell types and genotypes. This figure was plotted from the data shown in panel A. Statistical analysis was carried out with a 2-tailed t test.
The mutation spectrum at the lacI gene in macrophages, follicular B cells, and GC B cells from WT and Msh2−/− mice. (A) The mutation spectrum in the indicated cell types and genotypes showing the original nucleotide (left) and the mutation (top). Most of the mutations in macrophages and follicular B cells are G:C to A:T transition mutations, whereas in GC B cells, there is a high proportion of mutations at A:T base pairs. (B) The percentage of G:C and A:T mutations are plotted for the indicated cell types and genotypes. This figure was plotted from the data shown in panel A. Statistical analysis was carried out with a 2-tailed t test.
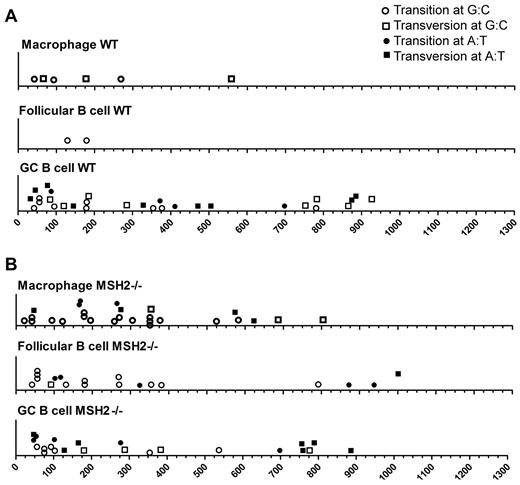
The locations of base substitutions in the lacI gene in the WT and Msh2−/− mice. (A) Position of mutations within the lacI gene in the indicated cell types from WT mice. (B) Position of mutations within the lacI gene in the indicated cell types from Msh2−/− mice.
The locations of base substitutions in the lacI gene in the WT and Msh2−/− mice. (A) Position of mutations within the lacI gene in the indicated cell types from WT mice. (B) Position of mutations within the lacI gene in the indicated cell types from Msh2−/− mice.
Within GC B cells, we observed that the spectrum of mutations was different from follicular B cells, with a greater proportion of mutations at A:T base pairs observed (Figures 3–4, Table 1). Combined with the 5-fold increase in the overall mutation frequency in GC B cells (Figure 1), these data suggest that mutations at both G:C and A:T base pairs have increased relative to follicular B cells but with a slight bias toward the latter. Ouchida et al21 previously observed an increase in the mutation frequency at A:T base pairs in GC B cells at a different reporter gene, and some of these mutations were caused by DNA polymerase η. Because SHM at A:T base pairs within the V region are because of Msh2 and its recruitment of DNA polymerase η,41-43 we examined whether mutations at A:T base pairs within the lacI gene were reduced in Msh2−/− mice. However, we found that the fraction of mutations at A:T and G:C base pairs at the lacI gene was not altered in Msh2−/− GC B cells (Figures 3–4, Table 1). Furthermore, we found that ∼ 30% of all mutations within the lacI gene in WT and Msh2−/− GC B cells occur within WA dinucleotides (Table 1), a motif that is preferentially mutated by DNA polymerase η. This value is greater than the expected frequency of 15% (151 WA motifs/kb in the lacI gene) for a mutation process that displays no sequence bias. Thus, in contrast to the V-region, some of the mutations at the lacI gene are likely because of polymerase η but independent of Msh2.
Discussion
The finding that the MMR pathway is required to produce mutations within the immunoglobulin V region during SHM, and hence do the exact opposite of what this DNA repair pathway is intended to do, is currently unexplained. What is clear is that mutations at A:T base pairs within the V-region require the actions of the Msh2/Msh6 heterodimer,14,16,17,19 Exo1,18 PCNA ubiquitination,44 and DNA polymerase η.41-43 In addition, non-Ig genes that were mutated by AID also showed a decrease in mutation frequency in the absence of Msh2.22 These data suggest either that (1) Msh2 produces mutations at genes mutated by AID or (2) Msh2 is mutagenic throughout the genome.
The authors of previous studies have shown that there is increased expression of MMR proteins in GC B cells45,46 and that proteins isolated from GC B cells can bind to a mismatched substrate and repair it in vitro.46 Although these results suggest that MMR proteins are actively participating in GC B-cell physiology, they do not address the specific question whether MMR has been co-opted to produce mutations throughout the genome. To resolve this issue, we examined whether mutations depend on Msh2 at a non-AID target in GC B cells. Indeed, we found that Msh2 does not induce mutations at the lacI gene in GC B cells and instead reduces the mutation load at this genetic locus. These results show that the MMR system functions normally in GC B cells at an AID-insensitive site, indicating that something interferes with normal MMR activity at some or all genes that are mutated by AID.
As to why MMR is mutagenic at sites of AID activity is still unknown. The finding that DNA polymerase η and Msh2 function together to induce SHM at A:T base pairs was surprising because this translesional polymerase is not normally recruited by MMR.47 This finding suggests that a replicative blocking lesion might stimulate recruitment of this translesional polymerase during repair of the AID-induced dU:dG mismatch. Indeed, we previously observed that mutations at A:T base pairs were closely linked to transversion mutations at cytidine molecules, suggesting that an abasic site was stimulating error-prone MMR.48 Furthermore, deficiency in UNG or MMR reduced this association, indicating that an abasic site generated by UNG within the MMR tract recruits DNA polymerase η, which then introduces mutations at A:T base pairs.48 A similar finding was reported by Krijger et al.49 As to why UNG interferes with MMR at the V-region and not elsewhere is still unclear, but one possibility is that Msh2/6 directs repair of the strand opposite to the AID-induced dU-containing strand, exposing the dU to UNG attack.48 This could come about by the asynchronous replication of the V-region in which Okazaki fragments preferentially occur on the top strand because of an origin of replication downstream of the V region.50 Because MMR preferentially excises the lagging strand,51 and if AID mutates the bottom strand, this would create the aforementioned scenario, leading to the concerted activity of MMR and UNG in stimulating mutations at A:T base pairs.
We observed that the background level of mutation increased by 5-fold from the follicular B-cell stage to the GC B-cell stage. Ouchida et al21 instead observed an approximately 2-fold increase in the background level of mutation from non-GC B cells to GC B cells. Differences in the reporter gene used and/or in the methodology used to analyze mutation frequencies might account for this discrepancy. For example, GC B-cell populations were pooled from different animals in the previous study21 whereas we examined GC B-cell populations from individual mice. Nevertheless, the increase in mutation frequency that we observed is remarkable when one considers that the level of mutations that accumulated from the fertilized egg, that is, when the lacI transgene was cloned, to the follicular B-cell stage from a 3-month-old mouse is 5 times less than the background mutation frequency that occurs in a 10-day GC reaction.
One possible explanation for this increase in background mutation frequency is the significant replication burst that occurs in GC B cells. However, considering that the replication error rate in mice is ∼ 10−9 mutations/base pair/cell division,52 if replication was the only source of error, a GC B cells would require ∼ 300 000 cell divisions in 10 days (to go from 0.86 × 10−4 in the follicular B-cell compartment to 3.94 × 10−4 in the GC B-cell compartment). This scenario is clearly impossible and suggests that the replication error-rate has increased or that an active non-AID mutagenic process is ongoing during the GC reaction (eg, reactive oxygen species). The high background mutation frequency in GC B cells coupled with AID-induced chromosomal translocations53 might explain why greater than 75% of all hematologic malignancies derive from GC or post-GC B cells.54
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Maribel Berru and Ashley Weiss for technical help, Dr Marc Shulman for critically reading the manuscript, and the Martin laboratory for helpful discussions. They also are grateful to Dr Tak Mak for the Msh2−/− mice.
This research is supported by studentships from the Canadian Institutes of Health Research to B.G. and B.B., and a grant from the Canadian Institutes of Health Research (MOP66965) to A.M., who is supported by a Canada Research Chair tier II award.
Authorship
Contribution: B.G. designed the research, analyzed data, and wrote manuscript; A.B., R.M.N., and B.B. designed the research and analyzed data; and A.M. designed the research and wrote manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alberto Martin, Department of Immunology, University of Toronto, Medical Sciences Building 7302, Toronto, ON, Canada, M5S 1A8; e-mail: alberto.martin@utoronto.ca.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal