Abstract
The class III receptor tyrosine kinase (RTKIII) Fms-like tyrosine kinase receptor 3 (Flt3) and its cytokine ligand (FL) play central roles in hematopoiesis and the immune system, by establishing signaling cascades crucial for the development and homeostasis of hematopoietic progenitors and antigen-presenting dendritic cells. However, Flt3 is also one of the most frequently mutated receptors in hematologic malignancies and is currently a major prognostic factor and clinical target for acute myeloid leukemia. Here, we report the structural basis for the Flt3 ligand-receptor complex and unveil an unanticipated extracellular assembly unlike any other RTKIII/V complex characterized to date. FL induces dimerization of Flt3 via a remarkably compact binding epitope localized at the tip of extracellular domain 3 of Flt3, and it invokes a ternary complex devoid of homotypic receptor interactions. Comparisons of Flt3 with homologous receptors and available mutagenesis data for FL have allowed us to rationalize the unique features of the Flt3 extracellular assembly. Furthermore, thermodynamic dissection of complex formation points to a pronounced enthalpically driven binding event coupled to an entropic penalty. Together, our data suggest that the high-affinity Flt3:FL complex is driven in part by a single preformed binding epitope on FL reminiscent of a “lock-and-key” binding mode, thereby setting the stage for antagonist design.
Introduction
Hematopoiesis is a finely regulated process during which diverse cell types originating from a limited and self-renewing population of hematopoietic stem cells (HSCs) are stimulated to proliferate and differentiate to create the cellular repertoire that sustains the mammalian hematopoietic and immune systems.1
The Fms-like tyrosine kinase receptor 3 (Flt3) is the most recent addition to the diverse family of hematopoietic receptors. Flt3 is activated on HSCs and early myeloid and lymphoid progenitors by its cognate ligand (FL), to initiate downstream signaling via the phosphatidylinositol 3-kinase/AKT and the Ras/Raf/extracellular signal-regulated kinase pathways.2,3 Consistent with the narrow expression profile of Flt3 in the bone marrow environment, signaling via the Flt3 ligand-receptor complex primarily impacts early hematopoiesis, particularly the proliferation and development of HSC and B-cell progenitors.2,4 In recent years, Flt3 and FL emerged as potent regulators of dendritic cell (DC) development and homeostasis,5-7 and DC-mediated natural killer cell activation,8 thereby gaining an important role at the interface of innate and acquired immunity and in cancer immunotherapy.9,10 Notably, Flt3/FL-driven DC generation yields both classic and plasmacytoid DCs from bone marrow progenitors regardless of myeloid or lymphoid commitment, a property that is currently unmatched by any other receptor-cytokine system relevant for DC physiology.11,12
Flt3 is a class III receptor tyrosine kinase (RTKIII) together with the prototypic platelet–derived growth factor receptors-α/β, colony-stimulating factor 1 receptor (CSF-1R), and KIT.13 Thus, Flt3 has been predicted to display a modular structure featuring an extracellular segment with 5 immunoglobulin (Ig)–like domains (residues 27-543), a single transmembrane (TM) helix (residues 544-563), a cytoplasmic juxtamembrane domain ([JM]; residues 572-603), and a split intracellular kinase module (residues 604-958). The RTKIII family is closely related to the RTKV family of vascular endothelial growth factor receptors (VEGFRs), which have 7 extracellular Ig-like domains. The hallmark of RTKIII/V signaling lies in the activation of the extracellular receptor segments on binding of the cognate cytokines, followed by intermolecular autophosphorylation and activation of the intracellular kinase domains.14
Besides the clear role of Flt3 signaling in hematopoiesis and immune system development, overexpression of wild-type or oncogenic forms of Flt3 have been implicated in several hematopoietic malignancies,2,15 and inflammatory disorders.16 In particular, internal tandem duplication in the JM region or point mutations in the kinase activation loop occur in 35% of patients with acute myeloid leukemia (AML), resulting in constitutive activation of the receptor and uncontrolled proliferation of hematopoietic precursors.2,3,17-19 Such mutation fingerprints have established Flt3 as the predominant prognostic factor in AML cases20 and have rationalized the targeting of Flt3 in a clinical setting.2,3,21
Although the cellular and physiologic role of the Flt3 ligand-receptor interaction has been featured prominently in the biomedical literature over the past 2 decades, the Flt3 signaling complex has remained uncharacterized at the molecular and structural levels. Such insights are the missing link to the structural and functional diversity of RTKIII/V extracellular complexes and would help provide a nearly complete picture of the entire Flt3 signaling complex given the available structure of the Flt3 intracellular kinase domains.22 A recent flurry of studies of RTKIII/V extracellular complexes led to a structural paradigm for RTKIII/V activation, whereby the receptors bind via their amino-terminal (N-terminal) Ig-like domains to the activating dimeric cytokine and concomitantly make homotypic contacts between their membrane-proximal domains.23-30 A universal feature of all characterized RTKIII/V complexes thus far is that the cytokine-binding epitope is distributed equally between extracellular domains 2 and 3, covering ∼ 2000 Å2 of surface area, and that homotypic receptor-receptor interactions are mediated by well conserved residues in the membrane-proximal domains (D4 in RTKIII and D7 in RTKV). Nonetheless, Flt3 seems to be an outlier among RTKIII/V receptors because of several unique features in its extracellular segment,31,32 thus raising the question whether the current structural paradigm could be extrapolated to Flt3. Notably, Flt3 exhibits intragenic homology relating extracellular domains 1 and 4, and domains 2 and 5, indicative of an ancient internal duplication event during evolution. Furthermore, Flt3 contains 12 additional cysteines that are not present in any of the homologous receptors, and it has a unique N-terminal sequence of 50 amino acids preceding Ig-like domain 1. Interestingly, a fully functional splice variant of murine Flt3 lacks extracellular domain 5 entirely, indicating that the domain most proximal to the cell membrane is not critical for receptor activation,33 contrary to other RTKIII/V receptors.28,34
Here, we provide the structural basis of extracellular complex formation between Flt3 and its cognate cytokine. Our study establishes the uniqueness of Flt3 within the RTKIII/V family and provides a reference platform for further structure-function studies and antagonist design.
Methods
Summary
To support structural and biophysical studies of human Flt3 ectodomain complexes, we designed a series of constructs for recombinant human Flt3 ectodomains (Flt3D1 [residues 27-161], Flt3D1-D2 [residues 27-244], Flt3D1-D3 [residues 27-346], Flt3D1-D4 [residues 27-434], and Flt3D1-D5 [residues 27-541]), based on intron-exon boundaries and sequence alignments with homologous receptors. Faced with prohibitively poor protein yields (100-200 μg/L medium), we established tetracycline-inducible cell lines in HEK293S cells deficient in N-acetylglucosaminyltransferase I (HEK293S GnTI−/−)35 that could secrete the target ectodomain variants with homogeneous glycosylation (Man5GlcNAc2) to milligram amounts.36 The yields and stability of Flt3D1-D3 and Flt3D1-D2 were much lower than for all other constructs. Ternary Flt3:FL complexes were prepared for structural studies by mixing purified Flt3 ectodomains36 with a molar excess of recombinant human FL37 followed by preparative size-exclusion chromatography (SEC). The occupancy of N-linked glycosylation sites and the disulfide bond network of Flt3D1-D5 were determined by mass spectrometry. Purified Flt3D1-D4:FL and Flt3D1-D5:FL complexes were crystallized,36 and the crystal structures were determined and refined to 4.3- and 7.8-Å resolution, respectively. Additional structural studies on Flt3D1-D5:FL were performed via negative-stain electron microscopy (EM) and small-angle X-ray scattering (SAXS). Binding studies of extracellular complex formation were carried out via isothermal titration calorimetry (ITC). A detailed account of all methods relevant to this study is provided in supplemental Data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Crystallographic data deposition
Coordinates and structure factors for the Flt3D1-D4:FL and Flt3D1-D5:FL complexes have been deposited in the Protein Data Bank ([PDB]; www.rcsb.org) and are accessible through accession numbers 3QS7 and 3QS9, respectively.
Results
Isolation of recombinant Flt3 ectodomain complexes and thermodynamic binding profile of complex formation
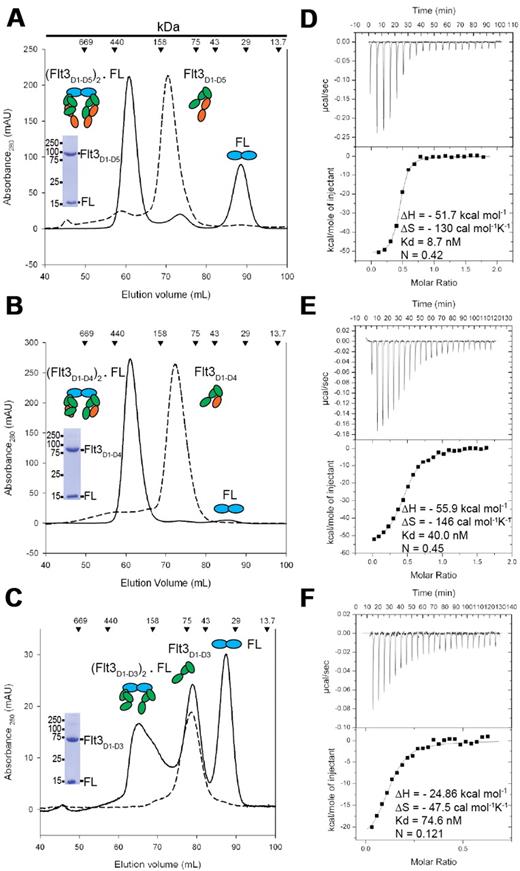
High-affinity complexes of purified glycosylated Flt3D1-D5, Flt3D1-D4, and Flt3D1-D3 with recombinant human FL produced in Escherichia coli,37 consistent with bivalent binding of FL to each of the ectodomain constructs, were initially established by analytical SEC. Subsequent batches for structural studies were obtained via preparative SEC in the presence of excess molar amounts of purified FL (Figure 1A-C). The elution profiles for all 3 ectodomain complexes were indicative of ligand-induced receptor dimerization. In contrast to Flt3D1-D5 and Flt3D1-D4, preparations of recombinant Flt3D1-D3 consistently contained a significant portion of receptor that was incapable of binding the ligand even in the presence of excess molar amounts of FL (Figure 1C). Conversely, excess molar amounts of Flt3D1-D3 did result in a complete titration of FL toward complex formation. Alternatively, we were not able to observe complex formation for Flt3D1 and Flt3D1-D2 via SEC, providing direct evidence that these ectodomain constructs do not carry a high-affinity ligand-binding site.
FL binds bivalently to Flt3 ectodomain variants to form high-affinity complexes. (A-B) Isolation of Flt3D1-D5:FL and Flt3D1-D4:FL by SEC. Also shown are Coomassie-stained sodium dodecyl sulfate–polyacrylamide gel electrophoresis strips corresponding to the peak fraction of the isolated complexes. (C) SEC on the Flt3D1-D3:FL mixture at the end of an ITC experiment, showing that a large amount of Flt3D1-D3 remains in the unbound form. Identical elution profiles were obtained in standard SEC experiments as well, in the presence of a large molar excess of FL. (D-F) Binding isotherms and thermodynamic parameters of FL binding to Flt3 ectodomains obtained by ITC. All ITC experiments were carried out by titrating recombinant human Flt3 ectodomain variants with FL.
FL binds bivalently to Flt3 ectodomain variants to form high-affinity complexes. (A-B) Isolation of Flt3D1-D5:FL and Flt3D1-D4:FL by SEC. Also shown are Coomassie-stained sodium dodecyl sulfate–polyacrylamide gel electrophoresis strips corresponding to the peak fraction of the isolated complexes. (C) SEC on the Flt3D1-D3:FL mixture at the end of an ITC experiment, showing that a large amount of Flt3D1-D3 remains in the unbound form. Identical elution profiles were obtained in standard SEC experiments as well, in the presence of a large molar excess of FL. (D-F) Binding isotherms and thermodynamic parameters of FL binding to Flt3 ectodomains obtained by ITC. All ITC experiments were carried out by titrating recombinant human Flt3 ectodomain variants with FL.
Characterization of Flt3 extracellular complexes by ITC led to several consensus observations (Figure 1D-F). First, all 3 characterized complexes exhibit high-affinity binding characterized by a strongly exothermic enthalpic term coupled to an entropic penalty. Second, FL exhibits bivalent binding to both Flt3D1-D5 and Flt3D1-D4 (Stoichiometry [n] = 0.5, 2 molecules of Flt3 to 1 molecule of FL). We note that the observed stoichiometry for the FL:Flt3D1-D3 interaction (n = 0.12) is consistent with our chromatographic data (Figure 1C). Furthermore, the ITC data clearly show that Flt3D1-D3 is capable of a high-affinity ternary complex just like the larger ectodomain constructs and that only ∼ 25% of recombinant Flt3D1-D3 may adopt an active conformation. Notably, the sequential exclusion of the membrane-proximal domains Flt3D4 and Flt3D5 leads to a very modest decrease in affinity (Kd [Flt3D1-D5:FL] = 8.7nM; Kd [Flt3D1-D4:FL] = 40nM; and Kd [Flt3D1-D3:FL] = 74.6nM), whereas the thermodynamic profiles remain similar (Figure 1D-F). Together, our data suggest that the membrane-proximal module Flt3D4-D5 does not contribute significantly to the overall stability of the complex.
Overall structure of the Flt3D1-D4:FL complex
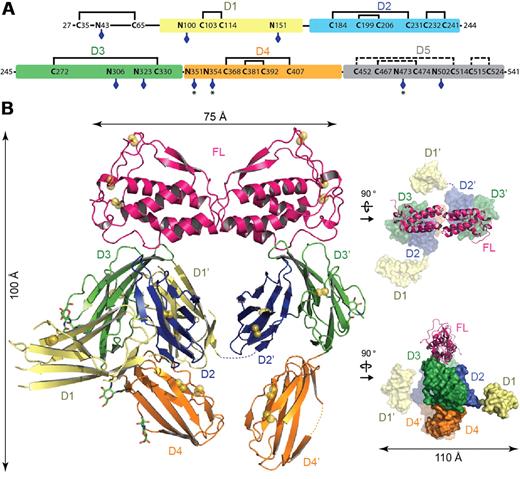
The crystal structure of the Flt3D1-D4:FL complex was determined to 4.3-Å resolution based on data obtained from a large-scale screening of crystals. Confronted with the recurring poor diffraction quality of Flt3D1-D4:FL crystals derivatized with heavy atoms and selenomethionine-labeled Flt3D1-D4, we successfully combined molecular replacement strategies relying on the high-resolution structure of human FL38 and a homology model for Flt3D330 (Table 1), with phase improvement protocols39 exploiting the noncrystallographic symmetry and high solvent content of the crystals. Such approaches combined with crystallographic refinement using information from high-resolution structures have recently emerged as a powerful option in macromolecular structure determination at low resolution.40-42 The ensuing electron density maps were exceptionally revealing and contained contiguous electron density for several unmodeled receptor domains, including direct crystallographic evidence for N-linked glycans (supplemental Figure 1A). To facilitate chain tracing we determined the atypical disulfide bond network of Flt3 as well as the actual number of N-linked glycosylation sites in extracellular Flt3 by mass spectrometry. We could confirm that all 9 N-linked glycosylation sites are at least partially occupied and that all cysteine residues present in Flt3D1-D4 are engaged in disulfide bonds (Figure 2A; supplemental Table 1).
X-ray data collection and refinement statistics
| . | Flt3D1-D4:FL . | Flt3D1-D5:FL . |
|---|---|---|
| Data collection | ||
| Source, wavelength (Å) | ESRF ID23-1, 0.9762 | ESRF ID23-1, 1.0762 |
| Detector | ADSC-Q315R | ADSC-Q315R |
| Resolution (Å) | 40.00-4.30 (4.45-4.30) | 35.00-7.80 (8.00-7.80) |
| Space group | P21 | P21 |
| Unit cell parameters | ||
| a | a = 103.89 | a = 124.75 |
| b | b = 146.26 | b = 153.55 |
| c | c = 105.95 | c = 133.87 |
| α = γ = 90°, β = 109.7° | α = γ = 90°, β = 94.6° | |
| Wilson B (Å2) | 135 | 401 |
| Unique reflections | 20 184 (1942) | 5656 (382) |
| Redundancy | 3.8 (3.8) | 3.4 (3.2) |
| Completeness (%) | 98.8 (98.9) | 96.6 (92.9) |
| Rmeas (%)* | 10.8 (75.9) | 12.8 (80.7) |
| Average I/σ(I) | 12.04 (2.08) | 9.1 (1.9) |
| Refinement | ||
| Resolution (Å) | 40.00-4.30 (4.53-4.30) | 35.00-7.80 (8.72-7.80) |
| Reflections working set/test set | 19 172/1010 (2763/141) | 5090/565 (1437/159) |
| Rwork, Rfree | 0.260/0.281 (0.268/0.268) | 0.337/0.346 (0.334/0.313) |
| Root-mean-square deviations | ||
| Bonds (Å) | 0.010 | n.a. |
| Angles (°) | 1.12 | n.a. |
| Average ADP (Å2) | 169 | 364 |
| Ramachandran analysis (%) | ||
| Favorable | 89.8 | n.a. |
| Outliers | 2.0 | n.a. |
| PDB access code | 3QS7 | 3QS9 |
| . | Flt3D1-D4:FL . | Flt3D1-D5:FL . |
|---|---|---|
| Data collection | ||
| Source, wavelength (Å) | ESRF ID23-1, 0.9762 | ESRF ID23-1, 1.0762 |
| Detector | ADSC-Q315R | ADSC-Q315R |
| Resolution (Å) | 40.00-4.30 (4.45-4.30) | 35.00-7.80 (8.00-7.80) |
| Space group | P21 | P21 |
| Unit cell parameters | ||
| a | a = 103.89 | a = 124.75 |
| b | b = 146.26 | b = 153.55 |
| c | c = 105.95 | c = 133.87 |
| α = γ = 90°, β = 109.7° | α = γ = 90°, β = 94.6° | |
| Wilson B (Å2) | 135 | 401 |
| Unique reflections | 20 184 (1942) | 5656 (382) |
| Redundancy | 3.8 (3.8) | 3.4 (3.2) |
| Completeness (%) | 98.8 (98.9) | 96.6 (92.9) |
| Rmeas (%)* | 10.8 (75.9) | 12.8 (80.7) |
| Average I/σ(I) | 12.04 (2.08) | 9.1 (1.9) |
| Refinement | ||
| Resolution (Å) | 40.00-4.30 (4.53-4.30) | 35.00-7.80 (8.72-7.80) |
| Reflections working set/test set | 19 172/1010 (2763/141) | 5090/565 (1437/159) |
| Rwork, Rfree | 0.260/0.281 (0.268/0.268) | 0.337/0.346 (0.334/0.313) |
| Root-mean-square deviations | ||
| Bonds (Å) | 0.010 | n.a. |
| Angles (°) | 1.12 | n.a. |
| Average ADP (Å2) | 169 | 364 |
| Ramachandran analysis (%) | ||
| Favorable | 89.8 | n.a. |
| Outliers | 2.0 | n.a. |
| PDB access code | 3QS7 | 3QS9 |
The values in parentheses refer to the highest resolution shell.
ADP indicates Atomic Displacement Parameters; and n.a., not applicable.
Rmeas = Σh√nh/(nh − 1) ΣhΣi| I(h,i) −<I(h)>| /ΣhΣiI(h,i), where nh is the multiplicity, I(h,i) is the intensity of the ith measurement of reflection h, and <I(h)> is the average value over multiple measurements.
Crystal structure of the Flt3D1-D4:FL complex. (A) Domain organization of the Flt3 extracellular segment. The 5 Ig-like domains of Flt3 (D1, residues 79-161; D2, residues 167-244; D3, residues 245-345; D4, residues 348-434; and D5, residues 435-533) are shown as colored boxes. The N-linked glycosylation sites are indicated with blue diamonds. Partially occupied glycosylation sites are indicated with an asterisk (*). Also shown is the disulfide bond network in Flt3D1-D4 as determined by mass spectrometry. The putative disulfide bridges in Flt3D5 are shown as dashed lines, based on homology with Flt3D2 and KITD5. (B) Overall structure of the Flt3D1-D4:FL complex. The crystal structure of the Flt3D1-D4:FL complex is shown in ribbon representation with the 2-fold symmetry axis of FL oriented along the vertical axis of the plane. Flt3 domains follow the coloring scheme in panel A. Disulfide bridges are shown as yellow spheres, and N-linked glycans as green sticks. The structural panels to the right show 2 alternative views of the complex with FL in ribbon representation and the receptor in surface representation.
Crystal structure of the Flt3D1-D4:FL complex. (A) Domain organization of the Flt3 extracellular segment. The 5 Ig-like domains of Flt3 (D1, residues 79-161; D2, residues 167-244; D3, residues 245-345; D4, residues 348-434; and D5, residues 435-533) are shown as colored boxes. The N-linked glycosylation sites are indicated with blue diamonds. Partially occupied glycosylation sites are indicated with an asterisk (*). Also shown is the disulfide bond network in Flt3D1-D4 as determined by mass spectrometry. The putative disulfide bridges in Flt3D5 are shown as dashed lines, based on homology with Flt3D2 and KITD5. (B) Overall structure of the Flt3D1-D4:FL complex. The crystal structure of the Flt3D1-D4:FL complex is shown in ribbon representation with the 2-fold symmetry axis of FL oriented along the vertical axis of the plane. Flt3 domains follow the coloring scheme in panel A. Disulfide bridges are shown as yellow spheres, and N-linked glycans as green sticks. The structural panels to the right show 2 alternative views of the complex with FL in ribbon representation and the receptor in surface representation.
The structure of the Flt3D1-D4:FL complex is unlike any of the structurally characterized RTKIII/V complexes to date and is characterized by several surprising features (Figure 2B; supplemental Figures 1 and 2A). The Flt3D1-D4:FL assembly can be described as a moderately open horseshoe ring structure measuring 100 × 75 × 110 Å, consisting of FL, Flt3D2, Flt3D3, and Flt3D4. FL binds to 2 receptor molecules bivalently and is accommodated by a binding epitope at the membrane-distal tip of Flt3D3, whereas Flt3D2 leans against the concave side of Flt3D3 and is stowed underneath FL in the ring opening (Figure 2B; supplemental Figure 1B). Intriguingly, the apparent 2-fold symmetry of the complex about the FL dimer interface only holds for the FL:Flt3D2-D3 subcomplex, because both Flt3D1 and Flt3D4 adopt asymmetric orientations compared with their tandem modules in the complex (Figure 2B). Remarkably, Flt3D4 does not engage in any obvious homotypic interactions as seen in the KIT structure.30 The N-terminal Flt3D1 exhibits significant disorder and domain plasticity manifested by at least 2 different orientations around the D1-D2 linker region (residues 162-166) and protrudes perpendicularly away from the plane of the ring assembly at the level of Flt3D2 without making any interactions with the rest of the complex (Figure 2B). Our electron density maps allowed us to reliably model only the core of the Flt3D1 structure (residues 79-161), but residual positive difference electron density extending away from the amino terminus of our model suggested that the atypical 50-amino acid module preceding Flt3D1 is probably associated with the core domain structure.
Flt3 uses a remarkably compact cytokine-binding epitope
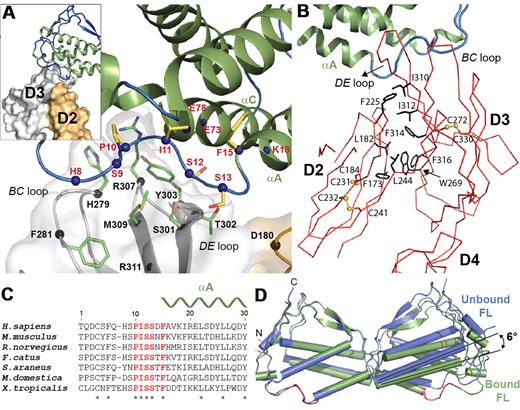
Perhaps the most unanticipated feature of the Flt3D1-D4:FL complex is that the ligand-binding epitope is almost exclusively contributed by Flt3D3 (Figure 3A), for which electron density was exceptionally clear, including information for some side chains. This module is a member of the “I-set” Ig domains and is structurally homologous to extracellular domain 3 of KIT25,30 and CSF-1R,23 featuring 8 β-strands making up the ABED and A′FGC β-sheets. However, the topology of Flt3D3 is unusual such that the polypeptide chain extending from Flt3D2 forms the N-terminal A strand in Flt3D3 (residues 246-249) by complementing strand B in a parallel manner, whereas the AA′ loop of Flt3D3 (residues 250-258) adopts an extended conformation (Figure 3B; supplemental Figure 1B). Flt3D2, which in all other RTKIII/V complexes contributes roughly half of the ligand-binding epitope, packs against the hydrophobic patch projected by the ABED-face of Flt3D3 centered around Trp269 burying ∼ 1000 Å2 (Figure 3B). Flt3D2 is homologous to KITD5 and is a member of the C2 subset of the Ig family (ABED/CFG topology), but it contains an additional solvent-exposed disulfide (Cys232-Cys241) bridging strands F and G. Although the AB and EF loops of Flt3D2 point in the direction of the ligand, they remain too far to engage in any interactions. The only point on Flt3D2 approaching FL within a distance that could mediate any form of interaction is centered on Asp180 on the AB loop of Flt3D2. However, it is not clear whether this interaction actually occurs because we were not able to model the side chain of Asp180 (Figure 3A).
Flt3-FL binding interface. (A) Close-up view of the Flt3-FL binding interface. FL is colored in green, Flt3D3 in gray, and Flt3D2 in orange. Residues that constitute the cytokine-receptor interface are shown as sticks protruding from spheres centered at their Cα positions. FL residues are colored in yellow and Flt3 residues are colored in green. (B) The unusual Flt3D2-Flt3D3 interface. Flt3D2-D4 (Cα trace in red) is shown together with FL in ribbon representation (green). Residues at the hydrophobic interface are shown as black sticks. Disulfide bonds in Flt3D2-D3 are shown as ball and sticks (yellow). (C) Structure-based alignment of diverse FL sequences revealing strict conservation of the PISSXF-segment (residues 10-15) within the N-terminal loop (colored in red). A complete alignment can be found in supplemental Figure 3. (D) Structural comparison of bound versus the unbound FL.
Flt3-FL binding interface. (A) Close-up view of the Flt3-FL binding interface. FL is colored in green, Flt3D3 in gray, and Flt3D2 in orange. Residues that constitute the cytokine-receptor interface are shown as sticks protruding from spheres centered at their Cα positions. FL residues are colored in yellow and Flt3 residues are colored in green. (B) The unusual Flt3D2-Flt3D3 interface. Flt3D2-D4 (Cα trace in red) is shown together with FL in ribbon representation (green). Residues at the hydrophobic interface are shown as black sticks. Disulfide bonds in Flt3D2-D3 are shown as ball and sticks (yellow). (C) Structure-based alignment of diverse FL sequences revealing strict conservation of the PISSXF-segment (residues 10-15) within the N-terminal loop (colored in red). A complete alignment can be found in supplemental Figure 3. (D) Structural comparison of bound versus the unbound FL.
The FL binding epitope on Flt3D3 engages in extensive interactions with the N-terminal loop (residues 8-13) of FL leading to αA and Lys18 on αA and is mainly contributed by the BC loop of Flt3D3 (residues 279-280) and strand D (residues 301-303). Additional interactions are mediated by the DE loop of Flt3D3 (residue 307) that contacts a small patch on the carboxyl-terminal region of helix αC of FL defined by residues 73 and 78. Therefore, the Flt3 ligand-receptor interaction results in a single contact site covering merely ∼ 900 Å2 of buried surface area. Structure-based alignments using diverse FL sequences revealed a remarkably strict conservation of the PISSxF cassette as well as Phe81 and Leu115, which help to lock the N-terminal loop in its observed conformation (Figure 3C; supplemental Figure 3).
Plasticity of FL on binding to Flt3
Comparison of FL in its unbound38 and now in its receptor-bound form reveals that the cytokine ligand does not undergo any significant local structural changes at its receptor binding epitope (Figure 3D). This is contrary to what has been observed in stem cell factor (SCF) in complex with KIT, whereby the cytokine ligand undergoes a cascade of structural rearrangements.25,30 However, the 2 FL subunits display a hinge-like rigid-body rearrangement about the dimer interface, which increases the tilt angle between the 2 protomers by 5-6° (Figure 3D). A similar motion was observed previously in the SCF:KITD1-D5 complex, although SCF, unlike FL, already seems to have significant variability in the receptor-free form, as shown by the range of its intersubunit tilt angles (2-6°).30
The Flt3D3-Flt3D4 domain elbow and the absence of homotypic receptor interactions
A second striking feature of the Flt3D1-D4:FL complex is the absence of any obvious specific homotypic receptor interactions. Based on the current paradigm of RTKIII activation, such interactions would be mediated by extracellular domain D4. Although Flt3D4 does point to its tandem Flt3D4′ in the complex, the 2 receptor domains stay clearly away from each other and deviate from the 2-fold symmetry of the complex. The inability of Flt3D4 to engage in homotypic interactions may also explain the observed disorder for this part of the structure, because we could only reliably model and refine a complete Flt3D4-Flt3D4′ tandem in only one of the 2 complexes in the asymmetric unit of the crystal, whereas for the second we could only place one of the 2 domains.
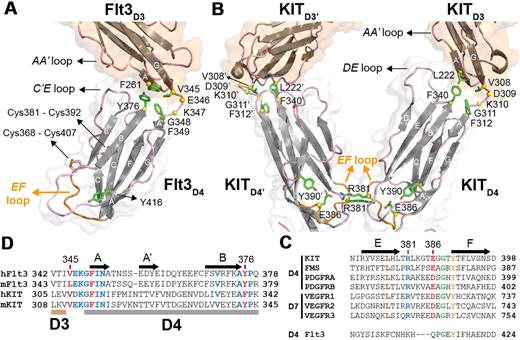
Closer inspection of the Flt3D4 topology and sequence reveals that Flt3D4 does not possess the conserved structure-sequence fingerprints seen in all other RTKIII/V homologs for this domain. For example, Flt3D4 has 2 additional disulfide bridges, a solvent-exposed cross-strand disulfide bridge (Cys368-Cys407) connecting strands B and E, and a second (Cys381-Cys392) connecting its unusual C′E loop with strand C. Most importantly, Flt3D4 displays an EF loop that drastically differs both in structure and sequence from all homologs (Figure 4A-C). The EF loop constitutes the otherwise conserved “tyrosine corner” motif in I-set Ig domains,43 and has been shown to mediate homotypic interactions in the case of KITD430 and VEGFRD728 (Figure 4B).
Flt3D3-Flt3D4 elbow and the absence of homotypic receptor contacts in the Flt3:FL complex. (A) The Flt3D3-Flt3D4 elbow. Flt3D3 (partially shown) and Flt3D4 are shown in ribbon representation. For clarity purposes only the locations of the atypical disulfide bridges in Flt3D4 (Cys368-Cys407 and Cys381-Cys391) are indicated. Residues mediating hydrophobic interactions between Flt3D3 and Flt3D4 are shown as green sticks. Residues in the Flt3D3-Flt3D4 linker (346-348) are shown as yellow spheres centered at their Cα positions. The side chains of residues mediating contacts between the AA′ loop of Flt3D3 and the C′E loop of Flt3D4 could not be modeled because of the low resolution of our analysis. The EF loop of Flt3D4 constituting the “tyrosine corner” around Tyr416 (green sticks) is shown in orange. (B) Sequence conservation of residues involved at the D3-D4 interface in KIT and Flt3 based on comparisons between human and murine Flt3 and KIT sequences. (C) KITD3-KITD4 orientation in the KIT:SCF complex. Homotypic receptor contacts between tandem ectodomain 4 modules in the KIT:SCF complex are mediated by salt bridges via residues Arg381 and Glu386 residing on the EF loops (orange; PDB entry 2E9W). Residues at the hydrophobic KITD3-KITD4 interface are shown as green sticks. Residues in the KITD3-KITD4 linker region (Asp309-Gly311) are shown as yellow spheres. (D) Flt3D4 displays an atypical EF loop within the RTKIII/V family. The pair of residues mediating the homotypic contacts in KITD4 and VEGFR-2D7 is well conserved in the corresponding domains of all RTKIII/V members but not in Flt3D4.
Flt3D3-Flt3D4 elbow and the absence of homotypic receptor contacts in the Flt3:FL complex. (A) The Flt3D3-Flt3D4 elbow. Flt3D3 (partially shown) and Flt3D4 are shown in ribbon representation. For clarity purposes only the locations of the atypical disulfide bridges in Flt3D4 (Cys368-Cys407 and Cys381-Cys391) are indicated. Residues mediating hydrophobic interactions between Flt3D3 and Flt3D4 are shown as green sticks. Residues in the Flt3D3-Flt3D4 linker (346-348) are shown as yellow spheres centered at their Cα positions. The side chains of residues mediating contacts between the AA′ loop of Flt3D3 and the C′E loop of Flt3D4 could not be modeled because of the low resolution of our analysis. The EF loop of Flt3D4 constituting the “tyrosine corner” around Tyr416 (green sticks) is shown in orange. (B) Sequence conservation of residues involved at the D3-D4 interface in KIT and Flt3 based on comparisons between human and murine Flt3 and KIT sequences. (C) KITD3-KITD4 orientation in the KIT:SCF complex. Homotypic receptor contacts between tandem ectodomain 4 modules in the KIT:SCF complex are mediated by salt bridges via residues Arg381 and Glu386 residing on the EF loops (orange; PDB entry 2E9W). Residues at the hydrophobic KITD3-KITD4 interface are shown as green sticks. Residues in the KITD3-KITD4 linker region (Asp309-Gly311) are shown as yellow spheres. (D) Flt3D4 displays an atypical EF loop within the RTKIII/V family. The pair of residues mediating the homotypic contacts in KITD4 and VEGFR-2D7 is well conserved in the corresponding domains of all RTKIII/V members but not in Flt3D4.
Structural comparisons of the 2 independent Flt3D1-D4:FL complexes in the crystal asymmetric unit revealed slight orientational plasticity of Flt3D4 around the Flt3D3-Flt3D4 linker region. This stretch of residues and strand A of Flt3D4 are well conserved in Flt3 and KIT and other RTKIIIs, suggesting a common functional role (Figure 4D). A comparison of KIT in the cytokine-bound and -unbound forms showed that the KITD3-KITD4 linker region acts as a hinge to reorient KITD4 for homotypic interactions on ligand binding. Despite the absence of such homotypic contacts in Flt3, the domain elbow defined by Flt3D3 and Flt3D4 is similar to KIT, suggesting preservation of this interdomain relationship in both forms of the receptor. Thus, the orientation of Flt3D4 seems to be restricted by a core of well-defined hydrophobic interactions mediated by Phe261 (A′ strand of Flt3D3), Val345 (Flt3D3-Flt3D4 linker), Phe349 (A strand of Flt3D4), and Tyr376 (BC loop of Flt3D4), as well as additional interactions between the AA′ loop of Flt3D3 and the C′E loop of Flt3D4 (Figure 4A).
Architecture of the complete extracellular assembly of the Flt3 signaling complex
Crystals of Flt3D1-D5:FL grew reproducibly from several crystallization conditions but proved to be of low diffraction quality despite repeated attempts to improve crystal quality by various methods including glycan shaving. Nonetheless, a robust dataset to 7.8-Å resolution proved sufficient to elucidate the architecture of the complete extracellular Flt3 complex by molecular replacement based on the Flt3D2-D3: FL subcomplex as refined in the Flt3D1-D4:FL crystal structure (Table 1). We could subsequently place into electron density and optimize by rigid-body refinement protocols Flt3D1 and Flt3D4, extracted from the crystal structure of the Flt3D1-D4:FL complex, as well as a conservative homology model of Flt3D5 derived from the structure of human KITD5.30 Although the low resolution of the Flt3D1-D5:FL structure does not allow discussion of structural details, it does provide a reliable and valuable depiction of the organization features of the complete extracellular Flt3 complex.
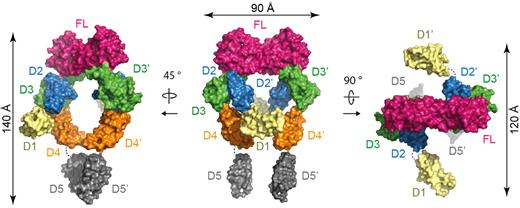
In the full-length ectodomain complex, the core structure observed in Flt3D1-D4:FL is mounted onto 2 membrane-proximal Flt3D5s facing each other to form an assembly resembling a hollow tennis racket (140 × 75 × 110 Å; Figure 5; supplemental Figure 2B). Remarkably, the asymmetry exhibited by the tandem Flt3D4 modules in Flt3D1-D4:FL is not present in the complete extracellular complex. Instead, the 2 Flt3D4 segments face each other nearly symmetrically according to the 2-fold symmetry of the Flt3D2-D3:FL core structure and approach to ∼ 20 Å from each other. Although this interreceptor separation is maintained at the ensuing Flt3D5 modules, the apparent 2-fold symmetry breaks down. Furthermore, the asymmetric projection of the N-terminal Flt3D1 domains perpendicularly out of the plane of the racket head occurs in a manner analogous to what we observed in the Flt3D1-D4:FL complex. Complementary studies of the full-length ectodomain complex by negative-stain EM and SAXS in solution corroborated the overall structural features revealed by the crystal structure (supplemental Figure 4A-B).
Assembly of the complete Flt3 ectodomain complex. Surface representations of the full-length Flt3 ectodomain complex. The central view shows the complex with the 2-fold symmetry axis of FL oriented vertically in the plane of the paper.
Assembly of the complete Flt3 ectodomain complex. Surface representations of the full-length Flt3 ectodomain complex. The central view shows the complex with the 2-fold symmetry axis of FL oriented vertically in the plane of the paper.
Discussion
Cytokine-mediated activation of hematopoietic cell surface receptors is central to developing and sustaining hematopoiesis and the immune system. The RTKIII receptor Flt3 and its cognate cytokine ligand FL are arguably the most exciting new addition to the repertoire of hematopoietic factors, because of their activity on hematopoietic progenitors and their pronounced impact on the development and homeostasis of antigen-presenting DCs. Because the importance of Flt3 signaling in early and late hematopoiesis continues to mount, we sought to elucidate the structural basis of the extracellular Flt3 receptor-ligand complex. The structural study we report here, complemented by a thermodynamic dissection of complex formation, shows that the Flt3:FL interaction is characterized by high-affinity bivalent binding of FL to Flt3 that does not invoke homotypic receptor interactions. The assembly showcases several unexpected features that now establish Flt3 as a structural outlier within the RTKIII/V family (Figure 6). The Flt3:FL interaction epitope is surprisingly a fraction of typical helical cytokine-receptor interaction and is dominated by contacts between a preformed N-terminal segment of FL and Flt3D3. Consistent with the polar receptor–cytokine interface, the thermodynamic blueprint of the interaction calls for an enthalpically driven binding event. The interaction carries a concomitant significant entropic cost that we now can rationalize in terms of the absence of a hydrophobic effect and the intrinsic entropy loss associated with bringing interaction partners together.
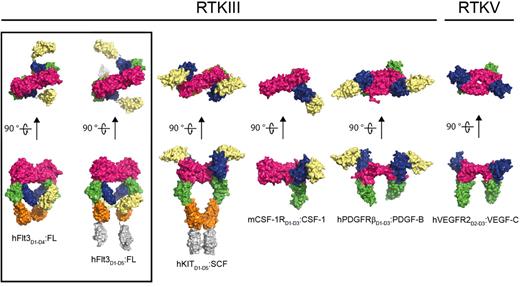
Comparison of representative RTKIII/V extracellular complexes. The structures shown represent the architecture of receptor-cytokine complexes for the different members of the RTKIII/V family: From left to right: human Flt3:FL (this study), human KIT:SCF (PDB 2E9W), mouse CSF-1R:CSF-1 (PDB 3EJJ), human platelet-derived growth factor receptor:platelet-derived growth factor (PDGFR:PDGF; PDB 3MJG), and human VEGFR2:VEGF (PDB 2X1X). The dimeric ligands are colored in magenta. Receptor ectodomains are colored as follows: D1 in pale yellow, D2 in blue, D3 in green, D4 in orange, and D5 in gray.
Comparison of representative RTKIII/V extracellular complexes. The structures shown represent the architecture of receptor-cytokine complexes for the different members of the RTKIII/V family: From left to right: human Flt3:FL (this study), human KIT:SCF (PDB 2E9W), mouse CSF-1R:CSF-1 (PDB 3EJJ), human platelet-derived growth factor receptor:platelet-derived growth factor (PDGFR:PDGF; PDB 3MJG), and human VEGFR2:VEGF (PDB 2X1X). The dimeric ligands are colored in magenta. Receptor ectodomains are colored as follows: D1 in pale yellow, D2 in blue, D3 in green, D4 in orange, and D5 in gray.
The Flt3-FL interaction interface covers a compact ∼ 900 Å2, which is at least 2 times less extensive than the buried surface area at the receptor-cytokine epitopes of all other RTKIII/V complexes, whereby the activating cytokine is harbored by a broad grapple defined by extracellular domains 2 and 323-25,27,30 (Figure 6). Comparison with diverse helical cytokine-receptor interactions44,45 shows that Flt3 is the only receptor for a helical cytokine that uses a single interaction site to bind its cognate ligand. Heterodimeric protein-protein interactions bury, on average, ∼ 1300 Å2 of surface area, which is strongly correlated with high-affinity binding.46 Flt3 and FL are clearly able to establish a tight interaction via a much more compact binding interface. A plausible explanation could be drawn from the rigidity of the receptor epitope on FL as a preformed binding platform, reminiscent of a classic “lock-and-key” binding mode observed in affinity-matured antibody-antigen interactions.47 Furthermore, a series of single amino-acid substitutions (His8Arg, Ser9Gly, Pro10Ser, Ser13Pro/Phe, and Phe15Leu) within the segment contributing almost the entire receptor binding epitope on FL, abolish receptor activation.48 This illustrates not only the possible individual contribution of each residue in this segment to binding but also the likely conformational stringency of the region. Indeed, a comparison of diverse FL sequences showed that not only the receptor-binding epitope is exquisitely conserved but also residues that help to lock the N-terminal loop both in its observed receptor-bound and receptor-free forms (Figure 3C; supplemental Figure 3).
Comparison of the Flt3:FL interaction and representative receptor-cytokine epitopes for all other RTKIII/Vs shows that engagement of Ig-like domain 3 (BC loop; DE loop and flanking residues) is the only common epitope feature of ligand binding (Figure 6). It thus seems that binding of D3 of RTKIII/V to the activating cytokine satisfies a geometric requirement that allows receptor molecules to approach to a critical distance of ∼ 60 Å from one another. This notion reinforces a fascinating aspect of RTKIII/V activation in that the cognate protein ligands are all dimeric with similar dimensions despite their grouping into 2 fundamentally different folds (4-helix bundles vs all-β cystine-knot scaffolds).23,24,27,30,38 Recently, interleukin-34 was identified as a second ligand to CSF-1R,49 thus adding a perplexing dimension to RTKIII signaling because interleukin-34 bears no sequence similarity to the currently known cytokine ligands for RTKIII/V or other proteins.
The uniqueness of the Flt3 extracellular complex is further highlighted by the absence of homotypic receptor interactions. Such interactions have recently emerged as an important aspect of RTKIII/V activation and are mediated by conserved structure-sequence fingerprints in the membrane-proximal domains.23,26,28-30 In both of our Flt3:FL complexes, the membrane-proximal modules remain separated by ∼ 20 Å at the D4-D5 junction, consistent with our comparative ITC data showing no significant contribution by the D4-D5 module. Flt3 is the only RTKIII/V family member that lacks the conserved set of residues involved in homotypic interactions in the homologous receptors, which now offers a strong rationale for the absence of such interactions in the extracellular Flt3 complex. Additional support comes from the existence of a fully active murine Flt3 isoform that lacks Flt3D5, demonstrating the dispensability of the membrane-proximal domain for receptor activation,33 contrary to the apparent importance of KITD5 in signaling.34 However, it is possible that homotypic interactions could be enhanced within the 2-dimensional spatial confinement of the cell membrane, and as a result of additional interactions between TM, cytosolic segments, or both of Flt3. In fact, the difference in the affinity for the complete ectodomain complex versus previously reported values for native Flt3 based on cell assays48,50 may reflect a combination of such factors. To this end, recent studies have highlighted the importance of homotypic interactions between TM and JM regions in RTKIII/V activation and pathology profiles.51,52 Interestingly, oncogenic variants of Flt3 carrying internal tandem duplication in the JM segment are constitutively active as homodimers or as heterodimers with wild-type receptor, indicating that enhanced intracellular receptor interactions can drive activation.18 Recently, several mutations in the extracellular segment of Flt3 were identified in AML patients,17,53 that we now can map onto the Flt3 ectodomain (supplemental Figure 5). However, the clinical relevance of these mutations awaits further study.
In the absence of structural information for unbound Flt3, we are left to wonder about any possible domain rearrangements in Flt3 on FL binding. Structural studies of KIT in the bound and unbound forms showed that KIT undergoes a large conformational switch at the KITD3-D4 junction, leading to homotypic receptor interactions.30 However, the extensive and unique hydrophobic interface observed between Flt3D2 and Flt3D3 (1000 Å2), the hydrophobic interface between Flt3D3 and Flt3D4, and additional contacts between loops thereof, provide evidence that the Flt3D2-D4 ectodomain segment would be too rigid to undergo significant domain rearrangements. This is further supported by our structural studies of the complete extracellular complex, which showed that the relative orientation of Flt3D3 and Flt3D4 only differs slightly from that observed in the Flt3D1-D4:FL complex. Nonetheless, Flt3 does exhibit significant domain plasticity at the 2 extremities of the extracellular assembly. This is most pronounced for Flt3D1, which emanates from the core of the assembly without making contacts with other complex components. We note that the stacking of Flt3D2 against Flt3D3 provides a fixed angle for projecting Flt3D1 from a point approximately halfway down the height of the complex. Flt3D1 is the largest and most atypical domain in Flt3 and the entire RTKIII/V family, but its role in Flt3 signaling is currently unknown. We are thus tempted to propose that Flt3D1 could mediate intermolecular contacts at the cell surface, stabilize the unbound receptor, or both.
The availability of structures for the complete extracellular Flt3 receptor-ligand complex, including a delineation of the receptor-cytokine interface, will probably have a significant impact on renewed efforts to antagonize Flt3 activity in a clinical setting. This is because wild type and mutated forms of Flt3 as well as autocrine secretion of FL have been implicated in the development of myeloid leukemias.2,15,21,54 Current strategies focus on inhibition of the intracellular kinase domains of Flt3, but they are faced with drug specificity issues and the emergence of primary and secondary resistance to treatment.21 More recently, an alternative therapeutic approach based on monoclonal antibodies directed against the extracellular domain of Flt355 indicated a possible momentum shift toward combined strategies in clinical targeting of Flt3. A daunting challenge in inhibiting protein-protein interactions is the extent of the interaction epitope that often covers > 1500 Å2 and the lack of prior knowledge of functional hot spots.56 In this regard, the compactness of the Flt3 ligand-receptor interface provides favorable perspectives for the druggability of the extracellular Flt3 binding epitope.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the European Synchrotron Radiation Facility (ESRF) and the Swiss Light Source (SLS) for synchrotron beam time allocation, and the staffs of beamlines ID-23 (ESRF) and X06SA/X06DA (SLS) for technical support.
K.V., J.E., B.V., and K.V.C. are research fellows of the Research Foundation Flanders, Belgium (Fonds voor Wetenschappelijk Onderzoek [FWO]). This research project was supported the FWO (grants 3G064307 and G059710) and Ghent University (Bijzonder Onderzoeksfonds instrument) to S.N.S. A.V.S. and D.I.S. were supported by Human Frontier Science Program Research grant RGP 55/2006. Access to these synchrotron facilities is supported by the European Commission under the 7th Framework Program: Research Infrastructures (grant 226716).
Authorship
Contribution: K.V. and M.J. expressed and purified recombinant proteins and performed ITC measurements and data analysis; K.V. crystallized the complexes; K.V., J.E., and S.N.S. carried out crystallographic experiments and analyzed data; K.V. and S.N.S. determined and refined crystal structures; B.V. assisted in the design and analysis of ITC data; K.V.C. assisted in tissue culture and mammalian protein expression; I.G. carried out EM imaging; A.D. and I.G. analyzed the EM data; G.V. collected and analyzed mass spectrometry data; D.I.S., A.V.S., K.V., and S.N.S. contributed to SAXS data collection and analysis; K.V., B.V., and S.N.S. wrote the manuscript; and S.N.S. designed and directed the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Savvas N. Savvides, Unit for Structural Biology, L-ProBE, Ghent University, K.L. Ledeganckstraat 35, 9000 Ghent, Belgium; e-mail: savvas.savvides@ugent.be.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal