Abstract
Chemokines and adhesion molecules up-regulated in lymphatic endothelial cells (LECs) during tissue inflammation are thought to enhance dendritic cell (DC) migration to draining lymph nodes, but the in vivo control of this process is not well understood. We performed a transcriptional profiling analysis of LECs isolated from murine skin and found that inflammation induced by a contact hypersensitivity (CHS) response up-regulated the adhesion molecules ICAM-1 and VCAM-1 and inflammatory chemokines. Importantly, the lymphatic markers Prox-1, VEGFR3, and LYVE-1 were significantly down-regulated during CHS. By contrast, skin inflammation induced by complete Freund adjuvant induced a different pattern of chemokine and lymphatic marker gene expression and almost no ICAM-1 up-regulation in LECs. Fluorescein isothiocyanate painting experiments revealed that DC migration to draining lymph nodes was more strongly increased in complete Freund adjuvant-induced than in CHS-induced inflammation. Surprisingly, DC migration did not correlate with the induction of CCL21 and ICAM-1 protein in LECs. Although the requirement for CCR7 signaling became further pronounced during inflammation, CCR7-independent signals had an additional, albeit moderate, impact on enhancing DC migration. Collectively, these findings indicate that DC migration in response to inflammation is stimulus-specific, mainly CCR7-dependent, and overall only moderately enhanced by LEC-induced genes other than CCL21.
Introduction
Lymphatic vessels play an important role in tissue homeostasis and immune surveillance, and they are also involved in pathologic conditions, such as tumor cell metastasis and chronic inflammation.1 Inflammation enhances the drainage of soluble mediators and the migration of leukocytes, particularly of dendritic cells (DCs), into afferent lymphatics, thereby stimulating the induction of adaptive immunity in draining lymph nodes (dLNs).2,3 Many elements of the response to inflammation can be attributed to changes at the transcriptional level. For example, it is well known that blood vascular adhesion molecules and chemokines are up-regulated during inflammation and mediate the recruitment of leukocytes. However, to date, the gene expression changes occurring in lymphatic endothelial cells (LECs) in vivo during tissue inflammation have not been comprehensively studied. Such experiments have generally been hampered by difficulties associated with the isolation of the rare LECs from tissues and the subsequent processing steps. In light of such technical constraints, most recent insights into the inflammatory response in LECs have come from in vitro experiments with cultured cells. Notably, in vitro treatment with inflammatory mediators was shown to up-regulate inflammatory chemokines2,4,5 and the adhesion molecules ICAM-1 and VCAM-12,6 in LECs. Both ICAM-1 and VCAM-1 were shown to mediate DC migration into lymphatic vessels under inflammatory conditions.2
A key molecule required for DC migration into lymphatics is the chemokine CCL21, which is constitutively expressed in LECs.7 Antigen uptake and activation enhance the expression of its receptor CCR7 in DCs, thereby inducing DC migration toward CCL21-expressing lymphatic vessels and onwards to dLNs. Several recent studies have reported that CCL21 is up-regulated in lymphatic vessels during inflammation.8-10 Besides inflammation-induced changes in adhesion molecule and chemokine expression in LECs, which are thought to favor DC migration, inflammatory mediators are also thought to boost migration by directly acting on DCs, for example, by increasing DC motility and responsiveness for CCL21.11 However, to date, the relative contribution of such DC- versus LEC-mediated pathways of enhancing DC migration have remained unclear.
In this study, we have performed a comprehensive ex vivo microarray-based analysis of inflamed and resting LECs isolated by FACS from murine ear skin. Surprisingly, we observed that inflammation induced by a contact hypersensitivity (CHS) response led to the down-regulation of the LEC lineage markers VEGFR3 and Prox-1, whereas inflammatory chemokines and adhesion molecules were strongly up-regulated in LECs. We also found that the pattern of chemokines and adhesion molecules induced in vivo depended on the nature of the inflammatory stimulus: ICAM-1 and certain inflammatory chemokines were only induced in LECs in the context of CHS, but not during inflammation induced by injection of complete Freund adjuvant (CFA). Unexpectedly, FITC painting experiments revealed that DC migration was more enhanced during CFA-induced inflammation than during CHS-induced inflammation, despite lower CCL21 and ICAM-1 induction in the latter model. Experiments in CCR7−/− mice revealed that DC migration during CFA-induced inflammation was mainly dependent on CCR7 expression. However, CCR7-independent signals also had a minor migration-enhancing effect, suggesting an additional contribution of LEC-expressed mediators other than CCL21. Collectively, our findings, for the first time, reveal that inflammatory gene expression in LECs in vivo is modulated in a stimulus-dependent manner. In addition, the DC migratory response is dependent on the type of inflammation induced and appears to be only moderately enhanced by LEC-expressed, inflammation-induced genes other than CCL21.
Methods
Mice
Wild-type (WT) FVB or C57BL/6 mice were purchased from Charles River Laboratories. H-2Kb-tsA58 (Immorto) mice on a C57BL/6 background12 (The Jackson Laboratory) and CCR7−/− mice,13 kindly provided by W. Hardt (ETH Zurich), were bred and housed in our facility. All experiments were approved by the Cantonal Veterinary Office Zurich.
CHS-induced ear skin inflammation
A CHS response toward oxazolone was induced in the ear skin of 6- to 12-week-old female mice as described.14 Briefly, mice were anesthetized by intraperitoneal administration of medetomidine (1 mg/kg) and ketamine (75 mg/kg) and sensitized by topical application of 2% oxazolone (4-ethoxymethylene-2-phenyl-2-oxazoline-5-one; Sigma-Aldrich) in acetone/olive oil (4:1 volume/volume) on the shaved abdomen (50 μL) and on each paw (5 μL). Five days later, 10 μL of a 1% oxazolone solution was applied topically to each side of the ears. Experiments were performed 24 to 48 hours after CHS challenge.
CFA-induced ear skin inflammation
CFA (Sigma-Aldrich) was mixed at a ratio of 1:1 with PBS and emulsified for 5 minutes (4°C, 30 Hz) on a QIAGEN TissueLyzer (Retsch); 10 μL of the emulsion was injected into the ear pinnae of anesthetized mice using a gastight 1705 syringe (Hamilton).
Protein extraction from mouse ears
Ears were harvested (n = 3 mice), cut into small pieces, and suspended in an Eppendorf tube in buffer containing 50mM Tris, 150mM NaCl, and protease inhibitor cocktail (Roche Diagnostics). A 5-mm steel bead (QIAGEN) was added, and tissue homogenization was performed on a QIAGEN TissueLyzer (4 × 1 minute, 4°C, 30 Hz). Supernatant was harvested after centrifugation (5 minutes, 4°C, 16 000g).
Analysis of chemokines in ear protein extracts and in immortalized LEC culture supernatants
Chemokine protein levels were measured at Cytolab by a multiplexed particle-based flow cytometric cytokine assay15 using kits purchased from Millipore, R&D Systems, and Bio-Rad. Analysis was performed on a FC500 MPL flow cytometer (Beckman Coulter).
FACS of LECs and RNA extraction
Twenty-four hours after induction of skin inflammation, mice (n = 3) were killed and the ears were harvested. Pooled ear halves were cut into small pieces and briefly digested by incubation for 20 minutes at 37°C in PBS containing 10 mg/mL of collagenase IV (Invitrogen), followed by passage through a 40-μm cell strainer (BD Biosciences). All subsequent steps were performed on ice. Resulting single-cell suspensions were rapidly stained with the following antibodies: allophycocyanin (APC)–labeled rat anti–mouse CD31; peridinin chlorophyll protein (PerCP)–labeled antimouse CD45 (both from BD Biosciences); hamster anti–mouse podoplanin (clone 8.1.1, Developmental Studies Hybridoma Bank, University of Iowa) and anti–hamster-phycoerythrin (Invitrogen). LECs were isolated on a FACSAria Cell Sorter (BD Biosciences) and sorted into RNAprotect Cell Protect reagent (QIAGEN). Total RNA was extracted using RNeasy plus micro kit (QIAGEN). The WT-Ovation Pico RNA amplification kit (Nugen Technologies) and the QIAQuick PCR purification kit (QIAGEN) were used for cDNA synthesis and a single round of isothermal amplification. The amplified cDNA was analyzed on a Bioanalyzer 2100 and RNA 6000 Nano LabChip (both from Agilent Technologies). Four paired samples (inflamed and control) were chosen for microarray experiments.
Analysis by quantitative PCR
Quantitative PCR analyses were performed on a 7900HT Fast Real-Time PCR System (Applied Biosystems), using TaqMan Gene Expression Master Mix (Applied Biosystems) and FastStart Universal SYBR Green Master Mix (Roche Diagnostics). The sequence identity of the quantitative PCR probes is provided in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Microarray hybridization, data processing, and analysis
See supplemental Methods. The microarray data have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus and are accessible through GEO Series accession numbers GSE26229 (FACS-sorted LECs) and GSE28234 (imLECs).
FACS analysis of ear single-cell suspensions
Uninflamed and CHS-inflamed ears of mice were digested with collagenase IV (Invitrogen) as previously described.14 Cell suspensions were stained with anti–mouse CD31-APC, antimouse CD45-PerCP (both from BD Biosciences), hamster anti–mouse podoplanin (clone 8.1.1.), and anti–hamster-phycoerythrin (Invitrogen) or anti–hamster-Alexa-488 (Invitrogen). The expression of candidate proteins was detected by additionally staining with biotinylated anti–mouse CD274, biotinylated anti–mouse CD44, anti-CD54-FITC (all from BioLegend), anti-CD62P-FITC and rat anti-CD74 (both from BD Biosciences), goat anti-CXCL9 (R&D Systems), and strepatividin–Alexa-488, donkey anti–rat-Alexa-488, and donkey anti–goat-Alexa-488 (all from Invitrogen). FACS analysis was performed on a BD FACSCanto (BD Biosciences) using the FACSDiva software. Data were analyzed with FlowJo Version 8.7.1 software (TreeStar).
In some experiments, ear single-cell suspensions were stained with anti-CD11c–APC and DCs were quantified with counting beads (Invitrogen).
Immunofluorescence analysis of ear sections
Immunofluorescence analyses of CHS-inflamed and control ears were performed as previously described.16 Sections were incubated for 2 hours at room temperature with the following primary antibodies in Antibody Diluent (Zytomed Systems): goat anti–mouse urokinase-type plasminogen activator receptor (uPAR; R&D Systems), rabbit anti–mouse neuron-glia-CAM-related cell adhesion molecule (NrCAM; Abcam), and rat anti–mouse VEGFR3 (eBioscience), in combination with rabbit anti–mouse LYVE-1 (AngioBio) or rat anti–mouse LYVE-1 (clone ALY717 ). Alexa488- or Alexa594-coupled secondary antibodies (Invitrogen) were used for detection. Slides were mounted with Vectashield mounting medium (Vector Laboratories).
Immunofluorescence analysis of ear whole mounts
Mice were killed and ears harvested after hair removal with VEET depilation cream. Subsequently, ears were split into 2 halves along the cartilage, and ear halves were fixed with 4% paraformaldehyde for 2 hours. Ears were washed with PBS and incubated for 2 hours in 12% bovine serum albumin/PBS. Subsequently, ears were incubated overnight with the following primary antibodies: rat anti–mouse I-A/I-E (BD Biosciences), goat anti–mouse CCL21 (R&D Systems), rabbit anti–mouse CCL2 (Santa Cruz Biotechnology), rabbit-anti–mouse CXCL1 (PeproTech), or corresponding isotype controls in combination with rabbit anti–mouse LYVE-1 (AngioBio) or rat anti–mouse LYVE-1 (clone ALY717 ). Next, ears were washed in PBS and incubated for at least 4 hours with Alexa-conjugated secondary antibodies (all from Invitrogen). Ears were washed in PBS and fixed for 10 minutes with 4% paraformaldehyde. Samples were mounted using Vectashield (Vector Laboratories).
Image acquisition
Stainings were examined using an Axioskop 2 MOT plus microscope equipped with a 20 × 0.50 NA Plan-Neofluar and a 40 × 1.3NA oil Plan-Apochromat objective and an AxioCam MRm monochrome digital camera (all from Carl Zeiss). Images were acquired using AxioVision Version 4.4 software (Carl Zeiss). Adobe Photoshop CS2 (Adobe System) was used for image processing. Some whole-mounted sections were analyzed on a Zeiss LSM 710-FCS confocal microscope equipped with a 20 × 0.8 NA Plan-Apochromat, a 40 × 1.2 NA water C-Apochromat, and a 63 × 1.4 NA oil DIC Plan-Apochromat objective (all from Carl Zeiss). Images were acquired using the Zeiss ZEN 2009 software (Carl Zeiss) and processed using Imaris version 7.1.1 software (Bitplane).
Isolation of immortalized LECs from Immorto mice and in vitro assays
See supplemental Methods.
FITC painting experiments
FITC (5 mg/mL, Thermo Scientific) was dissolved in acetone and dibutyl phthalate (1:1, Sigma-Aldrich), and 20 μL was applied to each side of the ear. Eighteen hours later, ear-draining auricular LNs were harvested and digested for 45 minutes by incubation in 0.4% collagenase IV (Invitrogen) at 37°C. Single-cell suspensions were passed through a 40-μm cell strainer (BD Biosciences). Total cell numbers were determined, and samples were stained with anti-CD11c–APC and anti-I-A/I-E-PerCP protein (both from BioLegend). The number of FITC+CD11c+IA/IE+ DCs per LN was calculated by multiplying the number of total LN cells with the fraction of FITC+CD11c+IA/IE+ DCs determined by FACS analysis.
Statistical analysis
Data were analyzed using the Student t test (unpaired, 2-tailed, or paired when indicated) and are presented as mean ± SEM. Differences were considered statistically significant when P < .05. All experiments were repeated 2 to 4 times.
Results
Gene expression analysis of LECs isolated from CHS-inflamed skin
To study how inflammation affects gene expression in LECs, a CHS response toward oxazolone was induced in the ear skin of mice. In this model, inflammation is caused by an adaptive T-cell recall response and is characterized by the production of proinflammatory and TH1 cytokines.18 One day after challenge with oxazolone, the ears of the mice were strongly inflamed (Figure 1A). Analysis of ear protein extracts detected elevated levels of TNF-α and IFN-γ (Figure 1B). Furthermore, high numbers of MHCIIhigh leukocytes were present in the inflamed ears (Figure 1C), some of which colocalized with lymphatic vessels (Figure 1C middle and right panels).
Isolation of LECs from uninflamed and CHS-inflamed mouse ears. (A) A CHS response toward oxazolone was induced in the ears of WT mice. Twenty-four hours after induction, CHS-challenged ears were markedly red and swollen. (B) IFN-γ and TNF-α protein levels were significantly increased in CHS-inflamed ears (n = 3 mice per group). **P < .01. ***P < .001. (C) Whole-mount immunofluorescence of inflamed and control ears detected large numbers of MHCIIhigh cells (red) around (middle panel) and within (right panel, orthogonal slice produced by confocal microscopy) LYVE-1-positive lymphatic vessels (green) in inflamed mouse ears. Scale bars represent 100 μm (left and middle) and 20 μm (right). (D) Ear tissues were digested enzymatically and stained for CD45, CD31, and podoplanin to differentiate between leukocytes (CD45+CD31−), BECs (CD45−CD31+podoplanin−), and LECs (CD45−CD31+podoplanin+). LECs were isolated from ear single-cell suspensions by FACS sorting. (E) Podoplanin was significantly down-regulated in inflamed LECs (INFL) compared with control LECs (CTR), as demonstrated by analysis of the median fluorescent intensity (MFI) of podoplanin expression found on cells in the LEC gate. *P < .05 (paired Student t test). (F) RNA was extracted from sorted LECs. cDNA was synthesized and subjected to one round of linear amplification. LEC preparations isolated from control ears or inflamed ears that were processed in parallel were treated as pairs. The induction of ICAM-1 and VCAM-1 in pairs of control and inflamed samples was analyzed by quantitative PCR.
Isolation of LECs from uninflamed and CHS-inflamed mouse ears. (A) A CHS response toward oxazolone was induced in the ears of WT mice. Twenty-four hours after induction, CHS-challenged ears were markedly red and swollen. (B) IFN-γ and TNF-α protein levels were significantly increased in CHS-inflamed ears (n = 3 mice per group). **P < .01. ***P < .001. (C) Whole-mount immunofluorescence of inflamed and control ears detected large numbers of MHCIIhigh cells (red) around (middle panel) and within (right panel, orthogonal slice produced by confocal microscopy) LYVE-1-positive lymphatic vessels (green) in inflamed mouse ears. Scale bars represent 100 μm (left and middle) and 20 μm (right). (D) Ear tissues were digested enzymatically and stained for CD45, CD31, and podoplanin to differentiate between leukocytes (CD45+CD31−), BECs (CD45−CD31+podoplanin−), and LECs (CD45−CD31+podoplanin+). LECs were isolated from ear single-cell suspensions by FACS sorting. (E) Podoplanin was significantly down-regulated in inflamed LECs (INFL) compared with control LECs (CTR), as demonstrated by analysis of the median fluorescent intensity (MFI) of podoplanin expression found on cells in the LEC gate. *P < .05 (paired Student t test). (F) RNA was extracted from sorted LECs. cDNA was synthesized and subjected to one round of linear amplification. LEC preparations isolated from control ears or inflamed ears that were processed in parallel were treated as pairs. The induction of ICAM-1 and VCAM-1 in pairs of control and inflamed samples was analyzed by quantitative PCR.
To isolate LECs from uninflamed or CHS-inflamed ears, the ear tissue was enzymatically digested, single-cell suspensions were stained for CD45, CD31, and podoplanin (Figure 1D),14,19 and LECs were isolated by FACS sorting. Interestingly, we observed that podoplanin expression was slightly reduced in inflamed LECs (Figure 1D-E). However, albeit this reduction, LEC and blood vascular endothelial cell (BEC) populations remained clearly separable during inflammation (Figure 1D), in agreement with previous results.14,19 Typically, 1500 to 6000 LECs (purity > 98%) were obtained from the pooled ears of 3 mice. To amplify the minute amounts of extracted total RNA, the RNA was transcribed into cDNA and subjected to a single isothermal amplification step, typically yielding 5 to 8 μg of amplified cDNA. Quantitative PCR analysis revealed a strong induction of ICAM-1 and VCAM-1 in LECs isolated from inflamed compared with control ears (Figure 1F).
Subsequently, microarray experiments were performed on the amplified cDNA samples. The analysis of the resulting array data revealed present call rates between 39.8% and 47.5%. Importantly, all control and inflammation-derived samples clustered among themselves (supplemental Figure 1A-B). Statistical analysis showed that 618 genes were induced at least 2-fold, whereas 689 genes were down-regulated at least 2-fold (P ≤ .01, supplemental Figure 1C). A significant up-regulation of ICAM-1 and VCAM-1 was detected in inflamed LECs (Table 1). Furthermore, the lymphatic endothelial origin of the hybridized samples was confirmed by present calls for many pan-endothelial marker genes and for most LEC marker genes (Figure 2A-B).
Differential expression of selected cell surface molecules
| Transcript . | Fold change . | P . |
|---|---|---|
| FcgRIIb (CD32)* | 438.5 | .000061 |
| CD62P (P selectin)* | 316.4 | .0012 |
| S1PR3 (Edg3)* | 288.9 | .000093 |
| CD53 | 246.3 | .0000027 |
| CD274 (PDL-1) | 153.0 | .000056 |
| C5aR1 (CD88) | 39.5 | .011 |
| Ly6a (Sca1) | 39.2 | .00070 |
| CD74 | 20.8 | .022 |
| CXCR4 | 18.1 | .0027 |
| VCAM1* | 13.9 | .033 |
| Cdh11 (OB-cadherin)* | 12.7 | .0073 |
| CD44* | 11.1 | .0026 |
| uPAR (Plaur) | 5.5 | .0044 |
| Cadm3 (Necl1)* | 3.7 | .052 |
| ICAM1 | 2.8 | .00072 |
| Vegfr3 (Flt4) | 0.758 | .054 |
| L1CAM | 0.404 | .204 |
| Fgfr3 | 0.379 | .043 |
| Tie2 (Tek)* | 0.354 | .0088 |
| Vegfr2 (KDR) | 0.299 | .0021 |
| Tie1 | 0.216 | .0064 |
| Itgb5* | 0.187 | .023 |
| NrCAM* | 0.186 | .013 |
| Leptinr (leptin receptor)* | 0.089 | .00090 |
| Itga8* | 0.011 | .0011 |
| Transcript . | Fold change . | P . |
|---|---|---|
| FcgRIIb (CD32)* | 438.5 | .000061 |
| CD62P (P selectin)* | 316.4 | .0012 |
| S1PR3 (Edg3)* | 288.9 | .000093 |
| CD53 | 246.3 | .0000027 |
| CD274 (PDL-1) | 153.0 | .000056 |
| C5aR1 (CD88) | 39.5 | .011 |
| Ly6a (Sca1) | 39.2 | .00070 |
| CD74 | 20.8 | .022 |
| CXCR4 | 18.1 | .0027 |
| VCAM1* | 13.9 | .033 |
| Cdh11 (OB-cadherin)* | 12.7 | .0073 |
| CD44* | 11.1 | .0026 |
| uPAR (Plaur) | 5.5 | .0044 |
| Cadm3 (Necl1)* | 3.7 | .052 |
| ICAM1 | 2.8 | .00072 |
| Vegfr3 (Flt4) | 0.758 | .054 |
| L1CAM | 0.404 | .204 |
| Fgfr3 | 0.379 | .043 |
| Tie2 (Tek)* | 0.354 | .0088 |
| Vegfr2 (KDR) | 0.299 | .0021 |
| Tie1 | 0.216 | .0064 |
| Itgb5* | 0.187 | .023 |
| NrCAM* | 0.186 | .013 |
| Leptinr (leptin receptor)* | 0.089 | .00090 |
| Itga8* | 0.011 | .0011 |
Microarray-based up-regulation/down-regulation of mRNAs encoding for selected cell surface molecules during CHS-induced inflammation. Examples represent individual probe IDs. P values were calculated using Student t test.
Multiple probe IDs annotated for this gene were among the present calls. The probe ID with the largest fold change is shown, but all other probe IDs displayed a similar up- or down-regulation.
Lymphatic marker genes are down-regulated in LECs during CHS-induced inflammation. Analysis of the normalized array intensity levels of various (A) pan-endothelial and (B) LEC specific marker genes. In the case of genes that were represented in the arrays by multiple probe sets, the average signal intensity of all present probes is displayed. (C) A significant, inflammation-induced down-regulation of LYVE-1, Prox-1, and VEGFR3 was confirmed by quantitative PCR analysis. *P < .05. ***P < .001. (D) Immunofluorescence was performed on cryosections, obtained from uninflamed (control) or CHS-inflamed ears 48 hours after challenge, and confirmed an inflammation-induced down-regulation (↓) of VEGFR3 (red) in LYVE-1-expressing lymphatics (green) at the protein level. Representative images from 2 different experiments are shown. Scale bar represents 50 μm.
Lymphatic marker genes are down-regulated in LECs during CHS-induced inflammation. Analysis of the normalized array intensity levels of various (A) pan-endothelial and (B) LEC specific marker genes. In the case of genes that were represented in the arrays by multiple probe sets, the average signal intensity of all present probes is displayed. (C) A significant, inflammation-induced down-regulation of LYVE-1, Prox-1, and VEGFR3 was confirmed by quantitative PCR analysis. *P < .05. ***P < .001. (D) Immunofluorescence was performed on cryosections, obtained from uninflamed (control) or CHS-inflamed ears 48 hours after challenge, and confirmed an inflammation-induced down-regulation (↓) of VEGFR3 (red) in LYVE-1-expressing lymphatics (green) at the protein level. Representative images from 2 different experiments are shown. Scale bar represents 50 μm.
CHS-induced inflammation leads to the down-regulation of lymphatic marker genes
Interestingly, the arrays indicated that the lymphatic marker genes Prox-1 and VEGFR3 were down-regulated in inflamed LECs (Figure 2B). A significant, inflammation-induced down-regulation of Prox-1 and VEGFR3 was confirmed by quantitative PCR analysis performed on the ex vivo sorted and processed LEC samples (Figure 2C). Furthermore, LYVE-1 was found to be significantly down-regulated in inflamed LECs (Figure 2C), in agreement with previously published reports.20,21 Down-regulation of VEGFR3 on lymphatics in inflamed ear tissue was confirmed by immunofluorescence (Figure 2D; supplemental Figure 2). Surprisingly, the observed inflammation-induced reduction in podoplanin protein expression (Figure 1D-E) did not coincide with a down-regulation of podoplanin mRNA (Figure 2B-C).
CHS-induced inflammation modulates the expression of cell surface molecules in LECs
Further analysis revealed that various LEC-expressed cell surface molecules were differentially expressed during CHS-induced inflammation (Table 1). Besides ICAM-1 and VCAM-1, other cell adhesion molecules, such as CD44, CD62P (P-selectin), and uPAR, were up-regulated during inflammation. Interestingly, several receptor tyrosine kinases implicated in the regulation of lymphangiogenesis (Tie-1, Tie-2, VEGFR2, and VEGFR3) were among the down-regulated cell surface molecules (Table 1; Figure 2A).22,23
Differential expression of selected cell surface molecules was validated at the protein level by FACS analysis, performed on ear tissue single-cell suspensions and by immunofluorescence (Figure 3). Interestingly, FACS analysis also confirmed an inflammation-induced up-regulation of CD274 (programmed death ligand-1) and of CD74 (the invariant chain of MHCII), molecules with well-described functions in antigen presentation (Figure 3A-B). A similar inflammation-induced up-regulation of these genes was also observed in BECs (supplemental Figure 3A-B). Immunofluorescence further confirmed the up-regulation of uPAR and the down-regulation of a NrCAM, a neuronal cell adhesion molecule24 with previously unreported expression in LECs (Figure 3C; supplemental Figure 2).
Validation of the differential expression of selected cell surface proteins in CHS-inflamed LECs. Differential expression of selected cell surface molecules was validated at the protein level by FACS analysis and by immunofluorescence. (A) FACS analysis was performed on single-cell suspensions obtained from enzymatically digested ear tissue. (Upper panels) The staining and gating scheme. (Lower 2 rows) Histograms of candidate gene expression in LECs (CD45−CD31+podoplanin+ cells). Black line indicates candidate gene; and gray, tinted line, isotype control. Representative plots from 3 or 4 different experiments are shown. The number in the top right of each image represents the ΔMFI, defined as the difference in the median fluorescent intensity between the candidate and the corresponding isotope control staining. (B) Summary of the ΔMFI values measured in all experiments. Data points (control [CTR] − inflamed [INFL]) from the same experiment are connected by a line. (C) Immunofluorescence was performed on cryosections, obtained from uninflamed (control) or CHS-inflamed ears 48 hours after challenge. Sections were stained for candidate genes (red) and costained with LYVE-1 (green) to outline lymphatic vessels. uPAR (red) was found to be up-regulated in CHS-inflamed ear skin (↑), whereas NrCAM (red) was down-regulated (↓). Representative images from 2 different experiments are shown. Scale bar represents 50 μm.
Validation of the differential expression of selected cell surface proteins in CHS-inflamed LECs. Differential expression of selected cell surface molecules was validated at the protein level by FACS analysis and by immunofluorescence. (A) FACS analysis was performed on single-cell suspensions obtained from enzymatically digested ear tissue. (Upper panels) The staining and gating scheme. (Lower 2 rows) Histograms of candidate gene expression in LECs (CD45−CD31+podoplanin+ cells). Black line indicates candidate gene; and gray, tinted line, isotype control. Representative plots from 3 or 4 different experiments are shown. The number in the top right of each image represents the ΔMFI, defined as the difference in the median fluorescent intensity between the candidate and the corresponding isotope control staining. (B) Summary of the ΔMFI values measured in all experiments. Data points (control [CTR] − inflamed [INFL]) from the same experiment are connected by a line. (C) Immunofluorescence was performed on cryosections, obtained from uninflamed (control) or CHS-inflamed ears 48 hours after challenge. Sections were stained for candidate genes (red) and costained with LYVE-1 (green) to outline lymphatic vessels. uPAR (red) was found to be up-regulated in CHS-inflamed ear skin (↑), whereas NrCAM (red) was down-regulated (↓). Representative images from 2 different experiments are shown. Scale bar represents 50 μm.
CHS-induced inflammation up-regulates chemokine expression in LECs
Various secreted proteins, including genes with a reported function in extracellular matrix remodeling and angiogenesis, were differentially expressed in LECs during CHS-induced skin inflammation (Table 2). Furthermore, many chemokines were highly up-regulated in inflamed LECs. Indeed, 11 of the top 100 inflammation-induced genes were chemokines (Table 2; Figure 4A; and data not shown). Most up-regulated chemokines were inflammatory chemokines (Figure 4A), the receptors of which are expressed on leukocytes found in inflamed tissues25 as well as on endothelial cells.26 Interestingly, no inflammation-induced up-regulation of CCL21 was observed. By contrast, a weak signal for CCL19, the second ligand of CCR7, was detected in the arrays of inflamed LECs (Figure 4A). The inflammation-induced up-regulation of several inflammatory chemokines was confirmed by quantitative PCR performed on amplified material from ex vivo isolated LECs (Figure 4B). Performing intracellular FACS analysis on ear tissue single-cell suspensions, an inflammation-induced up-regulation of CXCL9 protein was observed in LECs in 4 of 5 experiments performed (Figure 4C-D). By contrast, CXCL9 was consistently up-regulated in BECs (supplemental Figure 3A-B). Whole-mount immunofluorescence revealed an up-regulation of CXCL1 and CCL2 in lymphatic vessels of CHS-inflamed ears (Figure 4E; supplemental Figure 2). Interestingly, some baseline expression of CCL2, but not of CXCL1, was already detectable in lymphatic vessels in uninflamed control ears (Figure 4E).
Differential expression of selected secreted molecules
| Transcript . | Fold change . | P . |
|---|---|---|
| CXCL9 (MIG)* | 1135.8 | .0000052 |
| TIMP1 | 865.1 | .0000011 |
| CXCL5 (LIX) | 664.1 | .00050 |
| CXCL10 (IP-10) | 505.6 | .000034 |
| CXCL2 (MIP-2) | 225.5 | .00010 |
| S100a9 | 200.3 | .00067 |
| Saa3 | 144.0 | .00010 |
| MMP3 | 131.5 | .0015 |
| S100a8 | 125.5 | .00035 |
| CCL12 | 124.1 | .0026 |
| IL-1b | 115.3 | .000037 |
| Tnfaip6 | 104.0 | .000028 |
| CXCL14 (BRAK)* | 101.9 | .00057 |
| Tnc (tenascin C) | 68.3 | .000027 |
| CCL8 (MCP-2) | 62.8 | .00022 |
| CCL2 (MCP-1) | 58.0 | .00025 |
| CCL7 (MCP-3) | 48.2 | .000011 |
| MMP19* | 47.6 | .0069 |
| CCL9 (MIP-1g)* | 44.8 | .000017 |
| Adamts4* | 31.0 | .000076 |
| CCL19 (ELC) | 23.8 | .00075 |
| Adamts9* | 22.7 | .0202 |
| VCAN (versican)* | 21.8 | .086 |
| SRGN (serglycin) | 16.7 | .00096 |
| CSF1 (M-CSF* | 14.9 | .0061 |
| Upa (PLAU)* | 11.1 | .0082 |
| CXCL1 (KC)* | 10.3 | .03 |
| Adamts8 | 7.8 | .018 |
| Adamts5* | 4.0 | .0018 |
| CXCL12 (SDF1)* | 3.7 | .0086 |
| Adamts1 | 3.3 | .00029 |
| Sema3a* | 3.2 | .07 |
| Nid1* | 3.0 | .0087 |
| Col18a1* | 0.230 | .021 |
| Nid2 | 0.209 | .000018 |
| IL-7* | 0.198 | .0080 |
| Ntn1 (netrin 1)* | 0.087 | .0032 |
| TIMP4* | 0.053 | .0083 |
| Transcript . | Fold change . | P . |
|---|---|---|
| CXCL9 (MIG)* | 1135.8 | .0000052 |
| TIMP1 | 865.1 | .0000011 |
| CXCL5 (LIX) | 664.1 | .00050 |
| CXCL10 (IP-10) | 505.6 | .000034 |
| CXCL2 (MIP-2) | 225.5 | .00010 |
| S100a9 | 200.3 | .00067 |
| Saa3 | 144.0 | .00010 |
| MMP3 | 131.5 | .0015 |
| S100a8 | 125.5 | .00035 |
| CCL12 | 124.1 | .0026 |
| IL-1b | 115.3 | .000037 |
| Tnfaip6 | 104.0 | .000028 |
| CXCL14 (BRAK)* | 101.9 | .00057 |
| Tnc (tenascin C) | 68.3 | .000027 |
| CCL8 (MCP-2) | 62.8 | .00022 |
| CCL2 (MCP-1) | 58.0 | .00025 |
| CCL7 (MCP-3) | 48.2 | .000011 |
| MMP19* | 47.6 | .0069 |
| CCL9 (MIP-1g)* | 44.8 | .000017 |
| Adamts4* | 31.0 | .000076 |
| CCL19 (ELC) | 23.8 | .00075 |
| Adamts9* | 22.7 | .0202 |
| VCAN (versican)* | 21.8 | .086 |
| SRGN (serglycin) | 16.7 | .00096 |
| CSF1 (M-CSF* | 14.9 | .0061 |
| Upa (PLAU)* | 11.1 | .0082 |
| CXCL1 (KC)* | 10.3 | .03 |
| Adamts8 | 7.8 | .018 |
| Adamts5* | 4.0 | .0018 |
| CXCL12 (SDF1)* | 3.7 | .0086 |
| Adamts1 | 3.3 | .00029 |
| Sema3a* | 3.2 | .07 |
| Nid1* | 3.0 | .0087 |
| Col18a1* | 0.230 | .021 |
| Nid2 | 0.209 | .000018 |
| IL-7* | 0.198 | .0080 |
| Ntn1 (netrin 1)* | 0.087 | .0032 |
| TIMP4* | 0.053 | .0083 |
Microarray-based up-regulation/down-regulation of mRNAs encoding for selected secreted proteins during CHS-induced inflammation. Examples represent individual probe IDs. P values were calculated using Student t test.
Multiple probe IDs annotated for this gene were among the present calls. The probe ID with the largest fold change is shown, but all other probe IDs displayed a similar up- or down-regulation.
Up-regulation of chemokines in CHS-inflamed LECs. (A) The analysis of normalized array intensity levels revealed a strong up-regulation of chemokines in LECs isolated from inflamed skin. On the y-axis, the average signal intensities recorded in all replicates are shown. In the case of genes that were represented in the arrays by multiple probe sets, the average signal intensity of all probes is displayed. (B) The inflammation-induced up-regulation of selected chemokines was confirmed by quantitative PCR analysis performed on amplified cDNA derived from ex vivo isolated LECs. Data represent pooled values from 4 sample pairs. (C) The up-regulation of CXCL9 protein was analyzed by FACS. Histogram plots showing CXCL9 expression (intracellular staining) in LECs (gated on CD45−CD31+podoplanin+ cells) derived from CHS-inflamed or control ear single-cell suspensions. The number in the top right of each image represents the ΔMFI, defined as the difference in the median fluorescent intensity between the candidate and the corresponding isotope control staining. Black line represents CXCL9 staining; and gray, tinted line, isotype control staining. (D) Summary of the ΔMFIs of CXCL9 measured in all experiments. Data points (control [CTR] − inflamed [INFL]) from the same experiment are connected by a line. (E) The expression of CXCL1 and CCL2 (red) in CHS-inflamed ears was validated at the protein level by whole-mount immunofluorescence. Whole mounts were costained for LYVE-1 (green). White arrows indicate regions of chemokine expression and colocalization with lymphatic vessels. Scale bar represents 100 μm.
Up-regulation of chemokines in CHS-inflamed LECs. (A) The analysis of normalized array intensity levels revealed a strong up-regulation of chemokines in LECs isolated from inflamed skin. On the y-axis, the average signal intensities recorded in all replicates are shown. In the case of genes that were represented in the arrays by multiple probe sets, the average signal intensity of all probes is displayed. (B) The inflammation-induced up-regulation of selected chemokines was confirmed by quantitative PCR analysis performed on amplified cDNA derived from ex vivo isolated LECs. Data represent pooled values from 4 sample pairs. (C) The up-regulation of CXCL9 protein was analyzed by FACS. Histogram plots showing CXCL9 expression (intracellular staining) in LECs (gated on CD45−CD31+podoplanin+ cells) derived from CHS-inflamed or control ear single-cell suspensions. The number in the top right of each image represents the ΔMFI, defined as the difference in the median fluorescent intensity between the candidate and the corresponding isotope control staining. Black line represents CXCL9 staining; and gray, tinted line, isotype control staining. (D) Summary of the ΔMFIs of CXCL9 measured in all experiments. Data points (control [CTR] − inflamed [INFL]) from the same experiment are connected by a line. (E) The expression of CXCL1 and CCL2 (red) in CHS-inflamed ears was validated at the protein level by whole-mount immunofluorescence. Whole mounts were costained for LYVE-1 (green). White arrows indicate regions of chemokine expression and colocalization with lymphatic vessels. Scale bar represents 100 μm.
The gene expression response of LECs depends on the nature of the inflammatory stimulus
We next investigated whether the observed changes in chemokine, adhesion molecule, and LEC marker gene expression were a default response of LECs to inflammation or whether this response pattern varied depending on the nature of the inflammatory stimulus. Therefore, inflammation was induced by injection of CFA, which is composed of mycobacteria-derived constituents and induces inflammation by triggering innate immunity.27 CFA injection induced a similar degree of ear swelling as CHS-induced inflammation (data not shown). One day after CFA injection, LECs were isolated by FACS sorting from inflamed and control ears. Quantitative PCR analysis revealed a similar induction of VCAM-1 by both stimuli, whereas ICAM-1 was much less up-regulated during CFA-induced than during CHS-induced inflammation (Figure 5A). A lower CFA-induced up-regulation of ICAM-1 was also observed at the protein level when performing FACS analysis on ear tissue cell suspensions (Figure 5B). Furthermore, a striking reduction in the induction of CCL2, CCL8, CXCL9, and CXCL10 was observed in CFA-induced compared with CHS-induced inflammation (Figure 5A). In contrast to CHS-induced inflammation, LEC marker genes were not down-regulated by CFA-induced inflammation (supplemental Figure 4A). Other inflammation-induced genes, such as S100a9, TIMP1, or CD62P were similarly induced under both inflammatory conditions (supplemental Figure 4B).
The pattern of chemokine and ICAM-1 expression in LECs depends on the nature of the inflammatory stimulus. (A) Quantitative PCR analysis performed on amplified cDNA from ex vivo isolated LECs revealed marked differences in the pattern of ICAM-1 and chemokine production in response to CHS- or CFA-induced skin inflammation. Pooled data from 3 or 4 independent experiments are shown. (B) Up-regulation of ICAM-1 in LECs (gated on CD45−CD31+podoplanin+ cells) in response to CHS- but not CFA-induced inflammation was confirmed by FACS analysis performed on ear tissue single-cell suspensions. ΔMFI indicates difference in the median fluorescent intensity between the ICAM-1 signal and the isotype signal. Black line represents ICAM-1 staining; and gray, tinted line, isotype control staining. (C-E) Conditionally immortalized Immorto LECs (imLECs) were treated with combinations of IFN-γ/TNF-α or MDP/LTA to mimic CHS- or CFA-induced inflammation, respectively. (C) FACS analysis of the induction of ICAM-1 and VCAM-1 by the treatment. (D) Analysis of the mRNA chemokine expression profiles induced by the treatment. (E) Chemokines were measured in the cell culture supernatant of imLECs treated with IFN-γ/TNF-α or LTA/MDP using a FACS-based cytokine assay. *P < .05. **P < .01. ***P < .001.
The pattern of chemokine and ICAM-1 expression in LECs depends on the nature of the inflammatory stimulus. (A) Quantitative PCR analysis performed on amplified cDNA from ex vivo isolated LECs revealed marked differences in the pattern of ICAM-1 and chemokine production in response to CHS- or CFA-induced skin inflammation. Pooled data from 3 or 4 independent experiments are shown. (B) Up-regulation of ICAM-1 in LECs (gated on CD45−CD31+podoplanin+ cells) in response to CHS- but not CFA-induced inflammation was confirmed by FACS analysis performed on ear tissue single-cell suspensions. ΔMFI indicates difference in the median fluorescent intensity between the ICAM-1 signal and the isotype signal. Black line represents ICAM-1 staining; and gray, tinted line, isotype control staining. (C-E) Conditionally immortalized Immorto LECs (imLECs) were treated with combinations of IFN-γ/TNF-α or MDP/LTA to mimic CHS- or CFA-induced inflammation, respectively. (C) FACS analysis of the induction of ICAM-1 and VCAM-1 by the treatment. (D) Analysis of the mRNA chemokine expression profiles induced by the treatment. (E) Chemokines were measured in the cell culture supernatant of imLECs treated with IFN-γ/TNF-α or LTA/MDP using a FACS-based cytokine assay. *P < .05. **P < .01. ***P < .001.
When investigating the effects of inflammation on gene expression in BECs, we found that CHS- and CFA-induced inflammation led to similar differential chemokine response patterns in BECs (supplemental Figure 5A), as observed in LECs (Figure 5A). Furthermore, the induction of VCAM-1 was differentially regulated in BECs (supplemental Figure 5A). Therefore, a stimulus-specific response to inflammation appears to be a general feature of both endothelial cell types. Interestingly, CD62P was up-regulated at both the mRNA and protein level in LECs (Table 1; Figure 3A-B), but only at the protein level in BECs (supplemental Figures 3A-B, 5B). This indicates that, in LECs, CD62P is de novo synthesized, whereas in BECs, it may be up-regulated by release of presynthesized CD62P from Weibel-Palade bodies.28
To test whether differential inflammatory response patterns could also be induced in cultured LECs, we performed in vitro assays with conditionally immortalized LECs (imLECs), which we isolated from the skin of H-2Kb-tsA58 (Immorto) mice.12 imLECs expressed VEGFR3, Prox-1, LYVE-1, CD31, and podoplanin at the protein level (supplemental Figure 6A), and microarray-based analysis of these cells further confirmed their phenotypic similarities to FACS-sorted primary LECs (supplemental Table 1). Interestingly, the gene expression profile of imLECs more closely resembled FACS-sorted LECs from inflamed skin than LECs from control skin (supplemental Figure 6B; supplemental Table 1). imLECs were either treated with IFN-γ and TNF-α, to mimic tissue inflammation induced by a CHS response (Figure 1B), or with the (myco)bacterial cell wall components muramyl dipeptide (MDP) and lipoteichoic acid (LTA), to mimic CFA injection. Both inflammatory treatments resulted in a further up-regulation of ICAM-1 and VCAM-1 protein in imLECs (Figure 5C). Analysis of the gene expression response induced by the 2 treatments revealed several similarities to the chemokine induction pattern observed in LECs isolated from CHS- or CFA-inflamed tissue (Figure 5A,D-E). Most strikingly, CXCL9, CXCL10, and CCL8 were mainly induced on IFN-γ/TNF-α, but not on LTA/MDP treatment (Figure 5D-E). However, in the case of CCL2, CCL7, and CXCL5, the response patterns induced in imLECs did not correlate with the in vivo findings (Figure 5D-E), indicating that the in vitro conditions used did not completely mimic the inflammatory response of LECs to CHS- or CFA induced tissue inflammation.
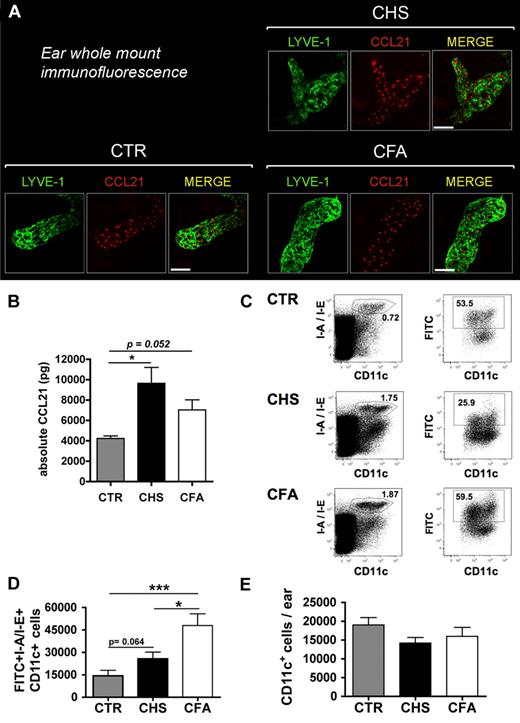
Inflammation-induced up-regulation of CCL21 protein does not correlate with DC migration. (A) Ear whole-mount immunofluorescence revealed abundant expression of CCL21 in lymphatic vessels in control and CHS- or CFA-inflamed ears. Scale bar represents 50 μm. (B) Quantification of CCL21 protein levels in ear lysates generated from uninflamed, CHS-inflamed, or CFA-inflamed ears of mice (n = 3 mice per group). Tissue inflammation led to a 2.3-fold (CHS) and 1.7-fold (CFA) increase in CCL21 protein. (C-D) FITC painting experiments were performed in uninflamed and CHS-inflamed or CFA-inflamed ears of mice. (C) LN single-cell suspensions were analyzed by FACS for the presence of I-A/I-E+CD11c+FITC+ cells. The number within each plot represents the percentage of gated cells. (D) Quantification of the total numbers of I-A/I-E+CD11c+FITC+ DCs detected stronger DC migration in response to CFA- compared with CHS-induced tissue inflammation. (E) CD11c+ cells in CHS or CFA-inflamed ears and control ears were analyzed by quantitative FACS analysis of ear single-cell suspensions. *P < .05. ***P < .001.
Inflammation-induced up-regulation of CCL21 protein does not correlate with DC migration. (A) Ear whole-mount immunofluorescence revealed abundant expression of CCL21 in lymphatic vessels in control and CHS- or CFA-inflamed ears. Scale bar represents 50 μm. (B) Quantification of CCL21 protein levels in ear lysates generated from uninflamed, CHS-inflamed, or CFA-inflamed ears of mice (n = 3 mice per group). Tissue inflammation led to a 2.3-fold (CHS) and 1.7-fold (CFA) increase in CCL21 protein. (C-D) FITC painting experiments were performed in uninflamed and CHS-inflamed or CFA-inflamed ears of mice. (C) LN single-cell suspensions were analyzed by FACS for the presence of I-A/I-E+CD11c+FITC+ cells. The number within each plot represents the percentage of gated cells. (D) Quantification of the total numbers of I-A/I-E+CD11c+FITC+ DCs detected stronger DC migration in response to CFA- compared with CHS-induced tissue inflammation. (E) CD11c+ cells in CHS or CFA-inflamed ears and control ears were analyzed by quantitative FACS analysis of ear single-cell suspensions. *P < .05. ***P < .001.
Taken together, our data indicate that the pattern of inflammatory chemokine production and the regulation of ICAM-1 and lymphatic marker gene expression in LECs highly depends on the nature of the inflammatory stimulus.
CCL21 protein levels are increased during CHS- and CFA-induced inflammation
Surprisingly, our microarray analysis did not reveal any inflammation-induced up-regulation of CCL21 mRNA expression in LECs (Figure 4A). Immunofluorescence performed in ear whole mounts detected abundant CCL21 expression in lymphatic vessels in uninflamed and in CHS- or CFA-inflamed skin (Figure 6A). Under all conditions, CCL21 expression was almost exclusively observed in LYVE-1-positive lymphatic vessels. Interestingly, we observed a granular staining pattern for CCL21 (Figure 6A), suggesting that most CCL21 is localized in intracellular vesicles, as recently reported.10 Quantification of CCL21 protein levels in ear extracts revealed a 2.3-fold increase in CHS-inflamed ears and a 1.7-fold increase in CFA-inflamed ears, compared with control ears (Figure 6B).
The magnitude of DC migration does not correlate with CCL21 and ICAM-1 expression in LECs
In a next step, we studied whether the stimulus-dependent differences observed in the inflammatory response in LECs (Figures 5A-B, 6B) had any functional consequences on DC migration to dLNs. To this end, FITC painting experiments were performed in the ears of mice with CHS- or CFA-induced skin inflammation. Application of FITC dissolved in dibutyl phthalate onto the skin of mice is known to induce DC mobilization to dLNs, where DCs can be quantified based on their green fluorescent signal.11 Eighteen hours after FITC painting, FITC+I-A/I-E (MHCII)+CD11c+ DCs were analyzed in the dLN by FACS (Figure 6C). This quantification revealed that DC migration to dLNs was markedly induced during inflammation (Figure 6D). Surprisingly, DC migration was 1.9-fold (P = .016) more enhanced during CFA-induced than during CHS-induced inflammation (Figure 6D), despite lower CCL21 and ICAM-1 induction in LECs (Figures 5B, 6A). The increase in DC migration was not simply the result of increased DC numbers in CFA-inflamed compared with CHS-inflamed tissue at the time of FITC painting (Figure 6E). This suggests that the enhancement of DC migration from CFA-inflamed tissue was the result of other inflammation-induced changes in LECs and/or in DCs.
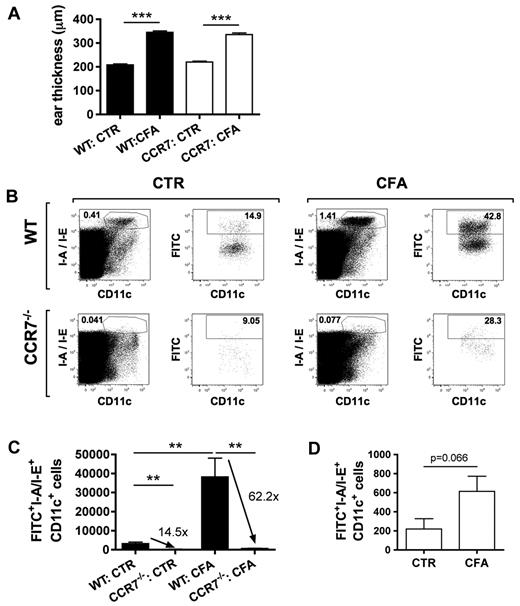
Inflammation enhances DC migration mainly by CCR7-dependent, but also by CCR7-independent mechanisms
In a next step, we wanted to investigate to which extent the observed enhancement of DC migration during CFA-induced inflammation was dependent on CCR7-CCL21 signaling. Because mice with defective CCL21 expression in lymphatics have not been generated or described,11 we instead made use of CCR7-deficient mice.7 FITC painting experiments were performed in uninflamed and in CFA-inflamed ears of CCR7−/− and WT mice. CFA injection induced a similar ear swelling response in WT and CCR7−/− mice (Figure 7A). Under uninflamed conditions, DC migration to dLNs was significantly reduced in CCR7−/− mice compared with WT mice, as previously reported7 (Figure 7B-C). Interestingly, the deficiency of CCR7−/− DCs to migrate to dLNs became even more pronounced during inflammation: Although approximately 14.5-fold more WT compared with CCR7−/− DCs had migrated to the dLN under uninflamed conditions, this ratio was elevated to 62.2-fold under inflammatory conditions (Figure 7C). This result indicates that CFA-induced inflammation enhanced DC migration in a mainly CCR7-dependent manner, either by increasing the availability of CCL21 on lymphatic vessels or by enhancing the responsiveness of DCs to CCR7 ligands. Interestingly, we observed that CFA-induced skin inflammation also moderately (2.2-fold, P = .066) enhanced DC migration in CCR7−/− mice (Figure 7D). Notably, CFA-induced inflammation did not increase DC numbers in the ears of CCR7−/− mice (data not shown), therefore excluding that the observed increase in CCR7-independent DC migration was a mere recruitment phenomenon. Importantly, these findings suggest that, besides CCL21, also other LEC-expressed chemotactic signals and/or adhesion molecules contribute to enhanced DC migration during inflammation, albeit to a moderate extent.
Inflammation enhances DC migration mainly by CCR7-dependent, and moderately by CCR7-independent, mechanisms. FITC painting experiments were performed in the uninflamed or CFA-inflamed ears of WT and CCR7−/− mice. (A) CFA injection induced a similar degree of ear swelling in WT and CCR7−/− mice. (B) LN single-cell suspensions were analyzed by FACS for the presence of I-A/I-E+CD11c+FITC+ cells. The number within each plot represents the percentage of gated cells. (C) Inflammation further enhanced the CCR7 dependence of DC migration, as demonstrated by the quantification of total numbers of I-A/I-E+CD11c+FITC+ cells. Numbers indicate the fold difference in DC migration observed in WT compared with CCR7−/− mice. (D) Inflammation also enhanced DC migration in CCR7−/− mice, as evidenced by the quantification of total numbers of I-A/I-E+CD11c+FITC+ cells in the dLNs. **P < .01. ***P < .001.
Inflammation enhances DC migration mainly by CCR7-dependent, and moderately by CCR7-independent, mechanisms. FITC painting experiments were performed in the uninflamed or CFA-inflamed ears of WT and CCR7−/− mice. (A) CFA injection induced a similar degree of ear swelling in WT and CCR7−/− mice. (B) LN single-cell suspensions were analyzed by FACS for the presence of I-A/I-E+CD11c+FITC+ cells. The number within each plot represents the percentage of gated cells. (C) Inflammation further enhanced the CCR7 dependence of DC migration, as demonstrated by the quantification of total numbers of I-A/I-E+CD11c+FITC+ cells. Numbers indicate the fold difference in DC migration observed in WT compared with CCR7−/− mice. (D) Inflammation also enhanced DC migration in CCR7−/− mice, as evidenced by the quantification of total numbers of I-A/I-E+CD11c+FITC+ cells in the dLNs. **P < .01. ***P < .001.
Discussion
In this study, we have analyzed the transcriptional changes that occur in LECs during skin inflammation. To our knowledge, our study represents the first description of the in vivo inflammatory response of LECs. Using this information, we have studied how inflammation impacts DC migration, a process thought to be indirectly controlled by inflammation-induced changes in gene expression in LECs.29,30
A first surprising finding of our study was that CHS-induced inflammation led to a marked down-regulation of various lymphatic marker genes in LECs. Besides LYVE-1, which was previously shown to be down-regulated during inflammation,20,21 we also observed a significant down-regulation of the transcription factor Prox-1 and of VEGFR3, a well-described Prox-1 target gene.31 In contrast, no significant down-regulation of LEC marker genes was observed during CFA-induced inflammation, indicating that this response is stimulus-specific. Interestingly, several pan-endothelial marker genes were also down-regulated during CHS-induced inflammation. It is presently unknown which factors induce this down-regulation and whether lymphatic markers remain down-regulated at later stages of the inflammatory response. Given that VEGFR3 and LYVE-1 are interesting diagnostic and therapeutic targets,32,33 inflammation-induced down-regulation of these genes could be of clinical relevance.
Another important finding of our study was that the up-regulation of chemokines and of ICAM-1 in LECs varied between different inflammatory stimuli, indicating a complex control mechanism of the inflammatory response LECs. Interestingly, ICAM-1 was recently shown to be up-regulated during CHS and to mediate DC migration from inflamed skin to draining LNs in this model.2 Similarly, we also observed a strong up-regulation of ICAM-1 during CHS. By contrast, CFA-induced inflammation was accompanied by almost no induction of ICAM-1. Nevertheless, DC migration was significantly more enhanced during CFA-induced than during CHS-induced inflammation. This suggests that ICAM-1 up-regulation does not enhance DC migration during every type of inflammation and may therefore not be a suitable target for blocking DC migration in the context of inflammation or tissue rejection.6
CHS- or CFA-induced inflammation led to a strong induction of inflammatory chemokines in both LECs and BECs. Moreover, striking differences were observed in the patterns of chemokines induced under these 2 inflammatory conditions. From the blood vascular system, it is well known that the composition of chemokines expressed or presented by the BECs during inflammation determines which leukocyte subsets are recruited into the tissue.25 Although not analyzed so far, it is possible that also the chemokines that are up-regulated in LECs during tissue inflammation mediate the migration of leukocytes, such as macrophages,34,35 neutrophils,36,37 or memory/effector T cells,38,39 into afferent lymphatics. In this regard, the stimulus-dependent differences in chemokine expression in LECs, which we observed in our study, could serve as an additional control mechanism to coordinate tissue retention of leukocytes or their exit by entry into afferent lymphatics.
Both CHS- and CFA-induced inflammation led to an up-regulation of CCL21 protein in the ear tissue, as determined by the analysis of ear protein extracts. Because LECs were virtually the only CCL21-producing cell type in the ear skin, we assume that this increase in CCL21 protein is the result of CCL21 production in LECs. Inflammation-induced up-regulation of CCL21 protein in LECs was recently also reported by other studies.8-10,40 In agreement with a recent report,10 we observed a granular expression pattern for CCL21, suggesting that CCL21 is stored mainly in intracellular compartments. The total amount of protein present in LECs may therefore not necessarily be indicative for the extracellular, chemoattractive fraction of CCL21. Intriguingly, we observed that CFA-induced inflammation was a stronger inducer of DC migration, despite slightly lower CCL21 protein levels present in ear extracts. Interestingly, CCL19 mRNA was also induced during both CHS- and CFA-induced inflammation (data not shown). However, we could not detect CCL19 protein expression in inflamed lymphatics by ear whole-mount immunofluorescence (data not shown), probably reflecting the low signal intensities for CCL19 detected in the arrays.
In agreement with a previous study,8 our FITC painting experiments in CCR7−/− mice demonstrated an overwhelming dependence of inflammation-induced DC migration on CCR7 signaling. Compared with the uninflamed state, the CCR7 dependence of DC migration became even more pronounced under inflammatory conditions, probably because of up-regulation of CCL21 in LECs and an increased responsiveness of DCs for CCR7 ligands. Furthermore, our FITC-painting experiments revealed that inflammation does not only enhance DC migration in WT mice but also moderately in CCR7−/− mice. This result indicates that enhanced migration is partially independent of CCR7 signaling and might additionally be mediated by the action of other, inflammation-induced factors, such as LEC-expressed inflammatory chemokines. Immature DCs reportedly express various receptors for inflammatory chemokines.41 Maturation induces the up-regulation of CCR7 and the concomitant down-regulation of inflammatory chemokine receptors in DCs.42,43 However, because DC maturation and migration are thought to be simultaneous processes,44 it is possible that inflammatory chemokines, which are up-regulated in LECs during inflammation, still contribute to DC migration into lymphatics. In this regard, CCL2 and CXCL12, which reportedly are also involved in DC migration from the skin to dLNs under uninflamed conditions, could be interesting candidates.45,46 However, compared with CCL21, this contribution appears to be rather modest.
In conclusion, our findings reveal that the inflammatory response of LECs and the migratory response of DCs during inflammation are stimulus-dependent. Our findings indicate that enhancement of DC migration during inflammation is only weakly stimulated by LEC-induced genes other than CCL21. The latter observation could have implications for new therapeutic approaches to modulate DC migration in the context of vaccination and of chronic inflammation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Claudia Ceranski, Carlos Ochoa, and Cornelius Fischer (all from ETH Zurich) for excellent technical assistance, Michal Okoniewski (Functional Genomics Center Zurich) for help with the array data analysis, and Veronique Angeli (National University of Singapore) for providing the protocol for CCL21 whole-mount immunofluorescence.
C.H. was supported by the Swiss National Fund (grant 310000-116128), the Prof Dr Max Cloëtta Foundation, the Swisslife Jubiläumsstiftung, and ETH Zurich.
Authorship
Contribution: B.V. designed and performed research, analyzed data, and wrote the paper; D.A., M.N., M.I., T.R., and O.A. performed research and analyzed data; and C.H. designed and performed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Cornelia Halin, Institute of Pharmaceutical Sciences, ETH Zurich, Wolfgang-Pauli Str 10, HCI H413, CH-8093 Zurich, Switzerland; e-mail: cornelia.halin@pharma.ethz.ch.



![Figure 3. Validation of the differential expression of selected cell surface proteins in CHS-inflamed LECs. Differential expression of selected cell surface molecules was validated at the protein level by FACS analysis and by immunofluorescence. (A) FACS analysis was performed on single-cell suspensions obtained from enzymatically digested ear tissue. (Upper panels) The staining and gating scheme. (Lower 2 rows) Histograms of candidate gene expression in LECs (CD45−CD31+podoplanin+ cells). Black line indicates candidate gene; and gray, tinted line, isotype control. Representative plots from 3 or 4 different experiments are shown. The number in the top right of each image represents the ΔMFI, defined as the difference in the median fluorescent intensity between the candidate and the corresponding isotope control staining. (B) Summary of the ΔMFI values measured in all experiments. Data points (control [CTR] − inflamed [INFL]) from the same experiment are connected by a line. (C) Immunofluorescence was performed on cryosections, obtained from uninflamed (control) or CHS-inflamed ears 48 hours after challenge. Sections were stained for candidate genes (red) and costained with LYVE-1 (green) to outline lymphatic vessels. uPAR (red) was found to be up-regulated in CHS-inflamed ear skin (↑), whereas NrCAM (red) was down-regulated (↓). Representative images from 2 different experiments are shown. Scale bar represents 50 μm.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/1/10.1182_blood-2010-12-326447/4/m_zh89991174180003.jpeg?Expires=1770852506&Signature=mv5CCbVMiNFwuxT1GUbR-~i1mIsyKJgyTy04CXCeUndNKep081BZKNe6vPuPqEtz2BvJsOduL0a8m1WerOSmkhXbhlYCk5ozOWRureBUqA5oBcc239E-ArzkfAcfqvdNmud48OlQc~oBCTVhtAsHQfcrHAZaxE6LkpAMAPHKLcPeNOBv7ObeZ0gbLishUJ3~KbfKC-Knat7CkY23sfpRkqb1xN9hEelN7V4gl-G3hFgiiV0FKOXhrlCwMB5smPTdNulrgEmkhVSuSUnM0F9GlKbUbiF56~OFB0~TJwU5a4flcv8DfzWLN4PB~ocKcvZrB9A-AKaX2SqH95BLpJSpew__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Up-regulation of chemokines in CHS-inflamed LECs. (A) The analysis of normalized array intensity levels revealed a strong up-regulation of chemokines in LECs isolated from inflamed skin. On the y-axis, the average signal intensities recorded in all replicates are shown. In the case of genes that were represented in the arrays by multiple probe sets, the average signal intensity of all probes is displayed. (B) The inflammation-induced up-regulation of selected chemokines was confirmed by quantitative PCR analysis performed on amplified cDNA derived from ex vivo isolated LECs. Data represent pooled values from 4 sample pairs. (C) The up-regulation of CXCL9 protein was analyzed by FACS. Histogram plots showing CXCL9 expression (intracellular staining) in LECs (gated on CD45−CD31+podoplanin+ cells) derived from CHS-inflamed or control ear single-cell suspensions. The number in the top right of each image represents the ΔMFI, defined as the difference in the median fluorescent intensity between the candidate and the corresponding isotope control staining. Black line represents CXCL9 staining; and gray, tinted line, isotype control staining. (D) Summary of the ΔMFIs of CXCL9 measured in all experiments. Data points (control [CTR] − inflamed [INFL]) from the same experiment are connected by a line. (E) The expression of CXCL1 and CCL2 (red) in CHS-inflamed ears was validated at the protein level by whole-mount immunofluorescence. Whole mounts were costained for LYVE-1 (green). White arrows indicate regions of chemokine expression and colocalization with lymphatic vessels. Scale bar represents 100 μm.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/1/10.1182_blood-2010-12-326447/4/m_zh89991174180004.jpeg?Expires=1770852506&Signature=pgA4Ut2JhFoyaFRxIl6BPqs5zSqgqdiNQtZWiT9uLghLefprd2dmlZl88CVlhm3XAfNd-V951xO02y1Q~Kfz1p72BMydphTxx6c9h24JwV4DF8a2elBVqLO90RrNHuJMv1BBpj41QoMOAI8Rws7KLWQeP27UIeAX-soDDBnV-qPPio2KNJIx25qTdxa3YeivSWYZ4XxVduFUDtRXDd-rS6m2bm4b~ndp~N6VMsui09xdW~YUn7MFa74qytLAnv~tigZpAnhyIX1FexLZ7iFU9dVCaykwGguIq1BEDh0uyV0oNydPaoOXS1zR6iSz48jpIZwreSlyXBiMGBpA699sNw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal